Abstract
Myalgic encephalomyelitis/chronic fatigue syndrome is an unexplained debilitating disorder that is frequently associated with cognitive and motor dysfunction. We analyzed cerebrospinal fluid from 32 cases, 40 subjects with multiple sclerosis and 19 normal subjects frequency-matched for age and sex using a 51-plex cytokine assay. Group-specific differences were found for the majority of analytes with an increase in cases of CCL11 (eotaxin), a chemokine involved in eosinophil recruitment. Network analysis revealed an inverse relationship between interleukin 1 receptor antagonist and colony-stimulating factor 1, colony-stimulating factor 2 and interleukin 17F, without effects on interleukin 1α or interleukin 1β, suggesting a disturbance in interleukin 1 signaling. Our results indicate a markedly disturbed immune signature in the cerebrospinal fluid of cases that is consistent with immune activation in the central nervous system, and a shift toward an allergic or T helper type-2 pattern associated with autoimmunity.
Similar content being viewed by others
Introduction
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a debilitating and heterogeneous disorder of an unknown cause characterized by severe fatigue that may be associated with cognitive and motor dysfunction. Assays of blood in some patients indicate altered T helper (Th)1 and Th2 cytokine profiles, natural killer cell function and activation of the 2-5A synthetase/RNase L antiviral pathway.1, 2, 3 Infection has been reported with a wide range of viruses and bacteria,4 including Epstein–Barr virus,5, 6, 7, 8 human herpes viruses 6 and 7,9, 10, 11 enteroviruses,7, 12 cytomegalovirus13, 14 and lentiviruses,15 as well as chlamydia,7 mycoplasma16 and borrelia species.17 An association of ME/CFS with central nervous system (CNS) dysfunction is supported by imaging studies documenting cerebral hypoperfusion and increased white matter T2 signal consistent with chronic inflammation18 in addition to regional differences in gray19 and/or white matter.19, 20
Routine cerebrospinal fluid (CSF) studies in patients with ME/CFS, including white blood cell count, glucose and protein, are typically unremarkable;21 however, high resolution mass spectrometry has shown abnormalities in the CSF proteome, including an increase in proteins related to the complement cascade.22, 23 PCR assays of CSF that target human adenoviruses, alphaviruses, herpes viruses (human herpesvirus-1, 2, 3, 4, 5 or 8), human parvovirus B19, dengue viruses 1–4, West Nile virus, Japanese encephalitis virus, Saint Louis encephalitis virus, enteroviruses A–D, coxsackieviruses and xenotropic murine leukemia virus-related virus have been negative;24 however, neither human herpesvirus 6 or 7 were assayed. Decreases in proteins related to axonal guidance pathways were also observed, specifically, proteins associated with integrin-mediated adhesion. These findings are intriguing in light of evidence that human herpesvirus-6 can interfere with cellular adhesion and differentiation.25, 26, 27 Other neurotropic viruses, including Borna disease virus and tick-borne encephalitis virus, interfere with neurite outgrowth and neural plasticity.28, 29 Here we report discrete immunological profiles in subjects with ME/CFS as compared with subjects with multiple sclerosis (MS) or no known disease that can be detected using efficient immunoassays that may enable diagnosis and provide insights into pathogenesis and treatment.
The current study compared immune signatures in the CSF of subjects with a diagnosis of ME/CFS, the neuroimmune, demyelinating disorder, MS, or without known disease (no disease (ND) controls) using multiplexed immunoassays (a total of 51 cytokines). Several factors drove our decision to use MS patients as a comparator group for ME/CFS: first, fatigue is common in both MS and in ME/CFS;30 second, white matter abnormalities, a core feature in MS,31 have recently been described in ME/CFS;19, 20 third, cytokine abnormalities are found in the CSF of MS patients.32
Materials and methods
Study population
The study population was comprised of ME/CFS case (n=32) and comparator/control (n=59) subjects who had contributed CSF samples to biobanks at either Sierra Internal Medicine (SIM), a private internal medicine clinic in Incline Village, Nevada, owned and operated by Daniel Peterson, MD, or the University of California Los Angeles (UCLA) Brain Bank. All case subjects met both the 1994 Fukuda criteria33 for a CFS diagnosis and the 2003 Canadian Consensus Criteria for ME/CFS15 at the time of initial assessment. The comparator/control group consisted of 40 subjects with MS (comparators), with 19 relapsing-remitting MS (RRMS) and 21 secondary progressive MS (SPMS), and 19 subjects who had undergone CSF sampling to rule out CNS infection or hemorrhage (ND controls) (Table 1). Comparator/control subjects were frequency-matched for age (±5 years) and sex to ME/CFS subjects.
