Abstract
Diagnosis of pancreatic ductal adenocarcinoma (PDAC) is associated with a dismal prognosis despite current best therapies; therefore new treatment strategies are urgently required. Numerous studies have suggested that epithelial-to-mesenchymal transition (EMT) contributes to early-stage dissemination of cancer cells and is pivotal for invasion and metastasis of PDAC1,2,3,4. EMT is associated with phenotypic conversion of epithelial cells into mesenchymal-like cells in cell culture conditions, although such defined mesenchymal conversion (with spindle-shaped morphology) of epithelial cells in vivo is rare, with quasi-mesenchymal phenotypes occasionally observed in the tumour (partial EMT)5,6. Most studies exploring the functional role of EMT in tumours have depended on cell-culture-induced loss-of-function and gain-of-function experiments involving EMT-inducing transcription factors such as Twist, Snail and Zeb1 (refs 2, 3, 7, 8, 9, 10). Therefore, the functional contribution of EMT to invasion and metastasis remains unclear4,6, and genetically engineered mouse models to address a causal connection are lacking. Here we functionally probe the role of EMT in PDAC by generating mouse models of PDAC with deletion of Snail or Twist, two key transcription factors responsible for EMT. EMT suppression in the primary tumour does not alter the emergence of invasive PDAC, systemic dissemination or metastasis. Suppression of EMT leads to an increase in cancer cell proliferation with enhanced expression of nucleoside transporters in tumours, contributing to enhanced sensitivity to gemcitabine treatment and increased overall survival of mice. Collectively, our study suggests that Snail- or Twist-induced EMT is not rate-limiting for invasion and metastasis, but highlights the importance of combining EMT inhibition with chemotherapy for the treatment of pancreatic cancer.
Similar content being viewed by others
Main
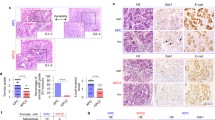
We crossed Twist1loxP/loxP (Twist1L/L) or Snai1loxP/loxP(Snai1L/L) mice with Pdx1-cre;LSL-KrasG12D;P53R172H/+ (KPC) to generate the Pdx1-cre;LSL-KrasG12D;P53R172H/+;Twist1L/L (KPC;TwistcKO) and the Pdx1-cre;LSL-KrasG12D;P53R172H/+;Snai1L/L (KPC;SnailcKO) mice, respectively. The resultant progeny were born in an expected Mendelian ratio, without overt phenotypic findings other than the anticipated emergence of spontaneous pancreatic cancer (Extended Data Fig. 1a). Genetic deletion of Snai1 or Twist1 did not significantly delay pancreatic tumorigenesis, alter tumour histopathology features or local invasion (Fig. 1a–c and Extended Data Table 1). KPC;TwistcKO and KPC;SnailcKO mice displayed similar tumour burden compared to KPC control mice (Extended Data Fig. 1b) and insignificant differences in overall survival (Fig. 1d). Loss of Twist1 or Snai1 expression in the pancreas epithelium was confirmed by in situ hybridization coupled with CK8 epithelial immunolabelling (Fig. 1e and Extended Data Fig. 1c) as well as immunolabelling for Twist and Snail (Extended Data Fig. 1d). Significant suppression of EMT was noted (Fig. 1f, g and Extended Data Fig. 1e, f). Lineage tracing (Fig. 1f and Extended Data Fig. 1e) and immunolabelling of the primary tumour (Fig. 1g) showed a significant decrease in the frequency of epithelial cells with expression of the mesenchymal marker αSMA (EMT+ cells) and a decrease in expression of the EMT-inducing transcription factor Zeb1 (Fig. 1h). Global gene expression profiling of tumours revealed a decrease in expression of EMT-associated genes (including Snai1 and Twist1) in KPC;SnailcKO and KPC;TwistcKO mice compared to KPC control (Extended Data Fig. 1f). Loss of Snail and Twist enhanced E-cadherin expression and suppressed Zeb2 and Sox4 expression in cancer cells (Extended Data Fig. 2a–c). Snai2 (Slug) expression was restricted to early pancreatic intraepithelial neoplasia (PanIN) lesions in all the experimental groups with no observed expression in advanced tumours and was significantly reduced in KPC;SnailcKO and KPC;TwistcKO mice compared to KPC control mice (Extended Data Fig. 2d).
a, Representative haematoxylin and eosin (H&E)-stained primary tumours (scale bar, 100 μm). b, Relative percentages of each primary tumour histological tissue phenotype. n = 31 (KPC), 14 (KPC;TwistcKO) and 30 (KPC;SnailcKO) mice; error bars represent s.d. c, Local invasiveness n = 31 (KPC), 14 (KPC;TwistcKO) and 30 (KPC;SnailcKO) mice; error bars represent s.d. d, Overall survival n = 29 (KPC), 12 (KPC;TwistcKO) and 33 (KPC;SnailcKO) mice. e, Twist1 or Snai1 in situ hybridization (black) with CK8 (red) immunolabelling in primary tumours (n = 3 mice for all groups; scale bar, 50 μm). Relative percentages of Twist1+CK8+ or Snai1+CK8+ double-positive cells are shown below (two-tailed t-test). f, αSMA immunolabelling in YFP lineage-traced primary tumours (n = 3 mice for both groups; scale bar, 50 μm; two-tailed t-test). g, αSMA (red), CK8 (green) and DAPI (blue) immunolabelling in primary tumours; white arrows indicate double-positive cells (n = 4 mice for all groups; scale bar, 20 μm). h, Zeb1 immunolabelling (n = 5 (KPC), 6 (KPC;TwistcKO) and 6 (KPC;SnailcKO) mice; scale bar, 50 μm; inset scale bar, 20 μm). i, Masson’s trichrome stain (MTS) (n = 8 (KPC), 7 (KPC;TwistcKO) and 7 (KPC;SnailcKO) mice; scale bar, 100 μm; error bars represent s.d.). Unless otherwise indicated error bars represent s.e.m., percentages represent per cent change from control and significance was determined by one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant.
