Abstract
A number of patient-specific and leukemia-associated factors are related to the poor outcome in older patients with acute myeloid leukemia (AML). However, comprehensive studies regarding the impact of genetic alterations in this group of patients are limited. In this study, we compared relevant mutations in 21 genes between AML patients aged 60 years or older and those younger and exposed their prognostic implications. Compared with the younger patients, the elderly had significantly higher incidences of PTPN11, NPM1, RUNX1, ASXL1, TET2, DNMT3A and TP53 mutations but a lower frequency of WT1 mutations. The older patients more frequently harbored one or more adverse genetic alterations. Multivariate analysis showed that DNMT3A and TP53 mutations were independent poor prognostic factors among the elderly, while NPM1 mutation in the absence of FLT3/ITD was an independent favorable prognostic factor. Furthermore, the status of mutations could well stratify older patients with intermediate-risk cytogenetics into three risk groups. In conclusion, older AML patients showed distinct genetic alterations from the younger group. Integration of cytogenetics and molecular mutations can better risk-stratify older AML patients. Development of novel therapies is needed to improve the outcome of older patients with poor prognosis under current treatment modalities.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is a clinically and biologically heterogeneous hematologic malignancy characterized by uncontrolled proliferation of hematopoietic precursors and loss of the ability to differentiate. Although the clinical outcome improves steadily in younger patients in the past 40 years, the survival for older patients remains very poor.1, 2
In addition to patient-specific factors, such as concomitant comorbidity, poor performance status and intolerance to intensive chemotherapy,3, 4 a number of leukemia-associated factors are related to the poor outcome in older AML patients.5, 6 Traditionally, cytogenetic findings establish the backbone for prognostic and therapeutical strategies in AML and are long used to risk-stratify AML patients and guide the treatment plan.7, 8 Appelbaum et al.5 demonstrated that older patients had less frequently favorable-risk but more commonly unfavorable-risk cytogenetics, particularly abnormalities in chromosomes 5, 7 and 17.
Many acquired gene mutations have been detected in AML patients, especially those with intermediate-risk cytogenetics, and some of them, such as mutations in NPM1, CEBPA, RUNX1, WT1, DNMT3A, ASXL1, IDH2 and FLT3 genes, have been shown to have prognostic significance.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 However, less is known about the clinical implications of gene mutations in older patients with AML. In this study, we aimed to comprehensively investigate the clinico-biological features and molecular genetic alterations and their clinical relevance in older AML patients. The findings from this study may pave ways for future targeted therapies in this group of patients with poor clinical outcome under the current treatment modality.
Materials and methods
Subjects
Totally, 462 adult patients who were newly diagnosed as having de novo non-M3 AML according to the FAB Cooperative Group Criteria23 at the National Taiwan University Hospital (NTUH) had cryopreserved cells for mutational analyses, and had complete clinical, cytogenetic and laboratory data were recruited for this study. Among them, 177 patients were 60 years or older. Patients with antecedent hematological diseases, history of cytopenia, family history of myeloid neoplasms or therapy-related AML were all excluded. Diagnosis and classification of AML were made according to the FAB Cooperative Group Criteria.23 This study was approved by the Research Ethics Committee of the NTUH and written informed consents were obtained from all participants in accordance with the Declaration of Helsinki. Among these patients, 329 (71.2%) received standard induction chemotherapy (Idarubicin 12 mg/m2 per day on days 1–3 and Cytarabine 100 mg/m2 per day on days 1–7) and then consolidation chemotherapy with two to four courses of high-dose Cytarabine (2000 mg/m2 q12h, total eight doses), with or without an anthracycline (Idarubicin or Mitoxantrone), after achieving complete remission (CR).20, 22 Because hypomethylating agents have not been reimbursed for the treatment of AML by the Taiwan government, only few patients received hypomethylating agents in this cohort; analysis of prognostic impact of hypomethylating agents was not carried out. The remaining patients received palliative therapy with supportive care and/or low-dose chemotherapy owing to underlying comorbidities or based on the decision of the physicians and patients.
Cytogenetics
Bone marrow cells were harvested directly or after 1–3 days of unstimulated culture as described previously.24 Metaphase chromosomes were banded by trypsin-Giemsa technique and karyotyped according to the International System for Human Cytogenetic Nomenclature.