Human subjects and ME/CFS biological sample collection
CSF collections were performed following the standardized office protocol at SIM. Potential side effects or adverse events associated with specific clinically indicated procedures such as lumbar puncture, as well as the clinical reason for these procedures, were discussed with patients in the course of their diagnostic work-up and treatment planning. In addition, informed consent was obtained at the same time to allow any excess sample to be de-identified for use in any future research investigations of SIM.
Selection of case and comparator samples
CSF samples were retrieved from repositories of SIM (ME/CFS) and the UCLA Brain Bank and shipped on dry ice to the Wisconsin Viral Research Group where they were kept frozen at −80 °C until preparation of aliquots. The UCLA Brain Bank specimen set included samples from comparator subjects with MS as well as from subjects free of any infectious or immunologic abnormalities on routine CSF testing to rule out such diagnoses (ND controls). Biobank specimens had been collected over time and maintained at –80 °C. CSF samples derived from the ME/CFS subjects had previously been clinically analyzed for oligoclonal banding, total protein, glucose and amino-acid profiles, helping to rule out other known contributory diagnoses such as MS. In addition, control subjects had no evidence of ongoing infectious or inflammatory processes on their routine clinical CSF analyses. All CSF specimens submitted for viral diagnostic testing to Wisconsin Viral Research Group represent archived diagnostic specimens exempt from HIPAA and Institutional Review Board consideration (46.101 (b)(4), Code of Federal Regulations). All samples were de-identified prior to shipment to the Center for Infection and Immunity at Columbia University for analysis.
Cytokine analyses
The CSF concentrations of the following immune molecules were determined using a magnetic bead-based 51-plex immunoassay: interleukin (IL)1 superfamily—IL2, IL4, IL7, IL13, IL15; type I IL/β chain family—IL5, GMCSF (CSF2); IL6 (gp130) family—IL6, LIF; IL12 family—IL12p40, IL12p70; IL10 family—IL10; IL17 family—IL17A, IL17F; type I interferons (IFNs)—IFNα2, IFNβ; type II IFN—IFNγ; tumor necrosis factor (TNF) superfamily—TNFα (TNFSF2), TNFβ (TNFSF1), CD40 ligand (CD40L), sFasL (TNFSF6), TRAIL (TNFSF10); CC chemokines—CCL2 (MCP1), CCL3 (MIP1α), CCL4 (MIP1β), CCL5 (RANTES), CCL7 (MCP3), CCL11 (eotaxin); CXC chemokines—CXCL1 (GROα), CXCL5 (ENA78), CXCL8 (IL8), CXCL9 (MIG), CXCL10 (IP10); PDGF family/VEGF subfamily—PDGFBB, VEGFA; cell adhesion molecules—sICAM1 (CD54), VCAM1 (CD106); serine protease inhibitor—serpin E1 (PAI1); adipose-derived hormones—leptin, resistin; and neurotrophic/growth/cellular factors—TGFα, TGFβ, FGFb, βNGF, HGF, SCF, MCSF (CSF1), GCSF (CSF3) (customized Procarta immunoassay, Affymetrix, Santa Clara, CA, USA). This cytokine panel was developed as an assay for investigating acute phase (‘sickness’) responses and neuroimmune dysregulation in neuropsychiatric disorders that are postulated to be triggered by immune/infectious factors. It includes a wide range of cytokines, chemokines and cellular factors that reflect key processes relating to systemic activation of inflammatory/immune signaling pathways involved in autoimmunity and antiinflammatory responses, as well as others implicated in CNS inflammation and neurovascular disruption.
Subject CSF samples were run in duplicate along with serial standards, buffer controls and in-house human control plasma samples.34 Mean fluorescence intensities of analyte-specific immunoassay bead sets were detected by a flow-based Luminex 3D suspension array system (Luminex, Austin, TX, USA).35 Cytokine concentrations were calculated by xPONENT build 4.0.846.0 (Luminex Corporation, Austin, TX, USA) and Milliplex Analyst software v.3.5.5.0 (EMD Millipore, Billerica, MA, USA) using a standard curve derived from the known reference concentrations supplied by the manufacturer. A five-parameter model was used to calculate final concentrations by interpolation. Values are expressed in pg ml−1. Concentrations obtained below the sensitivity limit of detection of the method were recoded to the mid-point between zero and the limit of detection for that analyte for statistical comparisons. Values obtained from the reading of samples that exceeded the upper limit of the sensitivity method were further diluted and cytokine concentrations calculated accordingly.
Statistical analyses
Descriptive statistics
Descriptive statistics were computed for demographic variables across diagnostic groups. Sex and age were included as possible confounders. Differences across diagnostic groups and subgroups in proportion of females and males and in year of sample acquisition were assessed by the χ2-test. Differences in median age were examined using the nonparametric Mann–Whitney U-test (two-group comparisons) and the Kruskal–Wallis (three-group comparisons) test.