While desmoplasia, including extracellular matrix (ECM) and myofibroblasts content (Fig. 1i and Extended Data Fig. 2e, f), tumour vessel density (Extended Data Fig. 2g), intratumoural hypoxia (Extended Data Fig. 2h), CD3+ T-cell infiltration (Extended Data Fig. 2i), and cancer cell apoptosis was unaffected with Twist/Snail deletion in KPC tumours (Fig. 2a), the proliferation of cancer cells in mice with suppressed EMT was significantly increased (Fig. 2b), as shown previously in mouse models of breast cancers11,12,13. Immunostaining experiments further revealed that EMT+ cancer cells are largely Ki67− (Extended Data Fig. 3a). Altogether, these data suggest that EMT driven by Twist/Snail transcription factors is dispensable for initiation and progression of primary pancreatic cancer.
a, b, Primary tumour immunolabelling for cleaved caspase-3 (a; n = 6 mice for all groups; scale bar, 50 μm) and Ki67 (b; n = 7 (KPC), 7 (KPC;TwistcKO) and 9 (KPC;SnailcKO) mice; scale bar, 100 μm). c, Percentage of YFP+ circulating tumour cells (CTCs) (n = 8 mice for both groups; two-tailed t-test; error bars represent s.d.). d, KrasG12D expression in whole blood cell pellets (n = 5 (KPC), 3 (KPC;TwistcKO) and 5 (KPC;SnailcKO) mice; error bars represent s.d.). e, Haematoxylin and eosin staining and CK19 immunolabelling of metastatic liver nodules. Metastatic tumour nodules (T) outlined by a dotted line (scale bar, 100 μm). A table presenting the number of positive tissues out of total tissues examined is shown below (χ2 analysis). f, Expression analysis of Twist1 and Snai1 in cultured primary tumour cell lines (n = 4 (KPC) and 5 (KPC;TwistcKO) individual cell lines (Twist1) or 4 (KPC) and 6 (KPC;SnailcKO) individual cell lines (Snai1); one-tailed t-test of ΔCt; error bars represent s.d.). g, Bright-field or YFP images and quantification of sphere number in cultured tumour cell lines (n = 3 (KPC), 2 (KPC;TwistcKO) and 3 (KPC;SnailcKO) individual cell lines; scale bar, 50 μm). h, Haematoxylin and eosin images (scale bar, 100 μm) of colonized lungs from intravenously injected cultured primary tumour cell lines KPC (n = 5 (cell line 1) and 5 (cell line 2) mice injected) and KPC;TwistcKO (n = 11 (cell line 1) and 4 (cell line 2) mice injected) and KPC;SnailcKO (n = 4 (cell line 1), 5 (cell line 2) and 5 (cell line 3) mice injected). A table presenting the number of colonized tissues out of total tissues examined is shown below (χ2 analysis). Unless otherwise indicated error bars represent s.e.m and significance was determined by one-way ANOVA. *P < 0.05, **P < 0.01, ****P < 0.0001; NS, not significant; ND, not detected.
Next, we investigated whether suppression of EMT impacts invasion and metastasis. The number of YFP+ circulating tumour cells from lineage-traced KPC and KPC;TwistcKO was found to be unchanged (Fig. 2c and Extended Data Fig. 3b), and expression of cancer-cell-specific KrasG12D mRNA in the blood from KPC, KPC;TwistcKO and KPC;SnailcKO mice was unaffected (Fig. 2d), suggesting that suppression of EMT in pancreatic tumours does not impact the rate of systemic dissemination of cancer cells. Extensive histopathological analyses, coupled with CK19 or YFP immunostaining of distant metastatic target organs, namely the liver, lung and spleen, indicated a similar frequency of metastasis in EMT-suppressed tumours when compared to control tumours (Fig. 2e, Extended Data Fig. 3c and Extended Data Table 1 and 2). The metastases were negative for Twist, Snail, Zeb1 and αSMA, with the exception of a few KPC metastatic cells that expressed αSMA or Zeb1 (Extended Data Fig. 3d–f), while being positive for E-cadherin and Ki-67 (Extended Data Fig. 3g, h). The proliferation rate of cancer cells in the metastases was similar in KPC, KPC;SnailcKO and KPC;TwistcKO mice (Extended Data Fig. 3h). Collectively, the results indicated that the deletion of Twist1 or Snai1 in genetically engineered mouse models of PDAC did not reduce metastatic disease.
To evaluate whether cancer cells from the pancreas with and without EMT program differentially benefited from impaired proliferation to form secondary tumours, we isolated cancer cells from KPC, KPC;TwistcKO and KPC;SnailcKO mice to assay their organ colonization potential. Twist1 was significantly reduced and Snai1 expression was undetectable in cancer cells isolated from Twist- and Snail-deleted tumours, respectively (Fig. 2f). Short-term potential to form tumour spheres (associated with putative cancer stem phenotype) appeared similar in TwistcKO and SnailcKO KPC cells when compared to control KPC cells (Fig. 2g)3,8,14,15,16. Lung colonization frequencies following i.v. injection of KPC cancer cells (Twist- or Snail-deleted) were similar to the control KPC cancer cells (Fig. 2h). These results suggest that a favoured epithelial phenotype of cancer cells (via suppression of EMT) did not impact the capacity to form tumour spheres or their ability for organ colonization17.
Cancer cell EMT is associated with gemcitabine drug resistance in PDAC patients and in the orthotopic mouse models of PDAC1,2,8,9,18,19,20,21,22,23. Moreover, enhanced frequency of EMT+ cancer cells in pancreatic tumours is associated with poor survival24,25. To determine whether EMT suppression enhances PDAC sensitivity to gemcitabine chemotherapy, we tested the gemcitabine sensitivity of cancer cells with suppressed EMT in KPC mice. Equilibrative nucleoside transporter (ENT1) and concentrating nucleoside transporter (Cnt3) were significantly upregulated in cancer cells lacking Snail and Twist, while ENT2 expression was unchanged (Fig. 3a–c). KPC, KPC;SnailcKO and KPC;TwistcKO mice were treated with gemcitabine and tumour burden was monitored by MRI (Extended Data Table 3). Tumour progression was suppressed in KPC;SnailcKO and KPC;TwistcKO mice when compared to treated KPC control mice (Fig. 3d). KPC;SnailcKO and KPC;TwistcKO mice treated with gemcitabine showed improved histopathology and increased survival (Fig. 3e–g).
