Abstract
Classical Hodgkin lymphoma (cHL) and anaplastic large cell lymphoma (ALCL) feature high expression of activator protein-1 (AP-1) transcription factors, which regulate various physiological processes but also promote lymphomagenesis. The AP-1 factor basic leucine zipper transcription factor, ATF-like 3 (BATF3), is highly transcribed in cHL and ALCL; however, its functional importance in lymphomagenesis is unknown. Here we show that proto-typical CD30+ lymphomas, namely cHL (21/30) and primary mediastinal B-cell lymphoma (8/9), but also CD30+ diffuse large B-cell lymphoma (15/20) frequently express BATF3 protein. Mass spectrometry and co-immunoprecipitation established interactions of BATF3 with JUN and JUNB in cHL and ALCL lines. BATF3 knockdown using short hairpin RNAs was toxic for cHL and ALCL lines, reducing their proliferation and survival. We identified MYC as a critical BATF3 target and confirmed binding of BATF3 to the MYC promoter. JAK/STAT signaling regulated BATF3 expression, as chemical JAK2 inhibition reduced and interleukin 13 stimulation induced BATF3 expression in cHL lines. Chromatin immunoprecipitation substantiated a direct regulation of BATF3 by STAT proteins in cHL and ALCL lines. In conclusion, we identified STAT-mediated BATF3 expression that is essential for lymphoma cell survival and promoted MYC activity in cHL and ALCL, hence we recognized a new oncogenic axis in these lymphomas.
Similar content being viewed by others
Introduction
Classical Hodgkin lymphoma (cHL) is one of the most common malignant lymphomas, particularly in young adults. In 30–40% of cHL cases the Hodgkin and Reed/Sternberg (HRS) tumor cells are latently infected by Epstein-Barr virus (EBV), which contributes to lymphoma pathogenesis in these cases.1 HRS cells most likely originate from preapoptotic germinal center (GC) B cells.2, 3 Anaplastic large cell lymphoma (ALCL) is an aggressive T-cell non-Hodgkin lymphoma, and two main subtypes are distinguished based on the presence or absence of translocations involving the ALK gene that result in constitutive ALK kinase activity. Despite the fact that they derive from different types of lymphocytes, HRS and ALCL cells share morphological, biological and phenotypical features, as well as oncogenic signatures, including constitutive activities of the JAK/STAT pathway and activator protein-1 (AP-1) transcription factors.4, 5 A striking feature of HRS cells is the lost expression of B-cell-specific molecules, and similarly, ALCL cells often lack expression of T-cell-specific proteins.1, 6 The tumor necrosis factor receptor CD30 is characteristically expressed by the tumor cells in cHL and ALCL, but also in a fraction of several other types of lymphomas, including primary mediastinal B-cell lymphoma (PMBL). CD30 is otherwise expressed by some activated B as well as T lymphocytes in normal lymphoid tissues.7
AP-1 factors regulate diverse cellular processes, including differentiation, proliferation and cell survival, but they are also critically involved in the development of various cancers.8 They contain a basic DNA binding domain and a leucine zipper dimerization domain, and dimers preferentially bind to consensus TGA(C/G)TCA or TGACGTCA DNA motifs.8 In HRS, ALCL and PMBL cells AP-1 family members JUN and JUNB are commonly expressed, and show high DNA binding activity.4, 5, 9, 10 Knockdown of JUN and JUNB, separately or in combination, reduced cell cycle progression and colony formation of HRS and ALCL cells, but their survival is barely affected. Ectopic expression of dominant-negative A-FOS, which inhibits the activity of AP-1 complexes, induced apoptosis in ALCL but not HRS cells.4 In a mouse model of NPM-ALK-mediated lymphomagenesis, JUN and JUNB supported tumor development and growth.11 Moreover, AP-1 induces immunomodulatory molecules such as galectin-1 in HRS and ALCL cells12 and programmed death ligand 1 in HRS cells,13 supporting the generation of the well-known immunosuppressive microenvironment in cHL.
We and others previously recognized high mRNA expression of the AP-1 family member basic leucine zipper transcription factor, ATF-like 3 (BATF3; alias 21 kDa small nuclear factor isolated from T cells (SNFT)) in primary HRS and PMBL cells.14, 15, 16 BATF3 mRNA levels are also higher in ALCL cells than in normal T lymphocytes,14, 17 and BATF3 (reported as SNFT) protein expression has been detected by immunohistochemistry in all ALCL subtypes.5 In normal cells BATF3 is expressed in T helper type 1 cells and plays a major role in the development and function of conventional dendritic cells.18, 19 Notably, loss of conventional dendritic cells in a BATF3-deficient background is restored by ectopic BATF expression, and enforced BATF3 expression restored BATF-dependent function in BATF-deficient lymphocytes.20 In a mouse model, CD2 promoter-driven BATF overexpression induced a lymphoproliferative disease.21 Given their functional redundancy,19 this suggests that also BATF3 has oncogenic potential.
Here, we analyzed the expression of BATF3 protein in several lymphomas and potential interaction partners of BATF3 in cHL and ALCL. To assess the functional importance of BATF3, we used short hairpin-mediated RNA interference, and we investigated the molecular mechanisms of BATF3 overexpression in these lymphomas.
Materials and methods
Cell culture
Detailed cell culture conditions are provided in the Supplementary Material and Methods. Cells were treated with JAK2 inhibitors AZD1480 and TG101348 (both Santa Cruz Biotechnology, Heidelberg, Germany) for 24 h with indicated concentrations, and with 5 ng/μl human recombinant IL-13 (PeproTech, Hamburg, Germany) for 30 min.