Data transformations
Because distributions deviated from normality, raw cytokine levels were first transformed using Box–Cox transformation defined as:

where for each cytokine variable, the optimal λ was identified to maximize the log-likelihood function.36 After transformation, all variables failed to reject the null hypothesis using the Kolmogorov–Smirnov test at the 0.05 level, confirming at the 95% confidence level that all transformed variables followed a normal distribution.
GLM and t-tests
General linear model (GLM) analyses were applied to examine both the main effect of diagnosis and the main and interaction effects of different fixed factors including diagnosis (three-level ME/CFS case vs MS comparator vs ND control comparisons) and sex, with age adjusted as a continuous covariate. Additional GLM analyses including storage time as an additional continuous covariate were also performed. Because GLM uses the family-wise error rate, no additional adjustments for multiple comparisons were applied. When the main effect of diagnosis (one-way analysis of variance/GLM or more complex models) or the interaction term of diagnosis × sex was significant (α=0.05), two-sample t-tests, also using a nominal α-level of 0.05, were conducted to compare the mean CSF levels of each transformed cytokine between diagnostic groups: cases vs ND controls, cases vs MS comparators and MS comparators vs ND controls.
Logistic regression modeling
Data for the 51 cytokines as a whole and all clinical covariates were also used to develop a logistic regression model for prediction of the binary response variable (cases vs ND controls, cases vs MS comparators and MS comparators vs ND controls). To eliminate multicollinearity among cytokines, we used a supervised feature selection method known as LASSO (least absolute shrinkage and selection operator)37 to guide selection of variables for inclusion in a final logistic regression model. The LASSO dimensionality reduction procedure regularizes the least squares by using the constraint that the L1 norm of the parameter vector is no greater than a given value, whereby increasing the penalty will cause more and more of the parameters to be driven to absolute zero. To avoid overfitting, we selected variables that had non-zero coefficients when the cross-validation deviance reached the minimum plus one s.e.m. The odds ratios (ORs), their associated 95% confidence intervals (CIs) and the P-values for the selected cytokines (the ‘features’ meeting LASSO criteria for inclusion) for all three pairwise comparisons were calculated accordingly.
NodeXL diagrams
The NodeXL platform was used to produce a network diagram of 51-plex assays within each subgroup.38 The platform provides a display of the relationships among the analytes, thereby facilitating discovery of different cytokine–cytokine structures across the different group populations. Bivariate Pearson’s correlations were first calculated between every pairwise combination of cytokine variables using their power-transformed (Box–Cox) values. We then used the Benjamini and Hochberg method to adjust for multiple comparisons with a 0.01 family-wise false discovery rate.39 Significantly correlated cytokine pairs were fed into the algorithm in NodeXL to produce the network diagram. Cytokines are represented by the ‘nodes’ in the diagram and significantly correlated cytokines are connected by ‘lines’ or edges. Red lines represent negative correlations and gray lines represent positive correlations.
Classification and regression tree (CART) analyses
The CART40 method was used to analyze differences among diagnostic subgroups (ME/CFS, MS and ND controls) by examining data from 51-plex cytokine assays from another perspective. CART is a nonparametric decision tree learning technique that produces a classification tree for dependent variables. It works by recursively splitting the feature space (the values of all the independent variables, or 'features,' included in the CART analysis) into a set of non-overlapping regions and then predicting the most likely value of the dependent variable within each region. The decision rule at each node of the tree is determined by searching over all independent variables and all possible values to provide the split that will best differentiate observations based on the dependent variable (here, diagnostic groups of ME/CFS, MS and ND controls). Once a rule is selected and splits each node into two, the same process is applied to each subsequent ‘child’ node in the classification tree. Partitioning stops when CART detects there is no further gain to be made: when diagnostic groups can be best distinguished by the criterion values associated with specific variables. To maximize predictive accuracy, the tree is pruned to optimize cross-validation. This is accomplished by repetitively resampling two subsets from the overall population, establishing one subset as a training set and allowing the other subset to function as a test set. This resampling procedure is repeated 100 times; selection of the final tree is based on the algorithm that best fits the data for the population in the test set.
Statistical analyses were run in SPSS version 22.0.0.0 (Chicago, IL, USA), MATLAB version R2013a (Natick, MA, USA) and R version 3.0.2 (Vienna, Austria).