a–c, Primary tumour immunolabelling for (ENT1 (a), ENT2 (b) and Cnt3 (c) (n = 6 (KPC), 5 (KPC;TwistcKO) and 4 (KPC;SnailcKO) mice; scale bar, 100 μm; error bars represent s.e.m., two-tailed t-test). d, MRI tumour volumes of KPC plus gemcitabine (+ gem.) (n = 13 mice, 10 died before day 19), KPC; TwistcKO + gem. (n = 15 mice, 5 died before day 19) and KPC; SnailcKO + gem. (n = 20 mice, 9 died before day 19). One-way ANOVA comparing mean tumour volumes on day 0 and day 19, error bars represent s.d. e, Survival on gemcitabine treatment to end point (day 21). f, Haematoxylin and eosin-stained primary tumours (scale bar, 100 μm). g, Relative percentages of each histological tissue phenotype of end-point mice (n = 3 (KPC + gem.), 9 (KPC; TwistcKO + gem.) and 11 (KPC; SnailcKO + gem.) mice; error bars represent s.d.; two-tailed t-test). *P < 0.05, **P < 0.01; NS, not significant.
Cancer cells isolated from the tumours of KPC;SnailcKO and KPC;TwistcKO mice showed epithelial morphology (Extended Data Fig. 4a) and reduced expression of mesenchymal genes compared to KPC cancer cell lines (Extended Data Fig. 4b). However, in tissue culture conditions (2D culture on plastic), equilibrative nucleoside transporters (ENT1/ENT2/ENT3) showed similar expression patterns (Extended Data Fig. 4b) and expression of concentrating nucleoside transporters (Cnt1/Cnt3) was not detected (data not shown). Increased proliferation of KPC;SnailcKO and KPC;TwistcKO cancer cells compared to KPC control cells (Extended Data Fig. 4c) probably accounted for the increased sensitivity to gemcitabine and erlotinib in this setting (Extended Data Fig. 4d).
Next, we crossed the Snai1L/L to the PDAC mouse model, Ptf1a (P48)-cre;LSL-KrasG12D;Tgfbr2L/L (KTC) to generate Ptf1a (P48)-cre;LSL-KrasG12D;Tgfbr2L/L;Snai1L/L (KTC; SnailcKO). The KTC model offers a reliable and penetrant disease progression rate with a consistent timeline of death due to PDAC. Similar to the KPC;SnailcKO mice, KTC;SnailcKO deletion exhibited suppression of EMT but did not affect primary tumour histopathology, lifespan, local invasion, desmoplasia or frequency of apoptosis (Fig. 4f and Extended Data Figs 5a–e and 6a). KTC;SnailcKO mice presented with significantly reduced Zeb1 expression in cancer cells but enhanced expression of Cnt3, ENT2 and proliferation (Extended Data Fig. 5e). ENT1 expression was unchanged in KTC;SnailcKO mice compared to KTC mice (Extended Data Fig. 6a). KTC;SnailcKO mice demonstrated enhanced response to gemcitabine therapy, with significant normal parenchymal area and reduced tumour tissue (Fig. 4a–c). Gemcitabine therapy in KTC;SnailcKO mice reduced tumour burden (Fig. 4d) and significantly improved overall survival (Fig. 4e) when compared to gemcitabine-treated control KTC mice. Gemcitabine therapy specifically increased cancer cell apoptosis and removed enhanced proliferation observed in EMT-suppressed tumours (Fig. 4g and Extended Data Fig. 5e), without impacting the desmoplastic reaction (Extended Data Fig. 6b). Overall, these results suggest an enhanced sensitivity of EMT-suppressed cancer cells to gemcitabine. Both ENT2 and Cnt3 were upregulated in EMT-suppressed tumours (Fig. 4g). These data support a possible mechanistic connection between EMT and resistance to chemotherapy in PDAC.
a, b, Haematoxylin and eosin-stained primary tumour (scale bar, 100 μm) and relative percentage of each histological tissue phenotype in KTC + gem. (n = 5) and KTC;SnailcKO + gem. (n = 7) mice (error bars represent s.d.). c, Local invasiveness (n = 5 (KTC + gem.) and 7 (KTC;SnailcKO + gem.) mice; error bars represent s.d.). d, Pancreatic mass (n = 3 (KTC + gem.) and 4 (KTC;SnailcKO + gem.) mice; error bars represent s.d.). e, Overall survival of KTC + gem. (n = 8) and KTC;SnailcKO + gem. (n = 4) mice. f, Overall survival of KTC (n = 6) and KTC;SnailcKO (n = 3) mice. g, αSMA (red), CK8 (green) and DAPI (blue) staining of primary tumours; white arrows indicate double-positive cells (n = 4 mice for both groups; scale bar, 20 μm), and immunolabelling for Zeb1 (n = 4 (KTC + gem.) and 5 (KTC;SnailcKO + gem.) mice; scale bar, 50 μm; inset scale bar, 20 μm), cleaved caspase-3 (n = 4 (KTC + gem.) and 5 (KTC;SnailcKO + gem.) mice; scale bar, 50 μm), Ki67 (n = 4 (KTC + gem.) and 5 (KTC;SnailcKO + gem.) mice; scale bar, 100 μm), ENT2 (n = 5 mice for both groups; scale bar, 100 μm), and Cnt3 (n = 5 mice for both groups; scale bar, 100 μm). Unless otherwise indicated error bars represent s.e.m. and significance was determined by two-tailed t-tests. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
Collectively, our studies provide a comprehensive functional analysis of EMT in PDAC progression and metastasis. Absence of either Twist1 or Snai1 did not alter cancer progression or the capacity for local invasion or metastasis to lung and liver in genetically engineered mouse models of PDAC. Metastasis occurs despite a significant loss of EMT with either the deletion of Snail or Twist, and in both settings, Zeb1, Sox4, Slug and Zeb2 are also significantly suppressed. Nevertheless, it is possible that other EMT-inducing factors may compensate for the loss of Snail or Twist to induce invasion and metastasis. While Pdx1 is expressed during the development of the pancreas (in early pancreatic buds and all three major lineages of the pancreas: ductal, acinar and β-islets), its expression is largely repressed in the adult exocrine pancreas26,27. Therefore, deletion of Snail or Twist occurs at the embryonic stage and mice are born normal and exhibit normal pancreas histology before the onset of cancer. The mice with Snail or Twist deletion develop PanIN lesions at the same frequency as the control mice. One could argue that suppression of EMT starting from the inception of cancer could have launched compensatory mechanisms to overcome EMT-dependent invasion and metastasis. However, such compensation is not observed with respect to chemoresistance, and previous studies have demonstrated that EMT and cancer cell dissemination are observed even before PDAC lesions are detected in KPC mice4.