Immunohistochemistry
Immunohistochemistry was performed on sections of formalin-fixed and paraffin-embedded cHL (n=30), diffuse large B-cell lymphoma (DLBCL, n=30), primary mediastinal B-cell lymphoma (PMBL, n=9) and reactive lymph nodes. Slides were stained with mouse anti-human BATF3 antibody (3H1; Abnova, Heidelberg, Germany), rabbit anti-human MYC antibody (ab31072; Abcam, Cambridge, UK) or anti-human phosphorylated STAT3 antibody (Y705, M9C6; Cell Signaling Technology, Leiden, Netherlands). For detection, the EnVision System-HRP (DAKO, Hamburg, Germany) was used. A cutoff of 50% positive tumor cells was applied to define sample positivity, but in positive cases almost all HRS cells were positive and in negative cases usually all HRS cells were negative.
Isolation and sorting of normal B-cell subsets from tonsils
Human tonsils were collected from children and adolescents undergoing routine tonsillectomy with donor’s informed consent, as approved by the local ethics committee of the Medical School of the University Duisburg-Essen. Antibodies and sorting strategies for fluorescence-activated cell sorting are given in the Supplementary Materials and Methods.
Production of lentiviral particles and transduction of lymphoma cell lines
Design and cloning of shRNAs into Lentiviral Gene Ontology (LeGO) vectors (lentigovectors.de22) is described in detail in the Supplementary Material and Methods.
Immunoblotting and co-immunoprecipitation
Details for immunoblotting and co-immunoprecipitation are provided in the Supplementary Material and Methods. For separation of nuclear and cytosolic cell fractions the BioVision (Milpitas, CA, USA) Nuclear/Cytosol Fractionation kit was used according to the manufacturer's instructions. Primary antibodies used are presented in the Supplementary Materials and Methods.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the Pierce Agarose ChIP Kit (Thermo Fisher Scientific, Darmstadt, Germany), following the manufacturer’s instructions. Details for ChIP are provided in the Supplementary Materials and Methods.
RNA isolation, reverse transcription quantitative PCR and gene expression analysis
Total RNA isolation, and reverse transcription and ChIP-qPCR are described in the Supplementary Material and Methods. TaqMan probes (Applied Biosystems, Darmstadt, Germany) and oligonucleotides are provided in the Supplementary Material and Methods. For gene expression analysis, samples were run on a GeneST2.0 platform and analyzed with the ANOVA test statistics implemented in Partek GS software. GeneChip data have been submitted to GEO (GSE93862).
Proliferation and apoptosis assays
Proliferation was measured using the cell membrane labeling dye Vybrant DiD (Thermo Fisher Scientific), according to the manufacturer’s instructions. Apoptosis was assessed by flow cytometry using annexin V-APC, propidium iodide (Becton Dickinson, Heidelberg, Germany) and CaspGLOW Red Active Caspase-3 Staining Kit (BioVision). Details are provided in the Supplementary Materials and Methods. Data were analyzed using Flowjo software (LLC, v7.6.5).
LC-MS/MS analysis and peptide identification and quantification
Five replicate samples of BATF3- and isotype-precipitated L-428 cells were subjected to mass spectrometry (MS) analysis. Details are described in the Supplementary Materials and Methods.
Statistical analysis
Data analysis of pairwise comparisons was tested for statistical significance by Student’s t-test with a 95% confidence interval (Prism 6; GraphPad Software Inc, La Jolla, CA, USA). P-values are as follows: *P<0.05; **P<0.01; ***P<0.001, and data are shown as mean values plus standard deviation (s.d.).
Results
BATF3 is expressed in many CD30-positive lymphomas and in CD30-positive normal B cells
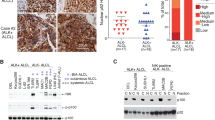
Previous studies showed that tumor cells from cHL, PMBL and ALCL express high levels of BATF3 mRNA.5, 14, 15, 16, 17, 23 A survey of gene expression profiles (GEPs) derived from a comprehensive set of HL and B-cell non-Hodgkin lymphoma entities revealed increased BATF3 expression in cHL and PMBL (Supplementary Figure S1a). HRS cells expressed approximately nine-fold higher levels of BATF3 than lymphocyte predominant tumor cells of NLPHL, and PMBL cells expressed about five-fold higher levels of BATF3 than DLBCL cells.14, 23 BATF2 transcripts were undetectable in GEPs of primary lymphomas (data not shown), and BATF mRNA levels were not significantly different between HRS cells and NLPHL or PMBL cells (Supplementary Figure S1b). In cHL, BATF3 was expressed two-fold higher on average in the group of EBV-negative cases but the difference was not statistically significant (P=0.08; Supplementary Figure S1c). The same tendency was observed in an independent GEP analysis of HRS cells (Supplementary Figure S1d).24 Using immunohistochemistry, the lymphoma cells in 21/30 (70%) cHL and in 8/9 (89%) PMBL expressed BATF3 protein (Figure 1a and Table 1). In line with the GEP data, BATF3 positivity significantly correlated with EBV negativity in cHL (P<0.002; Figure 1b). In DLBCL, BATF3 positivity was significantly associated with CD30 positivity (3/10 CD30− DLBCL vs 15/20 CD30+ DLBCL, P<0.05) but not with EBV negativity (Figure 1c and Table 1). We then assessed BATF3 protein expression and subcellular localization in various lymphoma cell lines using immunoblot analysis. Except for NLPHL-derived DEV, all cHL and ALCL lines analyzed expressed BATF3 protein (Supplementary Figure S1e), including CD30low L-1236 and CD30− SUP-HD1 cHL lines (data not shown). In non-Hodgkin lymphoma lines, BATF3 protein was expressed in PMBL lines and a DLBCL line of ABC subtype (Supplementary Figure S1f). In keeping with the subcellular localization of a functional transcription factor, BATF3 was predominantly identified in the nuclear fractions of cHL and ALCL lines (Figure 1d).