Results
Table 1 shows the demographic features of the overall study group, including 32 subjects with ME/CFS, 40 MS subjects and 19 ND controls. MS subjects were slightly younger (MS; 44.2±7.1 years) than the ME/CFS group (49.9±11.2 years, P=0.001) as well as in comparison with ND controls (50.5±8.5 years, P=0.004), but the mean age of the ME/CFS group did not differ from that of ND controls. CSF samples were collected from MS and ND control subjects at earlier time points as compared with the ME/CFS group (P<0.0001), but sample storage time was similar for the MS and ND control groups.
ME/CFS cases vs MS comparators vs ND controls
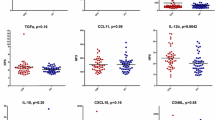
We compared cytokine levels of ME/CFS subjects with those of MS comparator and ND control subjects by one-way (diagnosis) and three-way (diagnosis, sex and age) GLM and found significant differences across the three groups in a large proportion of the 51 analytes (Supplementary Tables S1 and S2). Post hoc t-tests revealed prominent decreases in the levels of many pro- and anti-inflammatory cytokines in ME/CFS as compared with ND controls, including IL1ra, IL1β, IL5, IL6, IL8, IL10, IL12p40, IL17F, TNFβ, SCF (SF), CSF1 (MCSF), CSF2 (GMCSF), CSF3 (GCSF), PDGFBB, FGFb, VEGFA, LIF, resistin, serpin E1 (PAI1), sICAM1 (CD54) and VCAM1 (CD106). In contrast, only two analytes were increased in ME/CFS subjects relative to ND controls: CCL11 (eotaxin; P=0.0189) and CXCL10 (IP10; P=0.0261) (Figure 1 and Table 2).
Comparison of CSF levels of proinflammatory cytokine in ME/CFS, MS comparator and ND controls. Figures show mean ±s.e.m. for each cytokine. Only cytokines meeting significance criteria (P<0.05) in GLM analyses are represented. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 indicate P-values from two-sample t-test comparisons. (a) Cytokines with proinflammatory potential that are elevated in ME/CFS vs ND controls; (b) cytokines with antiinflammatory potential or proinflammatory cytokines that are decreased in ME/CFS vs ND controls. GLM, general linear model; ME/CFS, myalgic encephalomyelitis/chronic fatigue syndrome; MS, multiple sclerosis; ND, no disease.
Profiles of ME/CFS subjects also differed from those of MS subjects, with ME/CFS cases showing a markedly greater degree of CNS immune activation as compared with those with MS. Most notable among these were increases in IL2 (P=0.0062), IL7 (P=0.0029), IL17A (P<0.0001), IFNγ (P=0.0083), TNFβ (P=0.0001), sFasL (P=0.0017), TRAIL (TNFSF10; P=0.0171), CCL2 (MCP1, P<0.0001), TGFα (P=0.0003), TGFβ (P<0.0001) and CSF1 (MCSF) (P=0.0143). In addition, levels of some analytes were significantly reduced in the ME/CFS group as compared with MS comparators: IL1ra (P=0.0003), IL1α (P=0.0007), IL1β (P=0.0018), IL4 (P=0.041), IL6 (P=0.0274), IL10 (P=0.0002), IL12p40 (P=0.0001), CXCL10 (IP10, P=0.0004), SCF (SF, P=0.0012), HGF (P=0.0294), sICAM1 (CD54; P<0.0001) and VCAM1 (CD106; P<0.0001). Cytokine levels of RRMS and SPMS comparator subjects were similar except for slightly lower levels of IL5 in RRMS vs SPMS (P=0.0098) and higher leptin levels in RRMS as compared with SPMS (P=0.0227) (Supplementary Table S3).
Levels of immune molecules were largely decreased in MS subjects as compared with those of ND controls, with the exception of IL1α (P<0.0001), CCL11 (eotaxin; P=0.0003), CXCL10 (IP10; P<0.0001), HGF (P=0.0002) and leptin (P=0.0001), which were all increased (Table 2).
Logistic regression models
After data reduction through feature selection procedures, variables meeting LASSO criteria (Supplementary Tables S4–6) were included along with clinical covariates (age and sex) to construct the final logistic regression model and calculate the associated ORs, 95% CIs and P-values. Tables 3A,B,C show results for ME/CFS cases vs ND controls, ME/CFS cases vs MS and MS vs ND controls, respectively. In ME/CFS vs ND comparisons, higher levels of CCL11 (the allergy-associated CC chemokine, eotaxin) were strongly associated with ME/CFS caseness (OR, 33.3711; 95% CI, 2.0975, 530.9223; P=0.0130) as were lower levels of IL1β (OR, 0.0000; 95% CI, 0.0000, 0.9822; P=0.0498) (Table 3A). In ME/CFS vs MS comparisons, ME/CFS was associated with markedly higher levels of TGFβ (OR, 22.2495; 95% CI, 1.0910, 453.7519; P=0.0437), as well as elevated levels of CCL2 (OR, 1.1071; 95% CI, 1.0263, 1.1943; P=0.0085). ME/CFS was also associated with decreased levels of VCAM1 (OR, 0.9581; 95% CI, 0.9270, 0.9903; P=0.0112) (Table 3B). In MS vs ND comparisons, MS was associated with lower levels of CSF1 (MCSF) and higher levels of HGF and leptin (Table 3C).