Our study demonstrates that EMT results in suppression of cancer cell proliferation and suppression of drug transporter and concentrating proteins, therefore inadvertently protecting EMT+ cells from anti-proliferative drugs such as gemcitabine. The correlation of decreased survival of pancreatic cancer patients with increased EMT is probably due to their impaired capacity to respond to gemcitabine and chemotherapeutics, which is a standard of care for most patients28,29. A compromised response to chemotherapy probably also explains higher metastatic disease in association with decreased survival of patients with enhanced EMT signatures. Collectively, our study offers the opportunity to evaluate the potential of targeting EMT to enhance efficacy of chemotherapy and targeted therapies30.
Methods
Mice
Characterization of disease progression and genotyping for the Pdx1-cre;LSL-KrasG12D;P53R172H/+ (herein referred to as KPC) and Ptf1a (P48)-cre;LSL-KrasG12D;Tgfbr2L/L (herein referred to as KTC) mice were previously described31,32,33. These mice were bred to Snai1L/L (herein referred to as SnailcKO), Twist1L/L (herein referred to as TwistcKO), and R26-LSL-EYFP33. SnailcKO mice were kindly provided by S. J. Weiss. TwistcKO mice were kindly provided by R. R. Behringer via the Mutant Mouse Regional Resource Center (MMRRC) repository. The resulting progeny were referred to as KPC, KPC;SnailcKO, KPC;TwistcKO, KTC and KTC;SnailcKO mice and were maintained on a mixed genetic background. Both males and females were used indiscriminately. Mice were given gemcitabine (G-4177, LC Laboratories) via intraperitoneal injection (i.p.) every other day at 50 mg kg−1 of body weight. Hypoxyprobe was injected in a subset of mice i.p. at 60 mg kg−1 of body weight 30 min before euthanasia. For in vivo colonization assays, one million KPC, KPC;TwistcKO and KPC;SnailcKO tumour cells in 100 μl of PBS were injected intravenously via the retro-orbital venous sinus. Four to eleven mice were injected per cell line. All mice were euthanized at 15 days post injection. All mice were housed under standard housing conditions at MD Anderson Cancer Center (MDACC) animal facilities, and all animal procedures were reviewed and approved by the MDACC Institutional Animal Care and Use Committee. Tumour growth met the standard of a diameter less than or equal to 1.5 cm. Investigators were not blinded to group allocation but were blinded for the assessment of the phenotypic outcome by histological analyses. No statistical methods were used to predetermine sample size and the experiments were not randomized.
Histology and histopathology
Histology, histopathological scoring, Masson’s trichrome staining (MTS), and Picrosirius Red have been previously described19,33. Formalin-fixed tissues were embedded in paraffin and sectioned at 5 μm thickness. MTS was performed using Gomori’s Trichome Stain Kit (38016SS2, Leica Biosystems). Picrosirius red staining for collagen was performed using 0.1% picrosirius red (Direct Red80; Sigma) and counterstained with Weigert’s haematoxylin. Sections were also stained with haematoxylin and eosin (H&E). Histopathological measurements were assessed by scoring H&E-stained tumours for relative percentages of each histopathological phenotype: normal (non-neoplastic), PanIN, well-differentiated PDAC, moderately-differentiated PDAC, poorly-differentiated PDAC, sarcomatoid carcinoma, or necrosis. When tumour histology was missing or of poor quality, the mice were excluded from primary tumour histological analysis and this was determined blinded from genotype information. A histological invasion score of the tumour cells into the surrounding stroma was scored on a scale of 0 to 2, with 0 indicating no invasion and 2 indicating high invasion, where invasion is defined as tumour cell dissemination throughout the stroma away from clearly defined epithelial ‘nests’. Microscopic metastases were observed in H&E-stained tissue sections of the liver, lung and spleen. Positivity (one or more lesions in a tissue) was confirmed using CK19 and YFP immunohistochemistry. This data has been presented as a contingency table (Fig. 2e) and represented as the number of positive tissues out of the number of tissues scored. The ‘Any’ metastasis score is the number of mice positive for a secondary lesion found anywhere throughout the body out of the total number of mice scored.
