Abstract
Inhibition of the bromodomain and extra-terminal (BET) proteins is a promising therapeutic strategy for various hematologic cancers. Previous studies suggest that BET inhibitors constrain tumor cell proliferation and survival mainly through the suppression of MYC transcription and activity. However, suppression of the transcription of additional genes also contributes to the antitumor activity of BET inhibitors but is less well understood. Here we examined the therapeutic potential of CPI-0610, a potent BET inhibitor currently undergoing phase I clinical testing, in multiple myeloma (MM). CPI-0610 displays potent cytotoxicity against MM cell lines and patient-derived MM cells through G1 cell cycle arrest and caspase-dependent apoptosis. CPI-0610-mediated BET inhibition overcomes the protective effects conferred by cytokines and bone marrow stromal cells. We also confirmed the in vivo efficacy of CPI-0610 in a MM xenograft mouse model. Our study found IKZF1 and IRF4 to be among the primary targets of CPI-0610, along with MYC. Given that immunomodulatory drugs (IMiDs) stabilize cereblon and facilitate Ikaros degradation in MM cells, we combined it with CPI-0610. Combination studies of CPI-0610 with IMiDs show in vitro synergism, in part due to concomitant suppression of IKZF1, IRF4 and MYC, providing a rationale for clinical testing of this drug combination in MM patients.
Similar content being viewed by others
Introduction
Clonal expansion of terminally differentiated, immunoglobulin-secreting plasma cells in the bone marrow is a defining characteristic of multiple myeloma (MM).1 The transformation of a normal plasma cell to active myeloma is not a single-step process; instead, it is a lengthy process that involves the accumulation of multiple genetic aberrations. Genetic lesions such as inherited variation, hyperdiploidy and translocation have been implicated to have important roles in the initiation of the disease. Over time, the deregulated plasma cell clones acquire secondary genetic abnormalities, expand and evolve to become symptomatic myeloma.2
Overexpression of MYC is a recurrent oncogenic event in MM.3, 4 It has been reported that MYC overexpression occurs in about two-third of MM, and its activation is associated with hyperdiploidy and shorter survival.4 In human cancer, dysregulation of MYC is often a result of upstream oncogenic signaling or genomic changes.5, 6 In activated B cells and myeloma, MYC transcription is directly regulated by the transcription factor IRF4, whose expression is required for MM cell survival.7 The expression of IRF4 is, in turn, controlled by the lymphoid transcription factor IKZF1, which is a known target of the immunomodulatory drugs (IMiDs) thalidomide, lenalidomide and pomalidomide used in MM therapy.8, 9 Adding to this complexity, IRF4 is also a target of MYC transactivation, resulting in an auto-regulatory circuit in MM.7 The IKZF1–IRF4–MYC axis is critical for the survival of myeloma cells. This signaling cascade, therefore, presents multiple points of vulnerability, which can be exploited by novel agents that target these oncogenes.
Small-molecule inhibitors of the bromodomain and extra-terminal (BET) family of proteins provide a clinically applicable approach for inhibiting MYC and its downstream oncogenic signaling in various hematological malignancies.10, 11, 12, 13, 14 The BET family is comprised of four members as follows: BRD2, BRD3, BRD4 and BRDT. These proteins contain highly related tandem repeats of bromodomains that recognize acetylated lysine residues on histone tails and recruit transcriptional machinery to promote gene expression.15, 16, 17 Notably, BRD4 has been shown to be a therapeutic target in hematologic neoplasia.18 The prevailing consensus is that BET inhibitors suppress MYC transcription and activity by disrupting BRD4 binding to chromatin, thereby inhibiting proliferation and survival of tumor cells.10, 18, 19, 20 However, it appears that pharmacologic BET inhibitors do not solely target MYC: effects on the transcription of other genes likely contribute to the phenotypic consequences of BET inhibition.