Expression of BATF3 in lymphomas. (a) Representative images of BATF3 expression in sections of primary cHL (n=30), PMBL (n=9) and DLBCL (n=30)) by immunohistochemistry are shown. Bars indicate 50 μm. (b, c) Percentages of BATF3-positive cases are shown for the comparisons of EBV+ and EBV− cHL in (b) and CD30− and CD30+ DLBCL in (c). Statistical analysis was performed using a Student’s t-test. (d) BATF3 localization in cytoplasmic and nuclear fractions of cHL and ALCL lines by immunoblot analysis. Representative image of two independent experiments is shown. TATA box binding protein (TBP) and α-tubulin (TUB) served as references. Size of protein marker is indicated in immunoblot images.
By GEP, BATF3 expression was undetectable in normal B-cell subsets including memory, GC B and plasma cells.14 However, some blastoid cells in the GC and in the extrafollicular region of reactive lymph nodes stained weakly positive for BATF3 (Supplementary Figure S1g). Given the predominant expression of BATF3 in CD30+ lymphoma cells, we hypothesized that BATF3 is expressed in CD30+ normal B cells. To test this, we analyzed FACS-sorted CD30+ GC and non-GC B cells and CD30− memory B, GC B and plasma cells by reverse transcription quantitative PCR (RT-qPCR). BATF3 expression was significantly higher in CD30+ than CD30− B-cell subsets (Supplementary Figure S1h). For comparison, AP-1 factors JUN and FOS were included that showed highest expressions in plasma cells (Supplementary Figure S1h).25 Thus, BATF3 is predominantly expressed in CD30+ lymphomas, including cHL, PMBL, ALCL and a fraction of DLBCL, and the subset of normal CD30+ B cells.
BATF3 forms AP-1 complexes with JUN and JUNB in cHL and ALCL lines
To identify binding partners of BATF3 in lymphoma cells, we performed MS analysis after BATF3 immunoprecipitation (IP) using L-428 HRS cells. Peptides of 25 proteins were significantly enriched in the BATF3 IP fraction (⩾1.5-fold change; P<0.05) including BATF3, as expected, and JUN (Supplementary Figure S2a and Supplementary Table S1). The enrichment of JUNB and JUND by approximately 8- and 3.5-fold, respectively, did not reach statistical significance (Supplementary Figure S2a and Supplementary Table S1). In cHL and ALCL lines as well as primary tumor cells AP-1 factors JUN and JUNB are constitutively expressed (Supplementary Figure S2c).4, 9, 10, 26, 27 By co-IP, we identified JUN and JUNB in BATF3 IP fractions of L-428 and HDLM-2 cells, and BATF3 in corresponding JUN and JUNB IP fractions, validating the composition of BATF3-containing AP-1 complexes in cHL lines (Supplementary Figure S2b). These complexes were also detected in two ALCL lines (Supplementary Figure S2b). Apart from JUN proteins, the MS analysis identified a non-significant 1.3-fold enrichment of interferon regulatory factor (IRF) 4 and IRF5 in BATF3 IP fractions of L-428 cells (Supplementary Table S1). However, IRF4 did not coprecipitate with BATF3 in L-428 HRS and SR-786 ALCL cells (Supplementary Figure S2d), most likely due to the known weak interaction of BATF and IRF proteins at only few critical amino acids.28
BATF3 knockdown impairs proliferation and survival in cHL and ALCL lines
To assess the functional relevance of BATF3 in cHL, PMBL and ALCL, we used short hairpin (sh) RNA interference. Three of four shRNAs effectively reduced BATF3 protein expression in GFP+-sorted L-428 cells (Supplementary Figure S3a). We also monitored GFP+ cells in competitive culture conditions by flow cytometry. The proportion of shBATF3-transduced L-428 cells continuously decreased compared with the proportion of cells infected with a non-targeting shRNA (shNT), indicating selective disadvantage upon BATF3 knockdown (Supplementary Figure S3b). Transductions of cell lines from various lymphoma entities with two shRNAs against BATF3 established a toxic effect not only in cHL but also in ALCL lines and in one of two PMBL lines (Figures 2a and b and Supplementary Figure S3c). The fitness of a Burkitt lymphoma, a DLBCL and a T-cell leukemia line, which expressed very weak to moderate BATF3 levels (Supplementary Figure S3d), was not compromised, showing that the two shRNAs against BATF3 are not generally toxic. BATF3 downregulation did not affect surface CD30 expression (Supplementary Figure S3e). To evaluate an effect on cell survival, proportions of cells stained with annexin V and propidium iodide as well as of cells with active caspase-3 were determined by flow cytometry. Indeed, shBATF3-transduced cHL and ALCL lines showed significantly higher proportions of both annexin V-positive cells and caspase-3-active apoptotic cells than shNT-transduced lines (Figures 2c and d and Supplementary Figure S3f). We then measured cell division using a cell membrane labeling dye (Vybrant) to assess proliferation upon BATF3 downregulation. Over time, the Vybrant fluorescence intensities of shNT-transduced cHL and ALCL lines declined gradually, reflecting continuous proliferation. In contrast, cells transduced with shRNAs against BATF3 displayed a consistent reduction in their proliferative capacity (Figures 2e and f). Thus, knockdown of BATF3 is selectively toxic to cHL and ALCL cells, compromising both survival and proliferation.