Immunohistochemistry and Immunofluorescence
Tissues were fixed in 10% formalin overnight, dehydrated, and embedded in paraffin and 5-μm-thick sections were then processed for analyses. Immunohistochemical analysis was performed as described33. Heat-mediated antigen retrieval in 1 mM EDTA + 0.05% Tween20 (pH 8.0) for one hour (pressure cooker) was performed for Snail and Twist, 10 mM citrate buffer, pH 6.0, was used for one hour (microwave) for Ki67 or 10 min for all other antibodies. Primary antibodies are as follows: αSMA (M0851, DAKO, 1:400 or ab5694, Abcam, 1:400), cleaved caspase-3 (9661, Cell Signaling, 1:200), CD3 (A0452, DAKO, 1:200), CD31 (Dia310M, DiaNova, 1:10), CK8 (TROMA-1, Developmental Studies Hybridoma Bank, 1:50), CK19 (ab52625, Abcam, 1:100), Cnt3 (HPA023311, Sigma-Aldrich, 1:400), ENT1 (LS-B3385, LifeSpan Bio., 1:100), E-cadherin (3195S, Cell Signaling, 1:400), ENT2 (ab48595, Abcam, 1:200), Ki67 (RM-9106, Thermo Scientific, 1:400), Slug (9585, Cell Signaling, 1:200), Snail (ab180714, Abcam, 1:100), Sox4 (ab86809, Abcam, 1:200), Twist (ab50581, Abcam, 1:100), YFP (ab13970, Abcam, 1:1000), Zeb1 (NBP1-05987, Novus, 1:500), and Zeb2 (NBP1-82991, Novus, 1:100). Sections for pimonidazole adduct (HPI Inc., 1:50) or αSMA immunohistochemistry staining were blocked with M.O.M. kit (Vector Laboratories, West Grove, PA) and developed by DAB according to the manufacturer’s recommendations. Alternatively, for immunofluorescence, sections were dual-labelled using secondary antibodies conjugated to Alexa Fluor 488 or 594 or tyramide signal amplification (TSA, PerkinElmer) conjugated to FITC. Lineage-traced (YFP-positive) EMT analysis was performed on 8-μm-thick O.C.T. medium (TissueTek)-embedded frozen sections. Sections were stained for αSMA (ab5694, Abcam, 1:400) followed by Alexa Fluor 680 conjugated secondary antibody. Bright-field imagery was obtained on a Leica DM1000 light microscope or the Perkin Elmer 3DHistotech Slide Scanner. Fluorescence imagery was obtained on a Zeiss Axio Imager.M2 or the Perkin Elmer Vectra Multispectral imaging platform. The images were quantified for per cent positive area using NIH ImageJ analysis software (αSMA, Pimonidazole, Slug, and CD31), per cent positive cells using InForm analysis software (Ki67 and CD3), or scored for intensity either positive or negative (αSMA/CK8 dual staining, αSMA, CK19, YFP, Zeb1, Zeb2, Sox4, E-cadherin and cleaved caspase-3) or on a scale of 1–3 (E-cadherin) or 1–4 (ENT1, ENT2 and Cnt3).
In situ hybridization
In situ hybridization (ISH) was performed on frozen tumour sections as previously described34. In brief, 10-μm-thick sections were hybridized with antisense probes to Twist1 and Snai1 overnight at 65 °C. After hybridization, sections were washed and incubated with AP-conjugated sheep anti-DIG antibody (1:2,000; Roche) for 90 min at room temperature. After three washes, sections were incubated in BM Purple (Roche) until positive staining was seen. Digoxigenin-labelled in situ riboprobes were generated with an in vitro transcription method (Promega and Roche) using a PCR template. The following primers were used to generate the template PCR product. Twist1, forward, 5′-CGGCCAGGTACATCGACTTC-3′; reverse, 5′-TAATACGACTCACTATAGGGAGATTTAAAAGTGTGCCCCACGC-3′; Snai1, forward, 5′-CAACCGTGCTTTTGCTGAC-3′; reverse, 5′-TAATACGACTCACTATAGGGAGACCTTTAAAATGTAAACATCTTTCTCC-3′.
Gene expression profiling
Total RNA was isolated from tumours of KPC control, KPC;TwistcKO and KPC;SnailcKO mice (n = 3 in each group) by TRIzol (15596026, Life Technologies) and submitted to the Microarray Core Facility at MD Anderson Cancer Center. Gene expression analysis was performed using MouseWG-6 v2.0 Gene Expression BeadChip (Illumina). The Limma package from R Bioconductor35 was used for quantile normalization of expression arrays and to analyse differentially expressed genes between cKO and control sample groups. Gene expression microarray data have been deposited in GEO (Accession number GSE66981). Genes upregulated in cells acquiring an EMT program were expected to be downregulated in the TwistcKO and SnailcKO tumours compared to control tumours.
CTC assays
Blood (200 μl) was collected from KPC;LSL-YFP and KPC;TwistcKO;LSL-YFP (ROSA-LSL-YFP lineage tracing of cancer cells) mice and incubated with 10 ml of ACK lysis buffer (A1049201, Gibco) at room temperature to lyse red blood cells. Cell pellets were resuspended in 2% FBS containing PBS and analysed for the number of YFP+ cells by flow cytometry (BD LSRFortessa X-20 Cell Analyzer). The data was expressed as the percentage of YFP+ cells from gated cells, with 100,000 cells analysed at the time of acquisition. Whole blood cell pellets were also assayed for the expression of KrasG12D transcripts, using quantitative real-time PCR analyses (described below).
Primary pancreatic adenocarcinoma cell culture and analyses
Derivation of primary PDAC cell lines were performed as previously described36. Fresh tumours were minced with sterile razor blades, digested with dispase II (17105041, Gibco, 4 mg ml−1)/collagenase IV (17104019, Gibco, 4 mg ml−1)/RPMI for 1 h at 37 °C, filtered by a 70 μm cell strainer, resuspended in RPMI/20%FBS and then seeded on collagen I-coated plates (087747, Fisher Scientific). Cells were maintained in RPMI medium with 20% FBS and 1% penicillin, streptomycin and amphotericin B (PSA) antibiotic mixture. Cancer cells were further purified by FACS based on YFP or E-cadherin expression (anti-E-cadherin antibody, 50-3249-82, eBioscience, 1:100). The sorted cells, using BD FACSAriaTM II sorter (South Campus Flow Cytometry Core Lab of MD Anderson Cancer Center) were subsequently expanded in vitro. All studies were performed on cells cultivated less than 30 passages. As these are primary cell lines, no further authentication methods were applicable and no mycoplasma tests were performed.
MTT and drug sensitivity assays
MTT assay was performed to detect cell proliferation and viability by using Thiazolyl Blue Tetrazolium Bromide (MTT, M2128, Sigma) following the manufacturer’s recommendations with an incubation of two hours at 37 °C. For the drug treatment studies, a cell line derived from each of the KPC, KPC;SnailcKO and KPC;TwistcKO mice was treated with 20 μM gemcitabine (G-4177, LC Laboratories) or 100 μM erlotinib (5083S, NEB) for 48 h. The relative cell viability was detected using MTT assay with a cell line derived from each of the KPC, KPC;SnailcKO and KPC;TwistcKO mice. n is defined as the number of biological replicates of a single cell line. Control conditions included 1% DMSO vehicle for erlotinib. The relative absorbance was normalized and control (time 0 h or vehicle-treated) arbitrarily set to 1 or 100% for absorbance or drug survival, respectively.