Network associations
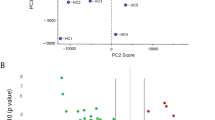
Network diagrams revealed unusual regulatory relationships among CSF cytokines in the case group as compared with ND controls (Figure 2). MS cytokine–cytokine correlations showed a pattern that differed from ND controls as well; however, these inter-cytokine structures were also distinct from the CSF profile of the case group. Whereas in ND controls no inverse relationships were found across any of the cytokines, several such associations were found in the CSF of case subjects with IL1ra (IL1ra-CSF1 (MCSF); IL1ra-CSF2 (GMCSF); IL1ra-IL17F) and in CSF of MS subjects in association with IL4 (IL4-PDGFBB, IL4-IL17F). We also found an unusual influence of PDGFBB on other cytokines in ME/CFS and MS that is not a factor in CSF cytokine regulatory networks in ND controls.
Network cytokine–cytokine associations differ for ME/CFS, MS comparators and ND controls. Network diagrams for ME/CFS subjects (a, n=32), MS comparators (b, n=40) and ND controls (c, n=19). Network diagrams of the 51 measured cytokines were created in NodeXL (http://nodexl.codeplex.com) using a 0.01 family-wise false discovery rate (FDR) to adjust for multiple comparisons (a, ME/CFS, P=0.0010; b, MS, P=0.0013; c, ND controls, P=0.0025). Red lines (edges) indicate negative correlations and gray lines indicate positive cytokine–cytokine correlations with associated P-values that fall below the FDR-corrected P-value criterion for each group. ME/CFS, myalgic encephalomyelitis/chronic fatigue syndrome; MS, multiple sclerosis; ND, no disease.
CART analysis
The CART decision tree machine-learning method was applied to cytokine and clinical covariate data (age and sex) to find predictors associated with diagnostic group (ME/CFS, MS or ND control). Predictor variables and cutoffs at each node in the decision tree are those with the maximum capacity to differentiate between the diagnostic groups. CART analysis produced no predictors relating to age or sex. The most informative cytokine classifiers in the CART-derived decision rules involve TGFβ, CXCL10, CXCL9, CCL7, sICAM1, IL12p70, LIF and IL7. Three ME/CFS immune phenotypes emerge from the CART analysis comparing ME/CFS subjects with MS comparators and ND controls: (1) an ME/CFS subgroup with lower TGFβ (<2.14 pg ml−1) and CXCL10 (<59.78 pg ml−1) with higher CXCL9 (⩾4.79 pg ml−1) and CCL7 (⩾3.50 pg ml−1); (2) another group with lower TGFβ (<2.14 pg ml−1) and LIF (<0.73 pg ml−1) but higher CXCL10 (⩾59.78 pg ml−1) and IL7 (⩾1.10 pg ml−1) and (3) a third subgroup with higher TGFβ (⩾2.14 pg ml−1) and lower sICAM1 (<207.50 pg ml−1) (Supplementary Figure S1).
Discussion
Others have reported cytokine abnormalities in CSF from subjects with ME/CFS;21 however, this study is the first to employ comprehensive cytokine panels, include both normal control and MS comparator subjects and apply logistic regression and network analysis models. Our results indicate a markedly disturbed immune signature in the CSF of ME/CFS subjects that is consistent with immune activation in the CNS. Persistent secretion of cytokines by activated microglia, brain immune cells of macrophage-monocyte lineage, may contribute to this pattern.
IL1ra, an endogenous downregulator of IL1 and its inflammatory cascades, has been used to address pathological fatigue in conditions other than ME/CFS, such as histiocytosis.41 Our finding that IL1ra has a negative influence on CSF1, CSF2 and IL17F in ME/CFS, without effects on IL1α or IL1β, suggests a disturbance in IL1 signaling pathways. The role of IL1ra in inhibition of allergic responses42, 43 is consistent with reports of an increased prevalence of allergic disorders in some studies of ME/CFS.44, 45 IL1ra-mediated downregulation of responses to allergic stimuli is associated with lower levels of CSF2 (GMCSF), a key regulator of granulocyte and macrophage maturational pathways.46 The inverse relationship we found between IL1ra and CSF2 in the CSF of cases using a network analysis approach suggests that neuroimmune responses may be shifted toward allergic or Th2 (autoimmune) patterns in the CNS of individuals with ME/CFS. One prior study of CSF cytokines in ME/CFS also found decreased levels of CSF2 (GMCSF),21 a molecule that shifts macrophages toward an M1-like, or inflammatory, phenotype.47 The presence of an allergic-type diathesis in the CNS of individuals with ME/CFS is further supported by the finding of increased CSF levels of CCL11 (eotaxin, a chemokine involved in the selective recruitment of eosinophils, an allergy-associated white blood cell subset48) in ME/CFS subjects relative to ND control subjects.