Here we investigated the anti-MM activity of CPI-0610, a novel BET inhibitor that is currently in a phase I clinical trial in relapsed MM (ClinicalTrials.gov Identifier: NCT02157636). In our study, CPI-0610 treatment resulted in MM cytotoxicity in vitro by inducing G1 cell cycle arrest and caspase-dependent apoptosis. CPI-0610 significantly delayed tumor growth and increased the survival of MM-bearing SCID mice in a xenograft model. Our findings indicate that BET inhibition not only results in a robust reduction of MYC transcription and activity but also suppresses the expression of IKZF1 and IRF4 in MM. Furthermore, we demonstrated that low-dose combinations of IMiDs and CPI-0610 displayed synergistic anti-MM activity.
Materials and methods
Gene expression profiling on plasma cells
Differential expression of IKZF1, IRF4 and MYC in CD138+ plasma cells during disease progression towards MM was analyzed using the publicly available GSE6477 microarray expression data set. This data set includes 15 normal, 21 monoclonal gammopathy of undetermined significance (MGUS), 23 smoldering myeloma and 103 active myeloma samples (75 newly diagnosed and 28 relapsed) profiled using Affymetrix Human Genome U133A Array.21 The expression of IKZF1, IRF4 and MYC was analyzed. Comparisons were done by t-test.
Cell viability and proliferation assays
The effect of CPI-0610 on the viability of MM cell lines and primary MM cells was determined by measuring MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Chemicon International, Temecula, CA, USA) dye absorbance as previously described.22 The effect of CPI-0610 on the viability of MM cell lines in the presence and absence of recombinant interleukin-6 (10 ng/ml), insulin-like growth factor-1 (50 ng/ml) or bone marrow stromal cell (BMSC)-conditioned media was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The effect of CPI-0610 on the proliferation status of MM cell lines in BMSC co-culture experiments was assessed using CyQUANT NF Cell Proliferation Assay Kit (Molecular Probes, Eugene, OR, USA) according to the manufacturer’s instructions.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed as previously described.23 Primer sequences for the analyzed genes are listed in Supplementary Table 1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on MM.1S cells cultured in the presence or absence of CPI-0610 (1 μM, 24 h) and/or lenalidomide (2 μM, 24 h) using EpiTect ChIP One-Day kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. BRD4 ChIP was performed using BRD4 antibody (Bethyl Labs, Montgomery, TX, USA). Normal rabbit IgG (Santa Cruz Biotechnology, Dallas, TX, USA) was used as a negative control. Quantitative detection of ChIP DNA was performed using SYBR Green real-time PCR. ChIP–qPCR assays were analyzed using Excel-based EpiTecChIP qPCR Data Analysis Template (Qiagen). Primer sequences for MYC, IKZF1 and IRF4 promoters are listed in Supplementary Table 2.
Lentivirus infection and short hairpin RNA sequences
The generation of lentiviral particles was previously described.24 The pLenti-C-mGFP-IKZF1 Human ORF Clone was obtained from OriGene (Rockville, MD, USA). Synthetic complementary oligonucleotides targeting the messenger RNA of interest were annealed and subcloned into Tet-pLKO-puro plasmid (Addgene, Cambridge, MA, USA). Targeting sequences are listed in Supplementary Table 3.
MM xenograft mouse model (plasmacytoma model)
The in vivo anti-MM activity of CPI-0610 was evaluated in a previously established MM xenograft model.25 In brief, 6–8-week-old CB17-SCID mice (Charles River Laboratories, Wilmington, MA, USA) were inoculated subcutaneously with 2.5 × 106 MM.1S LucNeo GFP+ cells 24 h after γ-irradiation (2 Gy). When tumors reached ~100 mm3, the mice were randomly assigned into two cohorts receiving subcutaneous injections with 10 mg/kg CPI-0610 b.i.d. or with control vehicle (n=8 per cohort). For in vivo studies, CPI-0160 was dissolved in 0.5% (w/v) methylcellulose (Sigma-Aldrich, St Louis, MO, USA) and stored at 4 °C until use. Mice were monitored for tumor volume by caliper measurements every 2–3 days. Tumor volume was measured using the following formula: ½ × (length) × (width)2. Tumor burden was also monitored by whole-body bioluminescence imaging weekly. Mice were killed when the tumor reached 2 cm. Tumor growth and survival were evaluated from the first day of treatment until death. The experimental protocol was approved by the Animal Care and Use Committee of the Massachusetts General Hospital.