Quantitative real-time PCR analyses (qPCR)
RNA was extracted from whole blood cell pellets following ACK lysis using the PicoPure Extraction kit as directed (KIT0214, Arcturus), or from cultured primary pancreatic adenocarcinoma cells using TRIzol (15596026, Life Technologies). cDNA was synthetized using TaqMan Reverse Transcription Reagents (N8080234, Applied Biosystems) or High Capacity cDNA Reverse Transcription Kit (4368814, Applied Biosystems). Primers for KrasG12D recombination are: KrasG12D, forward, 5′-ACTTGTGGTGGTTGGAGCAGC-3′; reverse, 5′-TAGGGTCATACTCATCCACAA-3′. 1/ΔCt values are presented to show KrasG12D expression in indicated experimental groups, statistical analyses were performed on ΔCt. Primer sequences for EMT-related genes are listed in Supplementary Table 1, GAPDH was used as an internal control. The data are presented as the relative fold change and statistical analyses were performed on ΔCt.
Tumour sphere assay
Tumour sphere assays were performed as previously described33. Two million cultured primary tumour cells were plated in a low-adherence 100-mm dish (FB0875713, Fisherbrand) with 1% FBS, Dulbecco’s modified Eagle’s medium, and penicillin/streptomycin/amphotericin. Cells were incubated for 7 days and formed spheres were counted at 100× magnification. Three, two and three cell lines were analysed for KPC control, KPC;TwistcKO and KPC;SnailcKO groups, respectively, five field of views per cell line were quantified.
MRI analyses
MRI imaging was performed using a 7T small animal MR system as previously described37. To measure tumour volume, suspected regions were drawn blinded on each slice based on normalized intensities. The volume was calculated by the addition of delineated regions of interest in mm2 × 1 mm slice distance. None of the mice had a tumour burden that exceeded 1.5 cm in diameter, in accordance with institutional regulations. All mice with measurable tumours were enrolled in the study (see Extended Data Table 3). Mice were imaged twice, once at the beginning of the enrolment (day 0), and a second time 20 days (day 19) afterwards. Surviving animals were euthanized at end point (day 21) for histological characterization.
Statistical analyses
Statistical analyses were performed on the mean values of biological replicates in each group using unpaired two-tailed or one-tailed t-tests (qPCR only), or one-way ANOVA with Tukey’s multiple comparisons test using GraphPad Prism, as stipulated in the figure legends. χ2 analyses, using SPSS statistical software, were performed comparing control to cKO groups for metastatic or colonization frequency across multiple histological parameters in all mice and mice ≥120 days of age in Extended Data Table 1. Fisher’s exact P value was used to determine significance. Results are outlined in Extended Data Table 2. Kaplan–Meier plots were drawn for survival analysis and the log rank Mantel-Cox test was used to evaluate statistical differences, using GraphPad Prism. Data met the assumptions of each statistical test, where variance was not equal (determined by an F-test) Welch’s correction for unequal variances was applied. Error bars represent s.e.m. when multiple visual fields were averaged to produce a single value for each animal which was then averaged again to represent the mean bar for the group in each graph. P < 0.05 was considered statistically significant.
References
Hotz, B. et al. Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 13, 4769–4776 (2007)
Arumugam, T. et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 69, 5820–5828 (2009)
Taube, J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl Acad. Sci. USA 107, 15449–15454 (2010)
Rhim, A. D. et al. EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012)
Kalluri, R. & Weinberg, R. A. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009)
McDonald, O. G., Maitra, A. & Hruban, R. H. Human correlates of provocative questions in pancreatic pathology. Adv. Anat. Pathol. 19, 351–362 (2012)
Guaita, S. et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 277, 39209–39216 (2002)
Wellner, U. et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biol. 11, 1487–1495 (2009)
Zhang, K. et al. Knockdown of snail sensitizes pancreatic cancer cells to chemotherapeutic agents and irradiation. Int. J. Mol. Sci. 11, 4891–4904 (2010)
Tsai, J. H., Donaher, J. L., Murphy, D. A., Chau, S. & Yang, J. Spatiotemporal regulation of epithelial–mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012)
Stockinger, A., Eger, A., Wolf, J., Beug, H. & Foisner, R. E-cadherin regulates cell growth by modulating proliferation-dependent β-catenin transcriptional activity. J. Cell Biol. 154, 1185–1196 (2001)
Muraoka-Cook, R. S., Dumont, N. & Arteaga, C. L. Dual role of transforming growth factor beta in mammary tumorigenesis and metastatic progression. Clin. Cancer Res. 11, 937s–943s (2005)
Hugo, H. J. et al. Direct repression of MYB by ZEB1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res. 15, R113 (2013)
Mani, S. A. et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008)
Liu, H. et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl Acad. Sci. USA 107, 18115–18120 (2010)
Wang, Z. et al. Activated K-Ras and INK4a/Arf deficiency promote aggressiveness of pancreatic cancer by induction of EMT consistent with cancer stem cell phenotype. J. Cell. Physiol. 228, 556–562 (2013)
Yang, J. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004)
Vega, S. et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18, 1131–1143 (2004)
Shah, A. N. et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 14, 3629–3637 (2007)
Yin, T. et al. Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J. Surg. Res. 141, 196–203 (2007)
Wang, Z. et al. Acquisition of epithelial–mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 69, 2400–2407 (2009)
Alagesan, B. et al. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin. Cancer Res. 21, 396–404 (2015)
Cursons, J. et al. Stimulus-dependent differences in signalling regulate epithelial-mesenchymal plasticity and change the effects of drugs in breast cancer cell lines. Cell Commun. Signal. 13, 26 (2015)
Javle, M. M. et al. Epithelial–mesenchymal transition (EMT) and activated extracellular signal-regulated kinase (p-Erk) in surgically resected pancreatic cancer. Ann. Surg. Oncol. 14, 3527–3533 (2007)