Mutation analysis
Analyses of relevant mutations in 21 genes, including Class I mutations, such as FLT3/ITD,25 FLT3/TKD,26 NRAS,27 KRAS,27 JAK2,27 KIT28 and PTPN11(ref. 28) mutations, and Class II mutations, such as CEBPA29 and RUNX1(ref. 17) mutations, as well as mutations in NPM1,12 WT1,30 TP53,31 Cohesin complex genes (including STAG1/2, SMC1A, SMC3, and RAD21),32 and those genes related to epigenetic modification, such as MLL/PTD,33 ASXL1,34 IDH1,35 IDH2,36 TET2(ref. 37) and DNMT3A20 were performed as previously described. Abnormal sequencing results were confirmed by at least two repeated analyses.
Statistical analysis
The discrete variables of patients with and without specific molecular alteration were compared using the Fisher exact test. If the continuous data were not normally distributed, Mann–Whitney U tests were used to compare continuous variables and medians of distributions. To evaluate the impact of age, molecular alterations and other variables on clinical outcome, only the patients who received conventional standard chemotherapy were included in analyses.20, 22 Overall survival (OS) was measured from the date of first diagnosis to the date of last follow-up or death from any cause, whereas relapse was defined as a reappearance of at least 5% leukemic blasts in bone marrow aspiration smears or new extramedullary leukemia in patients with a previously documented CR.38 Disease-free survival (DFS) was applied to patients receiving standard intensive chemotherapy and was measured from the date of CR until relapse from CR or death from any cause, whichever occurred first. Multivariate Cox proportional hazards regression analysis was used to investigate independent prognostic factors for OS and DFS. The proportional hazards assumption (constant hazards assumption) was examined by using time-dependent covariate Cox regression before conducting multivariate Cox proportional hazards regression. The variables including age, white blood cell counts at diagnosis, karyotype, NPM1/FLT3-ITD, WT1, CEBPA, RUNX1, MLL/PTD, ASXL1, TET2, IDH2, DNMT3A and TP53 mutations that showed prognostic implication with P value less than 0.1 in univariate analysis were used as covariates. Those patients who received hematopoietic stem cell transplantation (HSCT) were censored at the time of HSCT in survival analysis to ameliorate the influence of the treatment. A P value <0.05 was considered statistically significant. All statistical analyses were performed with the SPSS 20 (SPSS Inc., Chicago, IL, USA) and Statsdirect (Cheshire, England, UK).
Results
Comparison of clinical and laboratory features between older and younger patients
Among the 462 AML patients recruited, 261 were males and 201 were females (Table 1). One hundred and seventy-seven patients were 60 years or older with a median age of 71 years (range 60–90 years). There was no difference in gender, hemogram and lactate dehydrogenase level between younger patients and older patients. Older age was negatively associated with the expression of CD19 (P=0.022), CD15 (P=0.007) and CD34 (P=0.002) on the leukemic cells (Supplementary Table 1). There was no difference in the expression of other antigens.
Comparison of cytogenetic abnormalities and molecular gene mutations between older and younger patients
Chromosome data were available in 444 patients at diagnosis, including 166 older and 278 younger patients (Table 2). Compared with younger patients, the elderly had more frequently unfavorable-risk cytogenetic changes (21.1 vs 10.8%, P=0.004), but less commonly favorable-risk cytogenetics (4.2 vs 19.4%, P<0.001), such as t(8;21) (2.4 vs 13.7%, P<0.001) based on the Medical Research Council (MRC) classification.39 Specifically, the older patients had higher frequencies of complex chromosomal abnormalities (16.3 vs 8.6%, P=0.032), monosomy 5/5q deletion (8.4 vs 1.8%, P=0.001) and monosomy 7/7q deletion (8.4 vs 2.9%, P=0.012) but a lower incidence of t(7;11) (0 vs 3.6%, P=0.016). The distribution of simple chromosomal abnormalities with two or less changes involving chromosomes 8, 11, 13 and 21 was not different between the two groups.