BATF3 knockdown reduced proliferation and survival in cHL and ALCL lines. (a–f) Experiments were performed after lentiviral transduction with two shRNAs (sh1 and sh4) against BATF3 and a non-targeting shRNA (shNT) as control. (a) Reduced BATF3 protein expression in cHL and ALCL lines after transduction, assessed by immunoblot analysis. Whole-cell lysates were prepared from GFP+ (transduced) cells sorted 3 days after infection. A representative immunoblot of at least two experiments is shown. Percentages of BATF3 protein expression in shBATF3-transduced samples are indicated relative to shNT-transduced samples and β-actin (ACTB) that served as reference. Size of protein marker is indicated. (b) Cell lines of different origins, including cHL, ALCL, primary mediastinal B-cell lymphoma (PMBL), Burkitt lymphoma (BL), germinal center B-cell-like (GCB) DLBCL and a T-cell acute lymphoblastic leukemia (T-ALL) were transduced, and analyzed by flow cytometry to assess proportions of GFP+ cells over time. Percentages of GFP+ cells were normalized to day 3 and non-targeting control shRNA (shNT). Data are representative of at least two independent experiments. (c, d) Flow cytometric analysis of annexin V and propidium iodide (PI)-stained cHL and ALCL lines. GFP+ (transduced) cells were sorted 4 days after infection and maintained in culture for another 3–4 days before analysis to allow them to recover from cell sorting stress. Representative plots in (c) indicate gates and percentages of annexin V+ cells, and data summarized in (d) represent mean plus s.d. of at least three independent experiments. (e, f) Flow cytometric analysis of Vybrant-stained cHL and ALCL lines after transduction. Unsorted cells were stained with membrane integrating Vybrant dye 4 days after infection (day 0) and mean fluorescence intensities (MFI) of GFP+ (transduced) cells were determined at time points indicated. Histograms in (e) are representative of at least three independent experiments. MFI values for Vybrant in (f) are shown as mean plus s.d. and normalized to day 0. A paired t-test was performed for statistical analysis. *P<0.05; **P<0.01; NS, not significant.
BATF3 regulates MYC expression in cHL and ALCL lines and binds to the MYC promoter
To determine the molecular mechanism(s) critically regulated by BATF3, we investigated the global gene expression changes in L-428 cells after BATF3 knockdown. The expression of 107 annotated genes changed significantly (Supplementary Figure S4a and Supplementary Table S2). The regulation of several genes was verified by RT-qPCR in L-428 and two additional cHL lines (HDLM-2 and U-HO1) after transduction with sh4BATF3 (Supplementary Figure S4b). BATF3 downregulation did not affect BATF and BATF2 expression, hence they did not functionally compensate for BATF3 in this setting (Supplementary Figures S4c and d). To capture broader biological changes upon BATF3 downregulation, we performed gene set enrichment analysis (GSEA) using the hallmark collection (v5), which includes well-defined and non-redundant gene sets. GSEA identified four and two gene sets significantly upregulated and downregulated, respectively, in sh4BATF3-transduced L-428 cells (Supplementary Table S3). BATF3 knockdown significantly induced gene sets of interferon gamma and alpha response, JAK/STAT and NF-κB signaling pathways. These pathways are highly active in HRS cells, promoting proliferation and survival, hence their upregulation suggests a potential feedback mechanism in the event of shBATF3-induced toxicity.
Interestingly, only the two MYC gene sets in this collection were significantly associated with genes downregulated in shBATF3-transduced L-428 cells (Supplementary Table S3). Likewise, three MYC signatures derived from B-cell lymphoma lines29, 30 and murine MYC-positive GC B cells31 were significantly enriched among genes downregulated upon BATF3 knockdown (Figure 3a). While the reduction of MYC transcription did not reach statistical significance in sh4BATF3-transduced L-428 cells (Supplementary Figure S5a), MYC protein expression was markedly reduced by immunoblot analysis in cHL and ALCL lines after BATF3 knockdown (Figure 3b and Supplementary Figure S5b). Although BATF3 and MYC baseline protein levels did not indicate a strict association in cHL lines (Supplementary Figure S5c), we tested whether ectopically expressed BATF3 augments MYC expression. Enforced BATF3 expression in U-HO1 cells with lowest endogenous BATF3 protein levels slightly enhanced MYC expression (Supplementary Figure S5d); a stronger induction may be limited by the availability of cofactors, such as JUN and/or IRF proteins.19
BATF3 regulates MYC expression and binds to an AP-1 site in the MYC promoter in cHL and ALCL lines. (a) GSEA-derived enrichment plots of MYC signatures downregulated in shBATF3-transduced L-428 cells. MYC signatures were derived from human B-cell lymphoma cells and murine B cells.29, 30, 31 GSEA was performed with three paired biological replicates of shNT- and shBATF3-transduced L-428 cells. GFP+ cells were sorted 4 days after infection. Normalized enrichment scores (NES), P-values (p-val) and false discovery rates (FDR) are indicated. (b) Representative immunoblots of MYC protein reduction in cHL and ALCL lines transduced with sh1RNA and sh4RNA against BATF3 compared with control non-targeting shRNA (shNT) are shown. GFP+ (transduced) cells were sorted 3 or 4 days after infection. Percentages of BATF3 and MYC protein expression in shBATF3-transduced samples relative to shNT-transduced samples and β-actin (ACTB) are indicated. Size of protein marker is indicated. Graph shows the percentages of MYC protein expression of at least three independent transduction experiments as mean plus s.d. after normalization to shNT and ACTB. A paired t-test was performed for statistical analysis. *P<0.05; NS, not significant. (c) Schematic illustration of the MYC locus surrounding the transcription start site (TSS), including the PCR product that covers the AP-1 site (5′-TGAGTCA-3′) in the MYC promoter region. (d) BATF3 binding to the AP-1 site-containing sequence in the MYC promoter (but not upstream (−6 kb) or downstream (+3.3 and +13.3 kb) control sites) in cHL and ALCL lines, assessed by quantitative PCR (qPCR), following ChIP with anti-BATF3 or isotype control (IgG) antibodies. ChIP-qPCR signals were normalized to input DNA. Data are shown as mean plus s.d. of at least three independent experiments. (e) JUN binding to the AP-1 site-containing sequence in the MYC promoter (but not upstream (−6 kb) or downstream (+3.3 and +13.3 kb) control sites) of cHL lines L-428 and UHO-1, assessed by ChIP-qPCR with anti-JUN or isotype control (IgG) antibodies. ChIP-qPCR signals were normalized to input DNA. Data are shown as mean+s.d. of at least two independent experiments.