Results
Differential expression of IKZF1, IRF4 and MYC in different stages of MM development
We initially analyzed the gene expression profiles for IKZF1, IRF4 and MYC in various stages of plasma cell neoplastic states with a total of 162 samples, which include 15 normal samples, 21 MGUS samples, 23 smoldering myeloma samples and 103 active myeloma samples (75 newly diagnosed and 28 relapsed).21 Gene expression analysis shows a modest increase in IRF4 from normal to MGUS and then its expression flattens throughout disease progression. The transcript levels of MYC gradually increase as the disease progresses from normal plasma cells to active myeloma (Figure 1a). The increased IRF4 level at the MGUS stage could be an early event during myeloma development, whereas the gradual increase in MYC could either represent a secondary or tertiary mutation as a cell transforms or it could merely be a consequence of gene deregulation in upstream signaling. The IRF4 and MYC transcripts are constantly expressed at a higher level in patient-derived CD138+ cells compared with CD138− controls (Figure 1b), which are consistent with the previous findings that MYC and IRF4 are overexpressed in MM.4, 7 Gene expression analysis shows a decline in IKZF1 from normal to MGUS to smoldering myeloma, and there is no decrease in levels of IKZF1 in newly diagnosed MM and relapsed MM when compared with smoldering myeloma patients (Figure 1a). We found the transcript levels of IKZF1 varied among CD138+ cells derived from myeloma patients (Figure 1b). Nonetheless, Ikaros, the protein encoded by the gene IKZF1, is highly expressed in patient-derived CD138+ cells compared with their counterparts in CD138− cells (Figure 1c). It has been recently shown that thalidomide and the related IMiDs lenalidomide and pomalidomide target Ikaros for proteasomal degradation by the cereblon ubiquitin ligase in myeloma cells.8, 9 The IMiD-induced inhibition of myeloma cell growth is primarily via degradation of the Ikaros protein, suggesting that stabilization of the Ikaros protein rather than its gene upregulation confers a survival benefit to myeloma. Overall, our data confirmed the notion that increased levels of Ikaros, IRF4 and MYC are important events in myeloma development.
Differential expression of IKZF1, IRF4 and MYC in CD138+ plasma cells during disease progression in multiple myeloma. (a) The data set analyzed (GSE6477) includes 15 normal samples, 21 MGUS samples, 23 smoldering samples and 103 active myeloma samples.21 Increased IRF4 and MYC expression are observed in newly diagnosed and relapsed myeloma plasma cells compared with normal plasma cells (P=0.00407 and P=8.19 × 10−5, respectively, using t-test). Decreased IKZF1 expression is observed in myeloma cells compared with normal plasma cells (P=0.000318, using t-test). (b, c) Patient-derived CD138− and CD138+ cells were analyzed for transcript and protein levels of IKZF1, IRF4 and MYC by qPCR and immunoblot respectively. Pt-1, 3 and 5 are newly diagnosed MM patients. Pt-2 and 4 are relapsed MM patients.
Targeting IKZF1, IRF4 and MYC by short hairpin RNA results in MM cell death and reveals a positive-feedback mechanism that is essential for MM cell survival
To evaluate the individual contribution of IKZF1, IRF4 and MYC to the survival of myeloma cells, we used doxycycline-inducible short hairpin RNAs to separately knockdown the expression of these genes in the INA6 MM cell line. Depletion of IKZF1, IRF4 and MYC all led to induction of apoptosis in MM cells, with the most prominent induction of cell death being found with IKZF1 knockdown (Figure 2a). It is interesting to note that in cells that had lost MYC expression (shIKZF1 #3), probably due to an off-target effect of the short hairpin RNA-mediated knockdown or an unexpected side effect of lentiviral transduction, apoptosis was blocked despite a successful IKZF1 knockdown (Figure 2b).