Masugi, Y. et al. Solitary cell infiltration is a novel indicator of poor prognosis and epithelial-mesenchymal transition in pancreatic cancer. Hum. Pathol. 41, 1061–1068 (2010)
Park, J. Y. et al. Pdx1 expression in pancreatic precursor lesions and neoplasms. Appl. Immunohistochem. Mol. Morphol. 19, 444–449 (2011)
Offield, M. F. et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995 (1996)
Hidalgo, M. Pancreatic cancer. N. Engl. J. Med. 362, 1605–1617 (2010)
Kleger, A., Perkhofer, L. & Seufferlein, T. Smarter drugs emerging in pancreatic cancer therapy. Ann. Oncol. 25, 1260–1270 (2014)
Gore, A. J., Deitz, S. L., Palam, L. R., Craven, K. E. & Korc, M. Pancreatic cancer-associated retinoblastoma 1 dysfunction enables TGF-β to promote proliferation. J. Clin. Invest. 124, 338–352 (2014)
Hingorani, S. R. et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005)
Ijichi, H. et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-β signaling in cooperation with active Kras expression. Genes Dev. 20, 3147–3160 (2006)
Özdemir, B. C. et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 25, 719–734 (2014)
Keskin, D. et al. Targeting vascular pericytes in hypoxic tumors increases lung metastasis via angiopoietin-2. Cell Rep. 10, 1066–1081 (2015)
Smyth, G. K. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor (eds Gentleman, R., Carey, V., Dudoit, S., Irizarry, R. & Huber, W. ) (Springer, 2005)
Ying, H. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012)
Melo, S. A. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015)
Acknowledgements
We wish to thank D. Lundy, S. Yang, Z. Xiao, R. Deliz-Aguirre, T. Miyake and S. Lovisa for technical support and K. M. Ramirez and R. Jewell in the South Campus Flow Cytometry Core Laboratory of MD Anderson Cancer Center for flow cytometry cell sorting and analyses (partly supported by NCI grant no. P30CA16672). We also wish to thank E. Chang for scanning slides of histopathological specimens. This study was primarily supported by the Cancer Prevention and Research Institute of Texas. The research in the LeBleu laboratory is supported by UT MDACC Khalifa Bin Zayed Al Nahya Foundation.
Author information
Authors and Affiliations
Contributions
R.K. conceptually designed the strategy for this study and provided intellectual input. V.S.L. helped design experimental strategy, provided intellectual input, supervised the studies, performed immunohistochemistry and culture experiments, generated the figures and wrote the manuscript. X.Z. performed experiments to generate the genetically engineered mouse models and helped characterize the mouse phenotype, performed culture experiments, collected the tissue for analysis and contributed to the manuscript writing. J.L.C. characterized the mouse phenotype, analysed the data related to the genetically engineered mouse models, collected data, generated the figures and helped with manuscript writing and editing. H.S. performed experiments with mice and injected cancer cells and helped collect tissue, J.Ki., M.S., J.Ka., and C.-C.W. performed experiments and collected data. The data was analysed by J.L.C, V.S.L., X.Z., J.Ki, and C.-C.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 EMT inhibition is specific to tumour epithelium.
a, Representative images of haematoxylin and eosin-stained small intestine (SmInt), kidney, and heart (scale bar, 100 μm). b, Pancreatic mass of 29 (KPC), 13 (KPC;TwistcKO) and 28 (KPC;SnailcKO) mice, error bars represent s.d.; one-way ANOVA. c, Merge of Twist1 or Snai1 in situ hybridization (black) followed by CK8 (red) immunolabelling in tumours from KPC and KPC;TwistcKO or KPC;SnailcKO mice, respectively. White arrows highlight positive cells in the stroma, yellow arrows highlight negative epithelium (scale bar, 20 μm). d, Twist or Snail immunostaining in KPC and KPC;TwistcKO or KPC;SnailcKO tumours, respectively. Black arrows highlight positive cells in the stroma, red arrows highlight negative epithelium (scale bar, 20 μm). e, Channel separations of the representative images of αSMA immunolabelling in YFP lineage-traced tumours found in Fig. 1f (scale bar, 50 μm). f, EMT gene expression signature analysis in KPC, KPC;TwistcKO and KPC;SnailcKO cohorts (n = 3 mice). Red arrows indicate reduced Twist1 and Snai1 expression in KPC;TwistcKO and KPC;SnailcKO cohorts, respectively.
Extended Data Figure 2 General suppression of EMT markers does not affect desmoplasia.
a, E-cadherin immunolabelling and quantification of primary KPC (n = 5 mice), KPC;TwistcKO (n = 5 mice) and KPC;SnailcKO (n = 4 mice) (scale bar, 100 μm). b, Zeb2 immunolabelling and quantification of primary KPC (n = 6 mice), KPC;TwistcKO (n = 5 mice) and KPC;SnailcKO (n = 7 mice) (scale bar, 50 μm; inset scale bar, 20 μm). c, Sox4 immunolabelling and quantification of primary KPC (n = 7 mice), KPC;TwistcKO (n = 6 mice) and KPC;SnailcKO (n = 8 mice) (scale bar, 50 μm; inset scale bar, 20 μm). d, Slug immunolabelling and quantification of primary KPC (n = 4 mice), KPC;TwistcKO (n = 4 mice) and KPC; SnailcKO (n = 4 mice) tumours (scale bar, 50 μm; inset scale bar, 20 μm). e, Sirius Red staining and quantification of primary KPC (n = 21 mice), KPC;TwistcKO (n = 8 mice) and KPC;SnailcKO (n = 11 mice) (scale bar, 100 μm; error bars represent s.d.) f, αSMA immunolabelling and quantification of primary KPC (n = 5 mice), KPC;TwistcKO (n = 5 mice) and KPC;SnailcKO (n = 5 mice) (scale bar, 100 μm). g, CD31 immunolabelling and quantification of primary KPC (n = 4 mice), KPC;TwistcKO (n = 4 mice) and KPC;SnailcKO (n = 3 mice) (scale bar, 200 μm, inset scale bar, 100 μm). h, Pimonidazole staining and quantification of primary KPC (n = 4 mice), KPC;TwistcKO (n = 4 mice) and KPC;SnailcKO (n = 4 mice) (scale bar, 100 μm). i, CD3 immunolabelling and quantification of primary KPC (n = 5 mice), KPC;TwistcKO (n = 5 mice) and KPC;SnailcKO (n = 5 mice) (scale bar, 100 μm; inset scale bar, 25 μm). Unless otherwise indicated error bars represent s.e.m., and significance determined by one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
Extended Data Figure 3 EMT suppression does not alter epithelial characteristics of metastases.