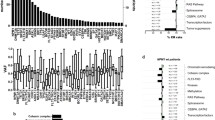
To investigate the difference of gene mutations in the pathogenesis of leukemia between older and younger AML patients, a complete mutational screening of 21 genes was performed. The most common molecular event in total cohort was FLT3/ITD (22.5%), followed by NPM1 (22.3%), DNMT3A (15.2%), TET2 (14.3%) and CEBPA mutations (14.3%). Among the elderly, the most prevalent molecular event was NPM1 (28.2%), followed by TET2 (24.3%), FLT3/ITD (22.6%), DNMT3A (20.9%) and RUNX1 mutations (19.8%) (Table 3). The median number of molecular gene mutations at diagnosis was higher in the older patients than the younger ones (2.0, range 0–5 vs 1.0, range 0–5, P<0.001). Older patients had significantly higher incidences of PTPN11, NPM1, RUNX1, ASXL1, TET2, DNMT3A and TP53 mutations than younger patients (6.2% vs 2.5%, P=0.050; 28.2% vs 18.6%, P=0.021; 19.8% vs 9.5%, P=0.002; 17.6% vs 6.7%, P<0.001; 24.3% vs 8.1%, P<0.001; 20.9% vs 11.6%; P=0.008; and 13.0% vs 4.2%, P=0.001, respectively). On the contrary, WT1 mutations were rarely seen in patients aged 60 years or older (3.4 vs 9.1%, P=0.023). Other genetic alterations were not significantly different between these two age groups. The distributions of molecular gene mutations in these two groups are distinct (Figure 1 and Supplementary Figure 1). Older patients had a higher frequency to harbor one or more adverse genetic alterations (including FLT3/ITD, WT1, RUNX1, ASXL1, DNMT3A and TP53 mutations)22, 31 than younger ones (69.5 vs 49.5%, P<0.001), and the difference remained similar between the two groups when two or more such gene mutations were counted (26.6 vs 13.3%, P=0.001). We further showed that 85 pairwise associations were significant with P<0.1 in the elderly cohort (Supplementary Figure 2).
The Circos plots depicted the relative frequency and pairwise co-occurrence of genetic alterations in the older (a) and younger AML patients (b). The length of the arc corresponds to the frequency of the first gene mutation, and the width of the ribbon corresponds to the proportion of the second gene mutation.
Prognostic impact of gene mutations in older patients
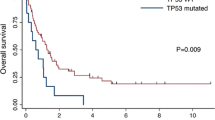
Fewer older patients received standard chemotherapy than younger patients (69/177, 40.0% vs 260/285, 91.2%, P<0.001); however, standard chemotherapy lead to a longer OS than palliative care only in this group of patients (median, 10.0 vs 3.0 months, P<0.001). Among the total cohort of 329 AML patients undergoing conventional intensive induction chemotherapy, 252 (76.6%) patients achieved CR. Older patients had a lower CR rate than younger population (56.5 vs 81.9%, P<0.001, Table 1). With a median follow-up of 69 months (ranges, 0.1–160), the elderly had significantly poorer OS and DFS than those aged below 60 years (median, 10.0 vs 61.0 months, P<0.001, Figure 2a, and median, 3.0 vs 9.0 months, P=0.001, Figure 2b, respectively). In multivariate Cox proportional hazards regression analysis for total cohort (Table 4), the independent poor risk factors for OS and DFS were older age, high white blood cell count >50 000/μl, and WT1, DNMT3A and TP53 mutations. On the other hand, NPM1+/FLT3-ITD− and CEBPAdouble-mutation were independent favorable prognostic factors. We also found that unfavorable-risk cytogenetics and RUNX1 mutations independently conferred poorer DFS and IDH2 predicted better OS.
The Kaplan–Meier survival curves for OS (a) and DFS (b) in 329 AML patients who received standard intensive chemotherapy. The older patients have significantly poorer OS and DFS than those aged below 60 years (median, 10.0 vs 61.0 months, P<0.001, and median, 3.0 vs 9.0 months, P=0.001, respectively).
In the multivariate Cox proportional hazards regression analysis for OS in the elderly, DNMT3A and TP53 mutations were independent poor prognostic factors, while NPM1+/FLT3-ITD− remained to have good prognostic impact. Intriguingly, the older patients harboring any unfavorable genetic alteration, including FLT3/ITD, DNMT3A or TP53 mutation, had more dismal survival compared with those not (median OS, 7.0 vs 14.0 months, P=0.042, Figure 3).
The poor prognostic impacts of some mutations, such as FLT3/ITD, RUNX1 and DNMT3A mutations were lost when the patients receiving HSCT were not censored on the date of transplantation, implying allogeneic HSCT might overcome the poor risk of the patients with these mutations. Unfortunately, none of the 69 elder patients underwent allogeneic HSCT.