Silencing of IKZF1, IRF4 and MYC is toxic to MM cells. (a) INA6 cells were transduced with doxycycline-inducible short hairpin RNAs (shRNAs). After a 4-day treatment with doxycycline (100 ng/ml), shRNA-mediated knockdown of MYC, IKZF1 or IRF4 all led to induction of apoptosis as determined by Annexin V staining. (b) Protein expression of Ikaros, IRF4 and MYC is examined in lentiviral transduced INA6 cells after a 3-day treatment with doxycycline (100 ng/ml). (c) Ikaros was overexpressed in Dox40 MM cells via lentiviral transduction, and protein expression of Ikaros, IRF4 and MYC was examined. (d) A proposed positive-feedback loop illustrates a molecular sequence of events going from IKZF1 to IRF4 to MYC and then back to IKZF1 via IRF4.
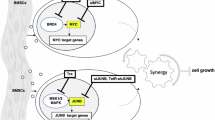
Ikaros is known to bind to the IRF4 promoter and regulates its gene expression in MM.8 Shaffer et al.7 have previously shown that IRF4 binds to the MYC promoter and regulates its expression at the transcriptional level. Thus, the IKZF1–IRF4–MYC axis has a central role in the transcriptional network of MM. With the use of doxycycline-inducible short hairpin RNAs targeting IKZF1, IRF4 and MYC, we identified a positive-feedback mechanism that is critical for the survival of MM. As shown in Figure 2b, suppression of IRF4 decreases both Ikaros and MYC protein expression (shIRF4 #1) and a decrease in IRF4 protein is observed upon MYC downregulation (shMYC #1 and shMYC #2). In addition, overexpression of Ikaros in MM cells led to downstream upregulation of IRF4 and MYC (Figure 2c). Together, these results implicate a feedback regulation from MYC to IRF4 and from IRF4 to IKZF1. Taken together with prior studies, our data illustrate a molecular sequence of events go from IKZF1 to IRF4 to MYC and then back to IKZF1 via IRF4, forming a positive-feedback loop in MM cells (Figure 2d).
BRD4 directly controls the expression of IKZF1, IRF4 and MYC in MM cells
CPI-0610 is a benzoisoxazoloazepine inhibitor of the BET family (Figure 3a).26 It blocks the function of BET proteins by disrupting their interactions with acetyl-lysine residues on chromatin, thereby attenuating the expression of key oncogenes. Recent studies of BET inhibition showed that depletion of BRD4 at the MYC promoter decreases MYC transcription.10, 18, 19, 20, 27 To test whether IKZF1 and IRF4 are under the same transcriptional control by the bromodomain protein, we performed a ChIP analysis using an anti-BRD4 antibody followed by qPCR in MM.1S cell line. We found that BRD4 localizes at IKZF1, IRF4 and MYC promoters, and its binding to the promoter elements decreases upon exposure to CPI-0610 (Figure 3b). These results demonstrated that BRD4 acts as a transcriptional co-activator that drives IKZF1, IRF4 and MYC expression in MM cells. Moreover, we showed that CPI-0610 treatment directly inhibits Ikaros, IRF4 and MYC expression in a panel of MM cell lines (Figure 3c). Together, our data demonstrated that CPI-0610 inhibits IKZF1, IRF4 and MYC expression and activity by interfering with their BRD4-mediated transcriptional activation.
BRD4 regulates expression of IKZF1, IRF4 and MYC at the levels of both transcription and protein in MM cells. (a) Chemical structure of CPI-0610. (b) ChIP assay showed the binding of BRD4 to the endogenous IKZF1, IRF4 and MYC promoters. Interaction of BRD4 with IKZF1, IRF4 and MYC promoters was assessed by qPCR. Data are shown as percent of input from two independent experiments. (c) Immunoblot analysis of protein levels of Ikaros, IRF4 and MYC in a panel of MM cell lines after a 24-h treatment with either dimethylsulphoxide or 800 nm of CPI-0610.