CSF, as a reflection of the microenvironment of the CNS, may provide unique clues to the pathogenesis of ME/CFS. Although increased CSF CCL11 and CXCL10 levels in ME/CFS subjects, along with dysregulation of IL1 signaling, suggest the possibility of an allergic process in central compartments,49, 50 such patterns may also be seen in a wide range of CNS infections.51, 52, 53, 54 Additional studies focused on delineating the relationships of clinical phenotypes of ME/CFS to immune profiles in both the CSF and the peripheral blood are of paramount importance to investigating potential causes and biomarkers for disease. Recent reports indicate regional changes in white matter volume19 and abnormalities in myelin in white matter tracts in ME/CFS.20 Accordingly, future work should also include comparisons with larger numbers of MS subjects representing different clinical subgroups, as well as additional control subjects with neurodegenerative disorders not primarily associated with white matter pathology such as Alzheimer disease or Parkinson disease. Better delineation of the neuroimmune signaling pathways that are specifically dysregulated in ME/CFS may accelerate discovery of effective treatments.
References
Lorusso L, Mikhaylova SV, Capelli E, Ferrari D, Ngonga GK, Ricevuti G . Immunological aspects of chronic fatigue syndrome. Autoimmun Rev 2009; 8: 287–291.
Lyall M, Peakman M, Wessely S . A systematic review and critical evaluation of the immunology of chronic fatigue syndrome. J Psychosom Res 2003; 55: 79–90.
Morris G, Anderson G, Galecki P, Berk M, Maes M . A narrative review on the similarities and dissimilarities between myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and sickness behavior. BMC Med 2013; 11: 64.
Komaroff AL, Cho TA . Role of infection and neurologic dysfunction in chronic fatigue syndrome. Semin Neurol 2011; 31: 325–337.
White PD, Thomas JM, Amess J, Crawford DH, Grover SA, Kangro HO et al. Incidence, risk and prognosis of acute and chronic fatigue syndromes and psychiatric disorders after glandular fever. Br J Psychiatry 1998; 173: 475–481.
Hickie I, Davenport T, Wakefield D, Vollmer-Conna U, Cameron B, Vernon SD et al. Post-infective and chronic fatigue syndromes precipitated by viral and non-viral pathogens: prospective cohort study. BMJ 2006; 333: 575.
Devanur LD, Kerr JR . Chronic fatigue syndrome. J Clin Virol 2006; 37: 139–150.
Lerner AM, Ariza ME, Williams M, Jason L, Beqaj S, Fitzgerald JT et al. Antibody to Epstein-Barr virus deoxyuridine triphosphate nucleotidohydrolase and deoxyribonucleotide polymerase in a chronic fatigue syndrome subset. PLoS One 2012; 7: e47891.
Komaroff AL . Is human herpesvirus-6 a trigger for chronic fatigue syndrome? J Clin Virol 2006; 37: S39–S46.
Komaroff AL, Jacobson S, Ablashi DV, Yamanishi K . Highlights from 5th International Conference on HHV-6 and -7. Herpes 2006; 13: 81–82.
Chapenko S, Krumina A, Kozireva S, Nora Z, Sultanova A, Viksna L et al. Activation of human herpesviruses 6 and 7 in patients with chronic fatigue syndrome. J Clin Virol 2006; 37: S47–S51.
Chia J, Chia A, Voeller M, Lee T, Chang R . Acute enterovirus infection followed by myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and viral persistence. J Clin Pathol 2010; 63: 165–168.
Lerner AM, Beqaj SH, Deeter RG, Fitzgerald JT . IgM serum antibodies to human cytomegalovirus nonstructural gene products p52 and CM2 (UL44 and UL57) are uniquely present in a subset of patients with chronic fatigue syndrome. In Vivo 2002; 16: 153–159.
Beqaj SH, Lerner AM, Fitzgerald JT . Immunoassay with cytomegalovirus early antigens from gene products p52 and CM2 (UL44 and UL57) detects active infection in patients with chronic fatigue syndrome. J Clin Pathol 2008; 61: 623–626.