Further, the older patients with intermediate-risk cytogenetics could be further separated into three risk groups according to the molecular genotype:22 mutations of NPM1 or IDH2 or CEBPAdouble-mutation in the absence of FLT3/ITD as a favorable genotype,9 mutations of RUNX1, WT1, ASXL1, DNMT3A or TP53 as an unfavorable genotype,9, 27 and the remaining mutation patterns as an intermediate-risk genotype. Among the older patients with intermediate-risk cytogenetics, those with a favorable genotype had a higher CR rate and a trend of lower relapse rate than those with intermediate-risk and unfavorable genotypes (CR, 81.8 vs 64.7 vs 30.3%, P=0.002 and relapse, 44.4 vs 72.7 vs 87.5%, P=0.150). These three groups also had distinct OS (median, 26.0 vs 15.0 vs 8.0 months, P<0.001, Figure 4a) and DFS (median, 12.0 vs 7.0 vs 0 months, P=0.002, Figure 4b). Patients with intermediate-risk cytogenetics but favorable genotype had OS and DFS similar to those with favorable-risk cytogenetics (P=0.349), while patients with intermediate-risk cytogenetics but unfavorable genotype had OS and DFS similar to those with unfavorable-risk cytogenetics (P=0.420).
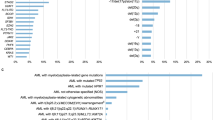
Genetic ontogeny was first proposed by Lindsley et al.40 Three types of mutations were defined: secondary-type mutations, TP53 mutations and de novo/pan AML mutations. Presence of secondary-type mutations predicted characteristic phenotype and poor outcome. We validated the impact of genetic ontogeny in our cohort. Five secondary-type genes, including three splicing factor genes (SRSF2, SF3B1 and U2AF1),41 ASXL1 and STAG2, were analyzed. In the 177 de novo AML patients aged 60 years or older, 34.5% had secondary-type mutations, 13.0% had TP53 mutation, 49.2% had de novo/pan AML mutations and 3.3% were undetermined. Among the 69 patients receiving standard chemotherapy, patients with secondary-type or TP53 mutations had a lower CR rate and a poor OS than those with de novo/pan AML mutations (43.5% vs 25.0% vs 66.7%, P=0.006, and median, 10.0 vs 3.0 vs 14.0 months, P=0.004, respectively).
Discussion
Most studies concerning prognostic factors in AML patients were focused on younger patients with less comorbidities and better performance status, and thus might not be representative of the general AML population in the real world.42 In this study, we recruited consecutively all de novo AML patients who had adequate samples for mutation analyses without restriction of age, so that we could compare the genetic alterations between older and younger patients and explored their clinical implications. We found that older AML patients had distinct clinico-biological features and genetic alterations from younger patients, and the status of mutations could predict the prognosis in this group of patients.
Traditionally, karyotype is one of the strongest prognostic factors in AML patients.43, 44 We showed that older patients had more frequently poor risk cytogenetics, such as complex chromosomal abnormalities, or aberrations involving chromosomes 5 and 7, but less frequently the core binding factor abnormalities. To better stratify AML patients into different risk groups, the European LeukemiaNet (ELN) panel first proposed a standardized classification according to both cytogenetics and molecular mutations in three genes, including FLT3/ITD, NPM1 and CEBPA mutations.7 Recently, several other genetic alterations were also found to have prognostic significance and were incorporated into risk stratification of AML patients.22, 31, 45, 46 However, there were only few reports in literature regarding the clinical impact of molecular alterations on older AML patients. In the studies from Cancer and Leukemia Group B (CALGB), RUNX1 and ASXL1 mutations were found to be more prevalent in the elder population with cytogenetically normal AML and were poor prognostic factors.14, 15 Older patients with WT1(ref. 47) or TP53(ref. 48) mutations had a shorter OS, while those with NPM149 mutations had a better CR rate and OS. Ostronoff et al.50 further depicted that NPM1 mutations in the absence of FLT3/ITD had a survival benefit among patients aged 55–65 years, but not in those older than 65 years. To our knowledge, this study is the first to comprehensively investigate the molecular genetic alterations of 21 genes among older patients with non-M3 AML. First, we showed that the distribution of genetic alterations and the burdens of gene mutations differed across the age groups. Second, older patients had higher incidences of PTPN11, NPM1, RUNX1, ASXL1, TET2, DNMT3A and TP53 mutations but less WT1 mutations. With the exception of NPM1 mutation, most of the other mutations that were more prevalent in older patients had unfavorable prognostic impact. Furthermore, older patients had a higher frequency to harbor one or more adverse genetic alterations (including FLT3/ITD, WT1, RUNX1, ASXL1, DNMT3A and TP53) than younger ones. Taken together, in addition to a higher incidence of adverse cytogenetics, the higher frequency and burdens of molecular mutations that are associated with poor prognosis in the elderly might explain the dismal outcome in this group of patients. Another possible cause to explain the dismal clinical outcome in older patients is that they are more vulnerable to the toxicity of chemotherapy agents, so may have higher treatment-related mortality.5, 51 However, our study demonstrated that the early mortality rate were comparable between the two age groups, similar to the national registration data of the United States52 and Swedish Acute Leukemia Registry.53, 54