CPI-0610 treatment results in decreased viability of MM cells in a dose-dependent manner
To evaluate the anti-MM potential of CPI-0610, we first examined the in vitro effect of CPI-0610 treatment in MM cell lines. A panel of MM cell lines was cultured in the presence of increasing doses of CPI-0610 (0–1500 nM) for 72 h. As shown in Figure 4a, CPI-0610 treatment resulted in dose-dependent cytotoxicity with EC50 ranging between 200 and 900 nM at 72 h for BET-sensitive cell lines (INA6, RPMI-8226, LR5, H929, MM.1S, MM.1R and U266). Three cell lines (Dox40, ANBL6 and ANBL6-BR) remained less sensitive to CPI-0610 treatment with ~35–40% decrease in viability at a dose of 1500 nM at 72 h. We next assessed the activity of CPI-0610 in primary CD138+ tumor cells isolated from patients with MM. CPI-0610 treatment resulted in 40% decrease in viability in primary cells isolated from a newly diagnosed patient (patient 5) and caused 50% cell death in primary cells isolated from a relapsed disease patient (patient 4), both at a dose of 2 μM at 72 h (Figure 4b). These results indicate that CPI-0610 although active in patient-derived MM cells will likely need to be studied in combination with other anti-MM therapies.
CPI-0610 induces dose-dependent cytotoxicity in MM cell lines and patient-derived CD138+ cells. (a) MM cells were treated with increasing doses of CPI-0610 (0–1500 nm) for 72 h, and cell viability was measured by MTT assays. (b) Patient-derived CD138+ MM cells were similarly tested in cytotoxicity (MTT) assays at 72 h. Pt-4 is a relapsed disease patient, and Pt-5 is a newly diagnosed myeloma patient.
CPI-0610 overcomes the protective effect of cytokines and BMSCs
It has been shown that the bone marrow microenvironment enhances MM cell growth and survival in MM.28, 29 We next treated INA6, MM.1S and MM.1R cells with increasing doses of CPI-0610 in the presence or absence of interleukin-6, insulin-like growth factor-1 or BMSCs. CPI-0610 inhibited cell growth induced by exogenous cytokines and decreased DNA synthesis of MM cells co-cultured with BMSCs in a dose-dependent manner (Figures 5a–c). These data suggest that CPI-0610 overcomes the protective effects conferred by cytokines and BMSCs in the BM milieu.
CPI-0610 induces dose-dependent cytotoxicity in MM cells alone or co-cultured with cytokines or BMSCs. INA6, MM.1S and MM.1R cells were cultured for 72 h in the presence of (a) interleukin-6 (10 ng/ml), insulin-like growth factor-1 (50 ng/ml), (b) BMSC-conditioned media or (c) BMSCs in the absence or presence of increasing doses of CPI-0610. (a, b) Cell viability was assessed by CellTiter-Glo Luminescent Cell Viability Assay. (c) Cell proliferation was evaluated by CyQUANT NF Cell Proliferation Assay.
CPI-0610 treatment leads to G1 cell cycle arrest and caspase-dependent apoptosis in MM cells
To characterize the molecular mechanisms underlying the cytotoxic effect of CPI-0610 in MM cells, we performed cell cycle analysis and apoptosis assay in INA6 and MM.1S cells cultured with dimethylsulfoxide control or CPI-0610 (800 nM). As demonstrated in Figure 6a, CPI-0610 treatment resulted in an increase in G1 cells at 48 h. We observed a significant increase in apoptotic MM cells after 72 h of CPI-0610 treatment (Figure 6b). This was associated with an increase in poly(ADP-ribose) polymerase cleavage (Figure 6c), caspase-3/7 and caspase-9 activities (Figure 6d). When we pretreated the cells with 2 μM of Z-DEVD-FMK (caspase-3 inhibitor) and Z-LEHD-FMK (caspase-9 inhibitor) an hour before CPI-0610 treatment, caspase-3 and -9 activities were blocked. No increase in caspase-8 activity was observed with CPI-0610 treatment in MM cells (data not shown). These results suggested that CPI-0610-induced apoptosis was predominantly activated via the intrinsic apoptotic pathway.