To establish MYC as a direct BATF3 target gene, we performed ChIP followed by quantitative PCR (ChIP-qPCR). Binding of BATF3 to a conserved AP-1 site in the MYC promoter32 but not to control sequences was readily detected in cHL and ALCL lines (Figures 3c and d). Concordant with previously reported binding of JUN at this site in NIH-3T3 fibroblasts,2 JUN bound to this AP-1 site at the MYC promoter also in L-428 and UHO-1 HRS cells by ChIP-qPCR (Figure 3e). MYC protein is expressed in tumor cells of most cHL (23/30; Table 2) and ALCL.30, 33 The functional relevance of MYC induction by BATF3 was tested by shRNA-mediated downregulation of MYC in two cHL and two ALCL lines. In line with and extending earlier reports, MYC knockdown was toxic in all four cell lines (Supplementary Figure S5e).30, 34 BATF3 expression was not reduced upon MYC downregulation (Supplementary Figure S5f). In ALCL cells MYC is an IRF4 target gene,30 but in HRS cells its expression did not change upon IRF4 knockdown (Supplementary Figure S5g), arguing against a role of IRF4 in BATF3-mediated regulation of MYC in cHL. Taken together, BATF3 binds to an AP-1 motif in the MYC promoter and controls MYC expression in cHL and ALCL lines.
BATF3 expression is regulated by the JAK/STAT pathway
Next, we sought to identify the mechanism(s) that promote BATF3 upregulation in lymphoma cells. Previous reports suggested that AP-1 factors are putative targets of the JAK/STAT signaling pathway, which is constitutively active in cHL, PMBL and ALCL.34, 35, 36, 37 To test whether JAK/STAT signaling regulates BATF3 expression, we treated cHL and PMBL cell lines with JAK2 inhibitors AZD1480 and TG101348 for 24 h. As a result, BATF3 protein expression decreased with increasing concentrations in PMBL lines and two of three cHL lines, along with a dephosphorylation of STAT6 (Figure 4a and Supplementary Figure S6a). Likewise, stimulation of JAK/STAT signaling with interleukin 13 (IL-13), which activates STAT6 in cHL lines,38 induced BATF3 protein expression in three cHL lines tested (Figure 4b).
JAK/STAT signaling regulates BATF3 expression in HRS cells and phosphorylated STAT binds to BATF3 promoter/enhancer sequences in cHL and ALCL lines. (a) Immunoblot analysis of reduced BATF3 expression in response to JAK2 inhibitors AZD1480 and TG101348 in cHL lines. Cells were treated with increasing concentrations of either inhibitor or vehicle control (DMSO) for 24 h. Phosphorylated STAT6 (p-STAT6), total STAT6 and β-actin (ACTB) served as references. Results are representative of two independent experiments. (b) Interleukin 13 (IL-13) treatment of cHL lines induced BATF3 protein expression, assessed by immunoblotting. Cell lines were serum-starved for 6 h before IL-13 stimulation (5 μg/ml) for 30 min. Representative blots of two independent experiments are shown. Size of protein marker is indicated. (a–c) Percentages of BATF3 and p-STAT6 protein expression in shBATF3-transduced samples relative to shNT-transduced samples and β-actin (ACTB) are indicated. (c) Schematic illustration of the BATF3 locus surrounding the transcription start site (TSS). PCR products covering STAT motifs (5′–3′) are shown. (d) Quantitative PCR (qPCR) of putative STAT-binding site products after ChIP with indicated anti-phosphorylated STAT in cHL and ALCL lines compared with isotype control antibody (IgG). ChIP-qPCR signals were normalized to input DNA. Data are shown as mean plus s.d. of at least three independent experiments.
To analyze whether STATs directly regulate BATF3, we performed ChIP with antibodies against phosphorylated STAT3 (p-STAT3), p-STAT5 and p-STAT6, followed by qPCR analysis for three potential STAT-binding sites in BATF3 (Figure 4c). Binding of p-STAT3 and p-STAT6, but not p-STAT5, to BATF3 sequences was readily detected in cHL lines L-428 and U-HO1 (Figure 4d). Consistent with unchanged BATF3 protein expression upon JAK2 inhibition, binding of p-STAT3 and p-STAT6 to BATF3 sequences was not detected in HDLM-2 cells (Supplementary Figure S6b). In ALCL lines Karpas 299 and SU-DHL-1 p-STAT3 directly bound to all three sites in BATF3 (Figure 4d). By immunohistochemistry, 15 of 16 cHL stained positive for p-STAT3, including 9/9 BATF3-positive but also 6/7 BATF3-negative cases (Supplementary Table S5), supporting the idea that constitutively active JAK/STAT signaling contributes to BATF3 overexpression also in primary HRS cells, and presumably in PMBL and ALCL.