Cytogenetic changes could well separate older AML patients into three risk groups in this study, similar to previous reports.55, 56, 57, 58 However, about 60–70% of the patients were in the intermediate-risk cytogenetic group which would hinder risk stratification of these patients for proper treatment. With the incorporation of nine gene mutations, including FLT3/ITD and mutations of CEBPA, NPM1, RUNX1, WT1, IDH2, ASXL1, DNMT3A and TP53, that are associated with prognosis,9, 27 we showed that older AML patients with intermediate-risk cytogenetics could be further stratified into three groups with different outcomes. The patients with favorable genotype (NPM1, IDH2 and CEBPAdouble-mutation in the absence of FLT3/ITD) had the longest survival, whereas those with unfavorable genotype (DNMT3A, ASXL1, WT1, RUNX1 or TP53) had the poorest outcome. The incorporation of the mutation status of these genes is helpful to stratify this highly heterogeneous population with intermediate-risk cytogenetics into distinct risk groups.
Genetic ontogeny, first proposed by Lindsley et al.,40 can help risk-stratify AML patients irrespective of their clinical assignment. In the 42 de novo AML patients aged 60 years or older in their study, 33.3% had secondary-type mutations, 21.4% had TP53 mutations and 45.2% had de novo/pan AML mutations. The frequencies of these three types of mutations in our cohort were not much different from those reported by Lindsley et al. The poor prognostic implication of secondary-type mutations in elderly patients with de novo AML was shown in this study as that of Lindsley et al.40 Similar to the high incidence of secondary-type mutations, elderly AML patients aged 60 years or older had higher incidence of dysplastic morphological features than those younger than 60 years (23.7 vs 14.4%; P=0.013); even none of them had a history of myelodysplastic syndrome, myeloproliferative neoplasm or other hematologic diseases.
In summary, we showed that older AML patients had distinct clinico-biological features, more frequently high-risk cytogenetics and gene mutations, and poorer prognosis. Integration of both cytogenetics and molecular alterations can better stratify older patients into different risk groups with distinct outcomes. It is warranted to develop novel therapies to improve the outcome of older patients with poor prognosis under current treatment modalities.
References
Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF et al(eds). SEER Cancer Statistics Review, 1975-2010 2013 National Cancer Institute: Bethesda, MD, USA, Available at: http://seer.cancer.gov/csr/1975_2010/.
Burnett A, Wetzler M, Lowenberg B . Therapeutic advances in acute myeloid leukemia. J Clin Oncol 2011; 29: 487–494.
Lowenberg B, Downing JR, Burnett A . Acute myeloid leukemia. N Engl J Med 1999; 341: 1051–1062.
Hiddemann W, Kern W, Schoch C, Fonatsch C, Heinecke A, Wormann B et al. Management of acute myeloid leukemia in elderly patients. J Clin Oncol 1999; 17: 3569–3576.
Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE et al. Age and acute myeloid leukemia. Blood 2006; 107: 3481–3485.
Erba HP . Prognostic factors in elderly patients with AML and the implications for treatment. Hematology Am Soc Hematol Educ Program 2007, 420–428.
Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010; 115: 453–474.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010; 116: 354–365.
Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia 2014; 28: 50–58.
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358: 1909–1918.
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363: 2424–2433.
Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005; 352: 254–266.
Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002; 100: 59–66.
Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood 2011; 118: 6920–6929.
Mendler JH, Maharry K, Radmacher MD, Mrozek K, Becker H, Metzeler KH et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol 2012; 30: 3109–3118.
Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrozek K, Maharry K et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 2008; 26: 4595–4602.
Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood 2009; 114: 5352–5361.
Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011; 118: 409–412.
Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2010; 28: 2348–2355.