Carruthers BM, Jain AK, DeMeirleir KL, Peterson DL, Klimas NG, Lerner AM et al. Myalgic Encephalomyelitis/chronic fatigue syndrome: clinical working case definition, diagnostic and treatments protocols. J Chr Fatigue Synd 2003; 11: 7–115.
Nijs J, Nicolson GL, De Becker P, Coomans D, De Meirleir K . High prevalence of Mycoplasma infections among European chronic fatigue syndrome patients. Examination of four Mycoplasma species in blood of chronic fatigue syndrome patients. FEMS Immunol Med Microbiol 2002; 34: 209–214.
Gustaw K . [Chronic fatigue syndrome following tick-borne diseases]. Neurol Neurochir Pol 2003; 37: 1211–1221.
Shungu DC, Weiduschat N, Murrough JW, Mao X, Pillemer S, Dyke JP et al. Increased ventricular lactate in chronic fatigue syndrome. III. Relationships to cortical glutathione and clinical symptoms implicate oxidative stress in disorder pathophysiology. NMR Biomed 2012; 25: 1073–1087.
Puri BK, Jakeman PM, Agour M, Gunatilake KD, Fernando KA, Gurusinghe AI et al. Regional grey and white matter volumetric changes in myalgic encephalomyelitis (chronic fatigue syndrome): a voxel-based morphometry 3 T MRI study. Br J Radiol 2012; 85: e270–e273.
Zeineh MM, Kang J, Atlas SW, Raman MM, Reiss AL, Norris JL et al. Right arcuate fasciculus abnormality in chronic fatigue syndrome. Radiology 2014; 274: 517–526.
Natelson BH, Weaver SA, Tseng CL, Ottenweller JE . Spinal fluid abnormalities in patients with chronic fatigue syndrome. Clin Diag Lab Immunol 2005; 12: 52–55.
Baraniuk JN, Casado B, Maibach H, Clauw DJ, Pannell LK, Hess SS . A chronic fatigue syndrome - related proteome in human cerebrospinal fluid. BMC Neurol 2005; 5: 22.
Schutzer SE, Angel TE, Liu T, Schepmoes AA, Clauss TR, Adkins JN et al. Distinct cerebrospinal fluid proteomes differentiate post-treatment lyme disease from chronic fatigue syndrome. PLoS One 2011; 6: e17287.
Schutzer SE, Rounds MA, Natelson BH, Ecker DJ, Eshoo MW . Analysis of cerebrospinal fluid from chronic fatigue syndrome patients for multiple human ubiquitous viruses and xenotropic murine leukemia-related virus. Ann Neurol 2011; 69: 735–738.
Knox KK, Carrigan DR . In vitro suppression of bone marrow progenitor cell differentiation by human herpesvirus 6 infection. J Infect Dis 1992; 165: 925–929.
Burd EM, Knox KK, Carrigan DR . Human herpesvirus-6-associated suppression of growth factor-induced macrophage maturation in human bone marrow cultures. Blood 1993; 81: 1645–1650.
Drobyski WR, Dunne WM, Burd EM, Knox KK, Ash RC, Horowitz MM et al. Human herpesvirus-6 (HHV-6) infection in allogeneic bone marrow transplant recipients: evidence of a marrow-suppressive role for HHV-6 in vivo. J Infect Dis 1993; 167: 735–739.
Gonzalez-Dunia D, Volmer R, Mayer D, Schwemmle M . Borna disease virus interference with neuronal plasticity. Virus Res 2005; 111: 224–234.
Wigerius M, Melik W, Elvang A, Johansson M . Rac1 and Scribble are targets for the arrest of neurite outgrowth by TBE virus NS5. Molec Cell Neurosci 2010; 44: 260–271.
Hanken K, Eling P, Hildebrandt H . The representation of inflammatory signals in the brain- a model for subjective fatigue in multiple sclerosis. Front Neurol 2014; 5: 264.
McDonald WI, Ron MA . Multiple sclerosis: the disease and its manifestations. Philos Trans R Soc Lond B Biol Sci 1999; 354: 1615–1622.
Navikas V, Link H . Review: cytokines and the pathogenesis of multiple sclerosis. J Neurosci Res 1996; 45: 322–333.
Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A . The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann Intern Med 1994; 121: 953–959.
Martins TB . Development of internal controls for the Luminex instrument as part of a multiplex seven-analyte viral respiratory antibody profile. Clin Diag Lab Immunol 2002; 9: 41–45.
Vignali DA . Multiplexed particle-based flow cytometric assays. J Immunol Methods 2000; 243: 243–255.
Box GEP, Cox DR . An analysis of transformations. J R Stat Soc Series B 1964; 26: 211–252.