Discussion
Transcription factor networks that normally drive lymphocytes through stages of differentiation are frequently hijacked during malignant transformation as a consequence of genetic lesions or increased expression due to constitutive signaling pathways. In HRS cells the repertoire of deregulated signaling pathways and transcription factors is exceptionally large, including constitutive activity of JAK/STAT, NF-κB and AP-1 factors.1 Here, we identified oncogenic properties of overexpressed BATF3 in HRS and ALCL cells, critically supporting their proliferation and survival. We show that BATF3 expression is regulated by JAK/STAT signaling and BATF3 in turn regulates the expression of MYC (Figure 5). These findings provide new insight in the pathogenic events of cHL and ALCL and perhaps other BATF3-positive lymphomas. In addition to its reported positivity in ALCL cells,5 BATF3 protein was frequently expressed not only by HRS cells but also by tumor cells of PMBL and CD30+ DLBCL. Thus, it emerged as predominantly expressed in CD30+ lymphomas, resembling the expression pattern of other AP-1 factors,9, 10 except for the inverse correlation with EBV in HRS cells. The reason for unnoticeable BATF3 protein in most EBV-positive cHL is presently unclear. This tendency is consistent with GEP data (Supplementary Figures S1b and c); however, Vrzalikova and colleagues reported higher frequencies of BATF3 positivity in EBV+ cHL by immunohistochemistry, likely due to a much lower cutoff for positivity in their analysis (P. Murray, personal communication). In normal immune cells BATF3 expression appeared to be restricted to CD8+ dendritic cells and Th1 T cells.18, 19 Indeed, in normal B-cell subsets BATF3 was only expressed by the rare CD30+ B cells, suggesting that it is constitutively but not aberrantly upregulated in CD30+ lymphoma cells. Notably, normal human tonsillar CD30+ T cells also expressed BATF3 by GEP (Supplementary Figure S1i).5 Hence, BATF3 and CD30 expression are closely associated in malignant and normal B and T cells; however, CD30 expression remained stable after BATF3 knockdown. Other AP-1 factors expressed in BATF3-positive cells possibly regulate the expression of CD30.26, 27, 39
Model of BATF3 linking oncogenic JAK/STAT signaling to MYC expression in cHL and ALCL. Interleukin 13 (IL-13) activates JAK2 in cHL lines, leading to the phosphorylation of STAT3/6. Phosphorylated STAT dimers bind to BATF3 in HRS and ALCL cells, promoting constitutively high BATF3 expression, resulting in the formation of AP-1 complexes with JUN and JUNB. In turn, BATF3/JUN dimers directly bind to MYC, contributing to high MYC protein levels in these lymphomas. BATF3-mediated expression of MYC and perhaps other target genes is essential for proliferation and survival of HRS and ALCL cells.
Increased BATF3 gene copy numbers were not apparent in primary cHL, but gains involving the BATF3 locus on chromosome 1q32.3 have been described in two cHL lines.24, 40, 41, 42 Gains of 1q in ALK-negative ALCL were also reported but the minimal region did not include 1q32.3.43 Three microRNAs potentially target BATF3: miR-22-3p, miR-140-3p1 and miR-214-5p (TargetScan v7.1). In ALK-negative but not ALK-positive ALCL miR-22 was downregulated,44 but BATF3 (reported as SNFT) protein was expressed in all ALCL subtypes.5 In agreement with previous reports suggesting that AP-1 factors are targets of the JAK/STAT pathway,35, 36, 37 we established a JAK/STAT-dependent expression of BATF3 in four of five lines analyzed. HRS cells express several cytokine receptors that support JAK/STAT signaling in an autocrine and/or paracrine fashion.38, 45, 46 Moreover, most if not all cases of cHL and probably PMBL feature intrinsically activated JAK/STAT signaling as a consequence of highly recurrent lesions of various genes, including JAK2 and the inhibitory molecules SOCS1 and PTPN1.47, 48, 49 HRS and PMBL cells express several phosphorylated STATs, and ALCL cells predominantly express STAT3(ref. 38) that is activated by NPM-ALK in ALK+ cases.50, 51 In ALK-negative ALCL, genes coding for JAK1 and STAT3 are frequently mutated, yet phosphorylated STAT3 is also expressed in the absence of mutations.35 By ChIP-qPCR, we demonstrated that phosphorylated STAT3 and STAT6 in cHL and STAT3 in ALCL lines directly bind to BATF3, supporting the concept that STAT activity is a major driver of BATF3 expression in cHL and presumably also in PMBL and ALCL. BATF3 is not only a JAK kinase but also a NF-κB signature gene,52 and Vrzalikova and colleagues report that BATF3 expression is also regulated by PI3K/Akt signaling (P. Murray, personal communication). Constitutive activity of these pathways in HRS cells therefore likely contributes to BATF3 expression, and may also account for unchanged expression or incomplete/moderate reduction of BATF3 in HDLM-2 cells upon JAK2 inhibition. Notably, BATF3 protein was also frequently expressed in CD30+ DLBCL. Mutations in the JAK/STAT pathway are uncommon in DLBCL but expression of phosphorylated STAT3 has been reported in a substantial fraction of cases often as a consequence of autocrine cytokine secretion.36, 37, 53 Therefore, an association of STAT(3) activity and BATF3 expression in a subset of DLBCL seems plausible but requires further investigation.