Hou HA, Kuo YY, Liu CY, Chou WC, Lee MC, Chen CY et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood 2012; 119: 559–568.
Hou HA, Tien HF . Mutations in epigenetic modifiers in acute myeloid leukemia and their clinical utility. Expert Rev Hematol e-pub ahead of print 9 February 2016, 1–23.
Chou SC, Tang JL, Hou HA, Chou WC, Hu FC, Chen CY et al. Prognostic implication of gene mutations on overall survival in the adult acute myeloid leukemia patients receiving or not receiving allogeneic hematopoietic stem cell transplantations. Leuk Res 2014; 38: 1278–1284.
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 1985; 103: 620–625.
Tien HF, Wang CH, Lin MT, Lee FY, Liu MC, Chuang SM et al. Correlation of cytogenetic results with immunophenotype, genotype, clinical features, and ras mutation in acute myeloid leukemia. A study of 235 Chinese patients in Taiwan. Cancer Genet Cytogenet 1995; 84: 60–68.
Chou WC, Hou HA, Liu CY, Chen CY, Lin LI, Huang YN et al. Sensitive measurement of quantity dynamics of FLT3 internal tandem duplication at early time points provides prognostic information. Ann Oncol 2011; 22: 696–704.
Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S . Prognostic relevance of FLT3-TKD mutations in AML: the combination matters—an analysis of 3082 patients. Blood 2008; 111: 2527–2537.
Hou HA, Kuo YY, Tang JL, Chou WC, Yao M, Lai YJ et al. Clinical implications of the SETBP1 mutation in patients with primary myelodysplastic syndrome and its stability during disease progression. Am J Hematol 2014; 89: 181–186.
Hou HA, Chou WC, Lin LI, Chen CY, Tang JL, Tseng MH et al. Characterization of acute myeloid leukemia with PTPN11 mutation: the mutation is closely associated with NPM1 mutation but inversely related to FLT3/ITD. Leukemia 2008; 22: 1075–1078.
Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res 2005; 11: 1372–1379.
Hou HA, Huang TC, Lin LI, Liu CY, Chen CY, Chou WC et al. WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood 2010; 115: 5222–5231.
Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, Tseng MH et al. TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J 2015; 5: e331.
Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood 2014; 123: 914–920.
Shiah HS, Kuo YY, Tang JL, Huang SY, Yao M, Tsay W et al. Clinical and biological implications of partial tandem duplication of the MLL gene in acute myeloid leukemia without chromosomal abnormalities at 11q23. Leukemia 2002; 16: 196–202.
Chen TC, Hou HA, Chou WC, Tang JL, Kuo YY, Chen CY et al. Dynamics of ASXL1 mutation and other associated genetic alterations during disease progression in patients with primary myelodysplastic syndrome. Blood Cancer J 2014; 4: e177.
Lin CC, Hou HA, Chou WC, Kuo YY, Liu CY, Chen CY et al. IDH mutations are closely associated with mutations of DNMT3A, ASXL1 and SRSF2 in patients with myelodysplastic syndromes and are stable during disease evolution. Am J Hematol 2014; 89: 137–144.
Chou WC, Lei WC, Ko BS, Hou HA, Chen CY, Tang JL et al. The prognostic impact and stability of Isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia 2011; 25: 246–253.
Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 2011; 118: 3803–3810.
Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 2003; 21: 4642–4649.
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood 1998; 92: 2322–2333.
Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015; 125: 1367–1376.
Hou HA, Liu CY, Kuo YY, Chou WC, Tsai CH, Lin CC et al. Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia. Oncotarget e-pub ahead of print 24 January 2016; doi:10.18632/oncotarget.7000.
Mengis C, Aebi S, Tobler A, Dahler W, Fey MF . Assessment of differences in patient populations selected for excluded from participation in clinical phase III acute myelogenous leukemia trials. J Clin Oncol 2003; 21: 3933–3939.
Keating MJ, Smith TL, Kantarjian H, Cork A, Walters R, Trujillo JM et al. Cytogenetic pattern in acute myelogenous leukemia: a major reproducible determinant of outcome. Leukemia 1988; 2: 403–412.
Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 2000; 96: 4075–4083.
Kihara R, Nagata Y, Kiyoi H, Kato T, Yamamoto E, Suzuki K et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia 2014; 28: 1586–1595.
Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366: 1079–1089.
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Margeson D et al. Mutations of the Wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood 2010; 116: 788–792.