CPI-0610 increases G1 phase and induces caspase-dependent apoptosis of MM cells. (a) Cell cycle analysis by propidium iodide (PI) staining was performed on INA6 and MM.1S cells cultured with dimethylsulphoxide (DMSO) or CPI-0610 (800 nM) for 48 h. CPI-0610 treatment resulted in an increase in G1 cells. (b) INA6 and MM.1S cells were cultured with DMSO or CPI-0610 (800 nM) for 72 h and analyzed for apoptosis using Annexin V/PI staining assay. CPI-0610 treatment resulted in an increase in apoptotic cell death. (c) This was preceded by an increase in poly(ADP-ribose) polymerase (PARP) cleavage after 24 h of CPI-0610 treatment (800 nM), as shown by immunoblot analysis. (d) INA6 cells were seeded at a density of 10 000 cells per well and incubated with CPI-0160 (800 nM) for 2 h for the detection of caspase-9 activity, and 4 h for the detection of caspase-3/7 activity. Pretreatment of INA6 cells with 2 μM of Z-DEVD-FMK and Z-LEHD-FMK for an hour blocked caspase-3 and -9 activities correspondingly.
CPI-0610 induces anti-MM activity in vivo in a plasmacytoma model
We next examined the in vivo efficacy of CPI-0610 using a human MM xenograft mouse model.30 Mice treated with CPI-0610 showed a significant delay in tumor growth (P=0.0438). We observed a decrease in tumor volume in CPI-0610-treated animals (890±133 mm3) vs controls (1354±162 mm3) at 17 days (Figure 7a). The decrease in tumor size was further confirmed by reduced bioluminescence signal in treated animals (395 vs 1169) imaged at 16 days (Figure 7b). Importantly, CPI-0610 was well tolerated in our animal studies. Mice treated with CPI-0610 had an average weight loss of 6.6% compared with control animals (data not shown). Mice were killed once the tumor reached 2 cm in diameter. The median overall survival of CPI-0610-treated animals was significantly prolonged compared with control animals (32 vs 22.5 days). A significant survival benefit was observed in CPI-0610-treated animals vs controls (P=0.0223) using log-rank analysis, as demonstrated by a Kaplan–Meier curve (Figure 7c). Following the death of the animals, tumors were collected and subjected to immunoblot analysis. We found increased poly(ADP-ribose) polymerase cleavage in tumor isolated from CPI-0610-treated animal compared with that of a control animal (Figure 7d). These results suggest that CPI-0610 constrains tumor growth in vivo by induction of apoptosis.
CPI-0610 is active in a MM xenograft model. (a) Tumor growth was significantly inhibited in the CPI-0610-treated animals compared with controls. A decrease in tumor volume in CPI-0610-treated animals (890±133 mm3, n=8) vs controls (1354±162 mm3, n=8) was noted on day 17 from the first day of treatment (P=0.0438) before control animals started to succumb to the tumor. (b) Antitumor activity was confirmed by decreased bioluminescence in CPI-0610-treated animals. (c) A Kaplan–Meier curve showed a significant survival benefit in CPI-0610-treated animals vs controls (P=0.0223), using log-rank analysis. (d) Tumors from CPI-0610-treated animals showed increased poly(ADP-ribose) polymerase (PARP) cleavage compared with controls.
CPI-0610 has synergistic anti-MM activity when combined with IMiDs
Based on the observations that depletion of MYC and IKZF1 are toxic to MM (Figure 2a), we combined CPI-0610 with lenalidomide or pomalidomide, IMiDs that stabilize cereblon and facilitate Ikaros degradation in MM cells.8, 9 Increasing doses of CPI-0610 (0, 0.25, 0.5 and 1 μM) were added to lenalidomide or pomalidomide (0, 1, 3 and 5 μM), and the cytotoxic effect of combined therapy was assessed by MTT assays in MM.1S cells at 48 h. A significant decrease in cell viability was observed with combined therapy compared with both drugs used alone (Figure 8a). Synergism was determined using the Chou and Talalay method.31 Isobologram analysis showed that low concentrations of CPI-0610 and IMiDs triggered synergistic anti-MM activity with a combination index <1 (Figure 8a). The same in vitro synergism was also observed in RPMI-8226 and H929 cells (Supplementary Figure 1). The enhanced cytotoxic effect of the combined therapy in MM cell lines is, in part, due to the suppression of IKZF1, IRF4 and MYC expression at the levels of transcription and protein in MM cells (Figures 8b and c).