BATF factors lack a transcriptional activation domain and thus require cofactors in order to positively regulate target gene expression.19 We established dimerization of BATF3 with JUN and JUNB in HRS and ALCL cells by MS and co-IP and found indication of an interaction with IRF transcription factors by MS. Although this could not be validated by co-IP, AP-1 and IRF factors reportedly co-localized to composite DNA motifs following ChIP-seq.28 They interact, however, at few critical amino acids only,20 and the complex seemed unstable when detached from DNA. IRF4 and IRF5 are highly expressed by HRS cells,54, 55 but unlike in ALCL and myeloma cells, where IRF4 binds to the MYC promoter and regulates MYC expression,30, 56 IRF4 seems unessential for MYC expression in cHL and ABC DLBCL.57
We show that BATF3 instructs an oncogenic program that is required for proliferation and cell survival of HRS and ALCL cells, in keeping with a major role of AP-1 activity in cHL and ALCL pathogenesis. A pathogenetic role of BATF3 in lymphomagenesis is supported by findings from Weiser and colleagues, demonstrating that ectopic BATF3 expression in B lymphocytes leads to the development of B-cell lymphomas in a mouse model (personal communication). Critically, BATF3 knockdown led to the downregulation of MYC signatures and consistently MYC protein expression in cHL as well as ALCL lines. Silencing of MYC is toxic for cHL and ALCL lines,30, 34 and although its downregulation in shBATF3-transduced cells points to MYC as a critical proliferation and/or survival factor in these cells, the relevance of other downstream targets cannot be excluded. For example, in PMBL cells ectopic MYC expression did not rescue JAK2 silencing-mediated toxicity.34 Histone modifications by JAK2 in PMBL and HRS cells and JAK1 in DLBCL cells keep the MYC locus in an open configuration.34, 53 MYC is expressed in many lymphomas, including cHL, PMBL, DLBCL, ALCL and Burkitt lymphoma, but with exception of the latter and some DLBCL this is infrequently the consequence of chromosomal rearrangements or increased gene copy numbers.33, 58 Hence, these lymphomas achieve MYC overexpression by other mechanisms. We discovered that BATF3 binds to a known AP-1 site in the MYC promoter,32 strengthening the idea that MYC is a primary target of BATF3-containing AP-1 dimers in cHL and ALCL.
Taken together, we established that constitutively high expression of BATF3 due to oncogenic JAK/STAT activity promotes survival and proliferation in cHL and ALCL. This oncogenic function of BATF3 is at least partly mediated through activation of MYC expression. Hence, BATF3 critically links JAK/STAT signaling and MYC activity in cHL and ALCL, defining a novel oncogenic axis in these lymphomas.
References
Schmitz R, Stanelle J, Hansmann M-L, Küppers R . Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annu Rev Pathol 2009; 4: 151–174.
Kanzler H, Küppers R, Hansmann ML, Rajewsky K . Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996; 184: 1495–1505.
Küppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci USA 1994; 91: 10962–10966.
Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J 2002; 21: 4104–4113.
Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia 2009; 23: 2129–2138.
Hapgood G, Savage KJ . The biology and management of systemic anaplastic large cell lymphoma. Blood 2015; 126: 17–25.
Stein H, Mason DY, Gerdes J, O'Connor N, Wainscoat J, Pallesen G et al. The expression of the Hodgkin's disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood 1985; 66: 848–858.
Eferl R, Wagner EF . AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868.
Drakos E, Leventaki V, Schlette EJ, Jones D, Lin P, Medeiros LJ et al. c-Jun expression and activation are restricted to CD30+ lymphoproliferative disorders. Am J Surg Pathol 2007; 31: 447–453.
Rassidakis GZ, Thomaides A, Atwell C, Ford R, Jones D, Claret FX et al. JunB expression is a common feature of CD30+ lymphomas and lymphomatoid papulosis. Mod Pathol 2005; 18: 1365–1370.
Laimer D, Dolznig H, Kollmann K, Vesely PW, Schlederer M, Merkel O et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med 2012; 18: 1699–1704.
Rodig SJ, Ouyang J, Juszczynski P, Currie T, Law K, Neuberg DS et al. AP1-dependent galectin-1 expression delineates classical hodgkin and anaplastic large cell lymphomas from other lymphoid malignancies with shared molecular features. Clin Cancer Res 2008; 14: 3338–3344.
Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O'Donnell E et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res 2012; 18: 1611–1618.
Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, van Noesel CJM et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 2008; 205: 2251–2268.
Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003; 198: 851–862.
Schwering I, Bräuninger A, Distler V, Jesdinsky J, Diehl V, Hansmann ML et al. Profiling of Hodgkin's lymphoma cell line L1236 and germinal center B cells: identification of Hodgkin's lymphoma-specific genes. Mol Med 2003; 9: 85–95.
Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol 2010; 28: 1583–1590.
Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008; 322: 1097–1100.
Murphy TL, Tussiwand R, Murphy KM . Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol 2013; 13: 499–509.
Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012; 490: 502–507.
Logan MR, Jordan-Williams KL, Poston S, Liao J, Taparowsky EJ . Overexpression of Batf induces an apoptotic defect and an associated lymphoproliferative disorder in mice. Cell Death Dis 2012; 3: e310.
Weber K, Bartsch U, Stocking C, Fehse B . A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther 2008; 16: 698–706.
Tiacci E, Döring C, Brune V, van Noesel CJ, Klapper W, Mechtersheimer G et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012; 120: 4609–4620.
Steidl C, Diepstra A, Lee T, Chan FC, Farinha P, Tan K et al. Gene expression profiling of microdissected Hodgkin Reed-Sternberg cells correlates with treatment outcome in classical Hodgkin lymphoma. Blood 2012; 120: 3530–3540.
Underhill GH, George D, Bremer EG, Kansas GS . Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood 2003; 101: 4013–4021.
Watanabe M, Ogawa Y, Ito K, Higashihara M, Kadin ME, Abraham LJ et al. AP-1 mediated relief of repressive activity of the CD30 promoter microsatellite in Hodgkin and Reed-Sternberg cells. Am J Pathol 2003; 163: 633–641.
Watanabe M, Sasaki M, Itoh K, Higashihara M, Umezawa K, Kadin ME et al. JunB induced by constitutive CD30-extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase signaling activates the CD30 promoter in anaplastic large cell lymphoma and reed-sternberg cells of Hodgkin lymphoma. Cancer Res 2005; 65: 7628–7634.
Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 2012; 490: 543–546.
Seitz V, Butzhammer P, Hirsch B, Hecht J, Gutgemann I, Ehlers A et al. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS ONE 2011; 6: e26837.
Weilemann A, Grau M, Erdmann T, Merkel O, Sobhiafshar U, Anagnostopoulos I et al. Essential role of IRF4 and MYC signaling for survival of anaplastic large cell lymphoma. Blood 2015; 125: 124–132.
Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 2012; 13: 1083–1091.
Iavarone C, Catania A, Marinissen MJ, Visconti R, Acunzo M, Tarantino C et al. The platelet-derived growth factor controls c-myc expression through a JNK- and AP-1-dependent signaling pathway. J Biol Chem 2003; 278: 50024–50030.
Chisholm KM, Bangs CD, Bacchi CE, Molina-Kirsch H, Cherry A, Natkunam Y . Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am J Surg Pathol 2015; 39: 294–303.
Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 2010; 18: 590–605.
Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015; 27: 516–532.
Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 2008; 111: 1515–1523.
Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008; 111: 3701–3713.
Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trümper L, Kapp U et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002; 99: 618–626.
Atsaves V, Lekakis L, Drakos E, Leventaki V, Ghaderi M, Baltatzis GE et al. The oncogenic JUNB/CD30 axis contributes to cell cycle deregulation in ALK+ anaplastic large cell lymphoma. Br J Haematol 2014; 167: 514–523.
Hartmann S, Martin-Subero JI, Gesk S, Husken J, Giefing M, Nagel I et al. Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma by array-based comparative genomic hybridization. Haematologica 2008; 93: 1318–1326.
Salipante SJ, Adey A, Thomas A, Lee C, Liu YJ, Kumar A et al. Recurrent somatic loss of TNFRSF14 in classical Hodgkin lymphoma. Genes Chromosomes Cancer 2016; 55: 278–287.
Feys T, Poppe B, De Preter K, Van Roy N, Verhasselt B, De Paepe P et al. A detailed inventory of DNA copy number alterations in four commonly used Hodgkin's lymphoma cell lines. Haematologica 2007; 92: 913–920.
Salaverria I, Bea S, Lopez-Guillermo A, Lespinet V, Pinyol M, Burkhardt B et al. Genomic profiling reveals different genetic aberrations in systemic ALK-positive and ALK-negative anaplastic large cell lymphomas. Br J Haematol 2008; 140: 516–526.
Liu C, Iqbal J, Teruya-Feldstein J, Shen Y, Dabrowska MJ, Dybkaer K et al. MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 2013; 122: 2083–2092.
Lamprecht B, Kreher S, Anagnostopoulos I, Johrens K, Monteleone G, Jundt F et al. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood 2008; 112: 3339–3347.
Scheeren FA, Diehl SA, Smit LA, Beaumont T, Naspetti M, Bende RJ et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5; evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood 2008; 111: 4706–4715.
Gunawardana J, Chan FC, Telenius A, Woolcock B, Kridel R, Tan KL et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet 2014; 46: 329–335.
Joos S, Küpper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000; 60: 549–552.
Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006; 25: 2679–2684.
Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 2005; 11: 623–629.
Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 2002; 21: 1038–1047.
Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011; 470: 115–119.
Rui L, Drennan AC, Ceribelli M, Zhu F, Wright GW, Huang DW et al. Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 2016; 113: E7260–E7267.
Kreher S, Bouhlel MA, Cauchy P, Lamprecht B, Li S, Grau M et al. Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc Natl Acad Sci USA 2014; 111: E4513–E4522.
Carbone A, Gloghini A, Aldinucci D, Gattei V, Dalla-Favera R, Gaidano G . Expression pattern of MUM1/IRF4 in the spectrum of pathology of Hodgkin's disease. Br J Haematol 2002; 117: 366–372.
Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W et al. IRF4 addiction in multiple myeloma. Nature 2008; 454: 226–231.
Yang Y, Shaffer AL 3rd, Emre NC, Ceribelli M, Zhang M, Wright G et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012; 21: 723–737.
Shaffer AL, Staudt LM . The case of the missing c-Myc. Nat Immunol 2012; 13: 1029–1031.
Acknowledgements
We thank Kerstin Heise, Kristin Rosowski, Philip Abstoß and Klaus Lennartz for excellent technical assistance. Cells were sorted using a FACS ARIA III at the imaging facility Essen IMCES. This work was supported by grants from the Deutsche Forschungsgemeinschaft (KU1315/7-1, KU1315/10-1, GRK1431/2) and the BMBF through the International Cancer Genome Consortium for malignant lymphomas (01KU1002F). The MS experiments were supported by PURE (Protein Research Unit Ruhr within Europe, Ministry of Science, North Rhine-Westphalia, Germany).
Author contributions
AL, RK and MAW designed research; AL, SH, MS, TB, ALW and MAW performed research; TB, LK-H and BS contributed to analytic methods and tools; SH, JA and M-LH contributed to clinical samples; AL, SH, TB, BS, LK-H, M-LH, RK and MAW analyzed data; AL, RK and MAW wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Lollies, A., Hartmann, S., Schneider, M. et al. An oncogenic axis of STAT-mediated BATF3 upregulation causing MYC activity in classical Hodgkin lymphoma and anaplastic large cell lymphoma. Leukemia 32, 92–101 (2018). https://doi.org/10.1038/leu.2017.203
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.203
- Springer Nature Limited
This article is cited by
-
siRNA-mediated downregulation of BATF3 diminished proliferation and induced apoptosis through downregulating c-Myc expression in chronic myelogenous leukemia cells
Molecular Biology Reports (2024)
-
Concise review: The heterogenous roles of BATF3 in cancer oncogenesis and dendritic cells and T cells differentiation and function considering the importance of BATF3-dependent dendritic cells
Immunogenetics (2024)
-
Challenges and new technologies in adoptive cell therapy
Journal of Hematology & Oncology (2023)
-
Transcriptional and epigenetic regulators of human CD8+ T cell function identified through orthogonal CRISPR screens
Nature Genetics (2023)
-
TET2 guards against unchecked BATF3-induced CAR T cell expansion
Nature (2023)