Tibshirani R . Regression shrinkage and selection via the Lasso. J R Stat Soc Series B 1996; 58: 267–288.
Smith M, Milic-Frayling N, Shneiderman B, Mendes Rodrigues E, Leskovec J, Dunne C NXL: a free and open network overview, discovery and exploration add-in for Excel 2007/2010. Social Media Research Foundation http://www.smrfoundation.org 2010. (accessed 1 August 2014) at http://nodexl.codeplex.com/.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 1995; 57: 289–300.
Breiman L, Friedman JH, Olshen RA, Stone CJ . Classification and Regression Trees. Wadsworth & Brooks/Cole Advanced Books & Software: Monterey, CA, USA, 1984.
Sevcikova S, Kubiczkova L, Sedlarikova L, Rihova L, Kryukov F, Szturz P et al. Impact of anakinra treatment on cytokine and lymphocytes/monocytes profile of an Erdheim-Chester patient. Klin Onkol 2014; 27: 276–282.
Pattaro C, Heinrich J, Werner M, de Marco R, Wjst M . Association between interleukin-1 receptor antagonist gene and asthma-related traits in a German adult population. Allergy 2006; 61: 239–244.
Amrol DJ, Hagaman DD, Sheller JR, Murray JJ . Soluble CD23 and interleukin-1 receptor antagonist in human asthmatics following antigen challenge. J Asthma 2005; 42: 73–76.
Endicott NA . Chronic fatigue syndrome in psychiatric patients: lifetime and premorbid personal history of physical health. Psychosom Med 1998; 60: 744–751.
Borish L, Schmaling K, DiClementi JD, Streib J, Negri J, Jones JF . Chronic fatigue syndrome: identification of distinct subgroups on the basis of allergy and psychologic variables. J Allerg Clin Immunol 1998; 102: 222–230.
Sim TC, Hilsmeier KA, Reece LM, Grant JA, Alam R . Interleukin-1 receptor antagonist protein inhibits the synthesis of IgE and proinflammatory cytokines by allergen-stimulated mononuclear cells. Am J Respir Cell Molec Biol 1994; 11: 473–479.
Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol 2012; 188: 5752–5765.
Pease JE . Asthma, allergy and chemokines. Curr Drug Targets 2006; 7: 3–12.
Lampinen M, Carlson M, Hakansson LD, Venge P . Cytokine-regulated accumulation of eosinophils in inflammatory disease. Allergy 2004; 59: 793–805.
Bisset LR, Schmid-Grendelmeier P . Chemokines and their receptors in the pathogenesis of allergic asthma: progress and perspective. Curr Opin Pulmon Med 2005; 11: 35–42.
Boulware DR, Bonham SC, Meya DB, Wiesner DL, Park GS, Kambugu A et al. Paucity of initial cerebrospinal fluid inflammation in cryptococcal meningitis is associated with subsequent immune reconstitution inflammatory syndrome. J Infect Dis 2010; 202: 962–970.
Chang EE, Yen CM . Eosinophil chemoattracted by eotaxin from cerebrospinal fluid of mice infected with Angiostrongylus cantonensis assayed in a microchamber. Kaohsiung J Med Sci 2004; 20: 209–215.
Kielian T . Microglia and chemokines in infectious diseases of the nervous system: views and reviews. Front Biosci 2004; 9: 732–750.
Hosking MP, Lane TE . The role of chemokines during viral infection of the CNS. PLoS Pathog 2010; 6: e1000937.
Acknowledgements
We are grateful for the support of the Chronic Fatigue Initiative/Hutchins Family Foundation and the Edward P Evans Foundation (WIL and MH). We appreciate the technical support of Wai Wong, Noel Pura and Shobun Baile.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
About this article
Cite this article
Hornig, M., Gottschalk, G., Peterson, D. et al. Cytokine network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome. Mol Psychiatry 21, 261–269 (2016). https://doi.org/10.1038/mp.2015.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2015.29
- Springer Nature Limited
This article is cited by
-
Proteomics and cytokine analyses distinguish myalgic encephalomyelitis/chronic fatigue syndrome cases from controls
Journal of Translational Medicine (2023)
-
Potential molecular mechanisms of chronic fatigue in long haul COVID and other viral diseases
Infectious Agents and Cancer (2023)
-
Treatment of 95 post-Covid patients with SSRIs
Scientific Reports (2023)
-
Polyphenols as possible alternative agents in chronic fatigue: a review
Phytochemistry Reviews (2023)
-
How mycobacterium tuberculosis infection could lead to the increasing risks of chronic fatigue syndrome and the potential immunological effects: a population-based retrospective cohort study
Journal of Translational Medicine (2022)