CPI-0610 in combination with IMiDs induces synergistic anti-MM activity. (a) MM.1S cells were cultured with increasing doses of lenalidomide or pomalidomide in the presence or absence of CPI-0610 (0–1000 nM) for 48 h. Cytotoxicity was assayed by MTT. The isobologram analysis confirms the synergistic effect. (b, c) MM.1S cells treated with CPI-0610 (1000 nM), lenalidomide (2 μM) or combined therapy were examined for transcript (4 h treatment) and protein (24 h treatment) levels by immunoblot and qPCR, respectively.
Discussion
The BET family of proteins are attractive drug targets because they control the transcription of a small subset of genes, including the oncogene MYC.10 Until the development of small-molecule BET inhibitors,10, 11 therapeutic strategies that modulate oncogenic MYC function did not exist. In MM, BET proteins occupy active enhancer and promoter regions to drive MYC transcription.32 BET inhibitors bind competitively to the acetyl-lysine recognition pockets to displace BET proteins from chromatin, thereby suppressing transcription of key oncogenic drivers.33, 34 It is widely accepted that the effects of BET inhibitors are primarily due to their ability to suppress MYC transcription in MYC-dependent cancers.10, 19, 20 Our study confirmed this notion, but we also found IKZF1 and IRF4 to be among the primary targets of the BET inhibitor CPI-0610, in addition to MYC, in MM. ChIP studies showed that transcription of IKZF1 and IRF4 are under the direct control of BRD4 and CPI-0610 treatment effectively inhibits their expression in MM cells.
Recent evidence in primary effusion lymphoma suggests a transcriptional network controlling the flow of information from IKZF1 to IRF4 to MYC.13 Interestingly, these transcriptional regulators are all found to be upregulated in patient-derived MM cells. We found that short hairpin RNA-mediated knockdowns of IKZF1, IRF4 or MYC were toxic to MM cells. These results prompted us to investigate the role of these transcriptional regulators in the anti-proliferative effect of CPI-0610 in MM. CPI-0610 displays pronounced anti-proliferative effect against the majority of MM cell lines within clinically achievable doses. The observed in vitro cytotoxicity is accompanied with concomitant downregulation of IKZF1, IRF4 and MYC. Upon CPI-0610 treatment, we observed a marked decrease in MYC expression at both transcript and protein levels. The decreases in Ikaros and IRF4 expression were still significant, but to a lesser extent compared with MYC. Knockdown studies of IKZF1, IRF4 and MYC illustrate a molecular sequence of events goes from MYC to IKZF1 via IRF4. Taken together with prior studies,7, 8, 9 we propose an auto-regulatory circuit centered on the IKZF1–IRF4–MYC axis in MM. The proposed transcriptional network presents multiple points of vulnerability to MM cells. In support of this premise, CPI-0610 potently inhibits MM cell growth in vitro and in vivo by simultaneously suppressing the expression of these three transcriptional regulators. In addition to the above mechanism, CPI-0610-induced apoptosis is caspase-dependent and is primarily activated via the intrinsic apoptotic pathway.
The use of combination therapy has become a standard practice in MM treatment. Combining different classes of drugs might improve patient outcomes by enhancing tumor apoptosis and overcoming drug resistance.35 We, therefore, studied CPI-0610 in combination with IMiDs, based on our observation that depletion of MYC and IKZF1 are toxic to MM. We observed striking synergy between low doses of CPI-0610 and IMiDs in MM cell lines. The enhanced cytotoxicity of CPI-0610 and IMiDs is due in part to concomitant suppression of IKZF1, IRF4 and MYC. As inhibition of IKZF1 is not complete with lower doses of CPI-0610, the addition of low doses of IMiDs can further suppress IKZF1 in MM cells. In conclusion, our data support evaluation of CPI-0610, alone and in combination with IMiDs, to improve patient outcome in MM.
References
Anderson KC, Carrasco RD . Pathogenesis of myeloma. Ann Rev Pathol 2011; 6: 249–274.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Kuehl WM, Bergsagel PL . MYC addiction: a potential therapeutic target in MM. Blood 2012; 120: 2351–2352.
Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011; 25: 1026–1035.
Dang CV . MYC on the path to cancer. Cell 2012; 149: 22–35.
Soucek L, Evan GI . The ups and downs of Myc biology. Curr Opin Genet Dev 2010; 20: 91–95.
Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W et al. IRF4 addiction in multiple myeloma. Nature 2008; 454: 226–231.
Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014; 343: 305–309.
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014; 343: 301–305.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917.
Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood 2014; 123: 697–705.
Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 2012; 120: 2843–2852.
Gopalakrishnan R, Matta H, Tolani B, Triche T Jr., Chaudhary PM . Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene 2012; 35: 1797–1810.
Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res 2013; 73: 3336–3346.
Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 2011; 31: 2641–2652.
LeRoy G, Rickards B, Flint SJ . The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol 2008; 30: 51–60.
Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM . Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999; 399: 491–496.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011; 478: 524–528.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674.
Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478: 529–533.
Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res 2007; 67: 2982–2989.
Santo L, Vallet S, Hideshima T, Cirstea D, Ikeda H, Pozzi S et al. AT7519, A novel small molecule multi-cyclin-dependent kinase inhibitor, induces apoptosis in multiple myeloma via GSK-3beta activation and RNA polymerase II inhibition. Oncogene 2010; 29: 2325–2336.
Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012; 119: 2579–2589.
Raje N, Hideshima T, Mukherjee S, Raab M, Vallet S, Chhetri S et al. Preclinical activity of P276-00, a novel small-molecule cyclin-dependent kinase inhibitor in the therapy of multiple myeloma. Leukemia 2009; 23: 961–970.
Cirstea D, Hideshima T, Santo L, Eda H, Mishima Y, Nemani N et al. Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia 2013; 27: 2366–2375.
Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y et al. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J Med Chem 2016; 59: 1330–1339.
Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM . Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene 2014; 33: 2928–2937.
Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC . Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer 2007; 7: 585–598.
Hideshima T, Anderson KC . Molecular mechanisms of novel therapeutic approaches for multiple myeloma. Nat Rev Cancer 2002; 2: 927–937.
Santo L, Hideshima T, Cirstea D, Bandi M, Nelson EA, Gorgun G et al. Antimyeloma activity of a multitargeted kinase inhibitor, AT9283, via potent Aurora kinase and STAT3 inhibition either alone or in combination with lenalidomide. Clin Cancer Res 2011; 17: 3259–3271.
Chou TC, Talalay P . Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984; 22: 27–55.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Filippakopoulos P, Knapp S . Targeting bromodomains: epigenetic readers of lysine acetylation. Nature Rev Drug Discov 2014; 13: 337–356.
Shi J, Vakoc CR . The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014; 54: 728–736.
Lonial S, Kaufman JL . The era of combination therapy in myeloma. J Clin Oncol 2012; 30: 2434–2436.
Acknowledgements
We thank members of Dr David Scadden’s laboratory: David Sykes, Youmna Kfoury, Francois Mercier, Dongjun Lee, Ninib Baryawno, Nicolas Sévère, Karin Gustafsson and Demetrios Kalaitzidis at Massachusetts General Hospital for helpful discussion. This work is supported by the Multiple Myeloma Research Fund at MGH. Constellation Pharmaceuticals, Inc., provided the CPI-0610 compound.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
JAM, RJS and MRC are employees and shareholders of Constellation Pharmaceuticals. NR is on advisory boards of Amgen, Bristol-Myers Squibb, Celgene Corporation and Takeda. NR has received research funding from AstraZeneca and Eli Lilly. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Siu, K., Ramachandran, J., Yee, A. et al. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia 31, 1760–1769 (2017). https://doi.org/10.1038/leu.2016.355
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.355
- Springer Nature Limited
This article is cited by
-
Targeting bromodomain-containing proteins: research advances of drug discovery
Molecular Biomedicine (2023)
-
Characterization and application of a lactate and branched chain amino acid metabolism related gene signature in a prognosis risk model for multiple myeloma
Cancer Cell International (2023)
-
The effects of MYC on tumor immunity and immunotherapy
Cell Death Discovery (2023)
-
Role of HOXA9 in solid tumors: mechanistic insights and therapeutic potential
Cancer Cell International (2022)
-
BRD4 drives esophageal squamous cell carcinoma growth by promoting RCC2 expression
Oncogene (2022)