Stirewalt DL, Kopecky KJ, Meshinchi S, Appelbaum FR, Slovak ML, Willman CL et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood 2001; 97: 3589–3595.
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Margeson D et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol 2010; 28: 596–604.
Ostronoff F, Othus M, Lazenby M, Estey E, Appelbaum FR, Evans A et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: a SWOG and UK National Cancer Research Institute/Medical Research Council report. J Clin Oncol 2015; 33: 1157–1164.
Kantarjian H, O'Brien S, Cortes J, Giles F, Faderl S, Jabbour E et al. Results of intensive chemotherapy in 998 patients age 65 years or older with acute myeloid leukemia or high-risk myelodysplastic syndrome: predictive prognostic models for outcome. Cancer 2006; 106: 1090–1098.
Oran B, Weisdorf DJ . Survival for older patients with acute myeloid leukemia: a population-based study. Haematologica 2012; 97: 1916–1924.
Juliusson G, Antunovic P, Derolf A, Lehmann S, Mollgard L, Stockelberg D et al. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood 2009; 113: 4179–4187.
Juliusson G . Older patients with acute myeloid leukemia benefit from intensive chemotherapy: an update from the Swedish Acute Leukemia Registry. Clin Lymphoma Myeloma Leuk 2011; 11 (Suppl 1): S54–S59.
Schoch C, Kern W, Krawitz P, Dugas M, Schnittger S, Haferlach T et al. Dependence of age-specific incidence of acute myeloid leukemia on karyotype. Blood 2001; 98: 3500.
Schoch C, Kern W, Schnittger S, Buchner T, Hiddemann W, Haferlach T . The influence of age on prognosis of de novo acute myeloid leukemia differs according to cytogenetic subgroups. Haematologica 2004; 89: 1082–1090.
Moorman AV, Roman E, Willett EV, Dovey GJ, Cartwright RA, Morgan GJ . Karyotype and age in acute myeloid leukemia. Are they linked? Cancer Genet Cytogenet 2001; 126: 155–161.
Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol 2006; 135: 165–173.
Acknowledgements
This work was partially sponsored by grants MOST 100-2628-B-002-003-MY3,103-2628-B-002-008-MY3, 103-2923-B-002 -001 and 104-2314-B-002-128-MY4 from the Ministry of Science and Technology (Taiwan), MOHW105-TDU-B-211-134005 from the Ministry of Health and Welfare (Taiwan), NTUH 102P06, from the Department of Medical Research, National Taiwan University Hospital, and Taiwan Health Foundation. We would like to acknowledge the service provided by the DNA Sequencing Core of the First Core Laboratory, National Taiwan University College of Medicine.
Author contributions
C-HT was responsible for data management and interpretation, statistical analysis and manuscript writing; H-AH was responsible for study design and plan, literature collection, data management and interpretation, statistical analysis and manuscript writing; C-YL was responsible for statistical analysis and interpretation of the statistical findings; Y-YK, L-IL was responsible for mutation analysis and interpretation; C-YC, W-CC, M-Y, S-YH, J-LT, B-SK, S-CH, C-TL, C-CL, S-JW, S-CC, WT and Y-CC contributed patient samples and clinical data; M-HT, C-FH, Y-CC, C-YL, F-YL and M-CL performed the gene mutation and chromosomal studies and H-FT designed and coordinated the study over the entire period and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Tsai, CH., Hou, HA., Tang, JL. et al. Genetic alterations and their clinical implications in older patients with acute myeloid leukemia. Leukemia 30, 1485–1492 (2016). https://doi.org/10.1038/leu.2016.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.65
- Springer Nature Limited
This article is cited by
-
Treatment of older adults with FLT3-mutated AML: Emerging paradigms and the role of frontline FLT3 inhibitors
Blood Cancer Journal (2023)
-
Poor prognostic implications of myelodysplasia-related mutations in both older and younger patients with de novo AML
Blood Cancer Journal (2023)
-
How Genetics Can Drive Initial Therapy Choices for Older Patients with Acute Myeloid Leukemia
Current Treatment Options in Oncology (2022)
-
Distinct clinico-biological features in AML patients with low allelic ratio FLT3-ITD: role of allogeneic stem cell transplantation in first remission
Bone Marrow Transplantation (2022)
-
Long-term survival after intensive chemotherapy or hypomethylating agents in AML patients aged 70 years and older: a large patient data set study from European registries
Leukemia (2022)