a, Immunolabelling of primary tumours (n = 3 mice) for αSMA (red), CK8 (green), Ki67 (white) and DAPI (blue); yellow arrows indicate EMT+ cells (scale bar, 20 μm). b, Representative dot plots of circulating YFP+ cells. c, Images of serial sections of KPC;LSL-YFP lung and liver metastasis stained for haematoxylin and eosin or immunolabelled for CK19 or YFP. Yellow dashed box represents magnified areas in panel below (scale bar, 200 μm; magnification scale bar, 50 μm). d, KPC metastatic tumours stained for Twist and Snail (n = 3 mice; scale bar, 20 μm; inset scale bar, 10 μm). e, Zeb1 immunolabelling and quantification of metastatic KPC (n = 4 mice), KPC;TwistcKO (n = 3 mice) and KPC;SnailcKO (n = 4 mice) (scale bar, 50 μm; inset scale bar, 20 μm). f, αSMA immunolabelling and quantification of metastatic KPC (n = 3 mice), KPC;TwistcKO (n = 3 mice) and KPC;SnailcKO (n = 3 mice) (scale bar, 50 μm; inset scale bar, 20 μm). g, E-cadherin staining on serial sections of αSMA immunolabelling and quantification of metastatic KPC (n = 4 mice), KPC;TwistcKO (n = 3 mice) and KPC;SnailcKO (n = 4 mice) (scale bar, 50 μm; inset scale bar, 20 μm). h, Ki67 immunolabelling and quantification of metastatic KPC (n = 7 mice), KPC;TwistcKO (n = 3 mice) and KPC;SnailcKO (n = 3 mice) (scale bar, 50 μm; inset scale bar, 20 μm). Unless otherwise indicated error bars represent s.e.m., percentages indicated represent per cent decrease from control, and significance was determined by one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
Extended Data Figure 4 EMT suppressed primary tumour cells have reduced mesenchymal markers and show resistance to chemotherapy in vitro.
a, Bright-field micrograph of cultured primary KPC, KPC;TwistcKO and KPC;SnailcKO cells (scale bar, 50 μm). b, EMT- and gemcitabine-transport-related gene expression shown by qPCR analysis in KPC (n = 3–4 cell lines), KPC;TwistcKO (n = 5 cell lines) and KPC;SnailcKO (n = 5–6 cell lines) (error bars represent s.d., one-tailed t-test, *P < 0.05, numbers list non-significant P values. nd, not detected, ns, not significant). c, MTT assay showing cell proliferation in KPC, KPC;TwistcKO and KPC;SnailcKO cells (n = 8, 8 and 8 biological replicates of a cell line for each genotype). d, Relative cell viability (MTT assay) in cultured KPC, KPC;TwistcKO and KPC;SnailcKO cells treated with gemcitabine or erlotinib (n = 8, 8 and 8 biological replicates of a cell line for each genotype). Unless otherwise indicated error bars represent s.e.m., significance was determined by one-way ANOVA. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Figure 5 EMT inhibition in KTC mice mirrors phenotype observed in KPC mice.
a, Representative images of haematoxylin and eosin-stained primary tumours (scale bar, 100 μm). b, Relative percentage of each histological tissue phenotype of KTC (n = 8 mice) and KTC;SnailcKO (n = 6 mice) primary tumours (error bars represent s.d.). c, Primary tumour invasiveness in KTC (n = 8 mice) and KTC;SnailcKO (n = 6 mice) (error bars represent s.d.). d, Pancreatic mass in KTC (n = 5 mice) and KTC;SnailcKO (n = 6 mice) (error bars represent s.d.). e, Immunolabelling and quantification of primary KTC (n = 5 mice), KTC;SnailcKO (n = 4 mice) for αSMA (red), CK8 (green) and DAPI (blue); white arrows indicate double-positive cells (scale bar, 20 μm), Zeb1 (scale bar, 50 μm; inset scale bar, 20 μm), cleaved caspase-3 (scale bar, 50 μm; n = 4 mice for both groups), Ki67 (scale bar, 100 μm), ENT2 (scale bar, 100 μm) and CNT3 (scale bar, 100 μm); error bars represent s.e.m. Significance was determined by two-tailed t-test. *P < 0.05, ***P < 0.001; ns, not significant.
Extended Data Figure 6 Desmoplasia is unaffected in EMT suppressed tumours with or without gemcitabine.
a, b, Staining and quantification of KTC (n = 5 or 6 mice), KTC;SnailcKO (n = 4 or 5 mice), KTC plus gemcitabine (+ GEM; n = 4 or 5 mice), KTC;SnailcKO + GEM (n = 5 mice) for Masson’s trichrome stain (MTS) (scale bars, 100 μm), Sirius Red staining (scale bars, 100 μm), and ENT1 (scale bars, 100 μm). Error bars represent s.d. (MTS and Sirius Red) or s.e.m. (ENT1), and significance was determined by two-tailed t-test. ns, not significant.
Supplementary information
Supplementary Table
This file contains Supplementary Table 1. (PDF 71 kb)
Rights and permissions
About this article
Cite this article
Zheng, X., Carstens, J., Kim, J. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015). https://doi.org/10.1038/nature16064
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16064
- Springer Nature Limited
This article is cited by
-
ZDHHC5-mediated S-palmitoylation of FAK promotes its membrane localization and epithelial-mesenchymal transition in glioma
Cell Communication and Signaling (2024)
-
A milestone in epithelial–mesenchymal transition
Nature Cell Biology (2024)
-
GABRP Promotes the Metastasis of Pancreatic Cancer by Activation of the MEK/ERK Signaling Pathway
Biochemical Genetics (2024)
-
Phenotypic Transitions the Processes Involved in Regulation of Growth and Proangiogenic Properties of Stem Cells, Cancer Stem Cells and Circulating Tumor Cells
Stem Cell Reviews and Reports (2024)
-
GATA6-AS1 suppresses epithelial–mesenchymal transition of pancreatic cancer under hypoxia through regulating SNAI1 mRNA stability
Journal of Translational Medicine (2023)