Abstract
Objectives:
The aim of this study was to explore the phenotypic differences underpinning obesity susceptibility or resistance based on the metabolic and transcriptional profiling of C57BL/6J mice fed a high-fat diet (HFD).
Methods:
The mice were fed either a normal diet or HFD for 12 weeks. After 6 weeks, the mice on HFD were classified as either obesity-prone (OP) or obesity-resistant (OR) depending on the body weight gain.
Results:
Lipid profiles from plasma and liver significantly improved in OR mice relative to the OP group. Energy expenditure was greater in OR mice than in OP mice, with a simultaneous decrease in body fat mass. Epididymal white adipose tissue (eWAT) and liver were enlarged in OP mice (with visible immune-cell infiltration), but these effects were attenuated in OR mice compared with OP mice. Overall glucose metabolism was enhanced in OR mice compared with OP mice, including homeostasis model assessment for insulin resistance, plasma glucose and insulin concentrations, glucokinase activity and hepatic glycogen. Plasma adipokines and proinflammatory cytokines were upregulated in OP mice, and these changes were attenuated in OR mice. Transcriptomic profiles of eWAT and liver revealed common and divergent patterns of transcriptional changes in OP and OR mice, and pointed to differential metabolic phenotypes of OP and OR mice. There were substantial differences between OP and OR mice in molecular pathways, including atherosclerosis signaling, sperm motility, cAMP-mediated signaling in eWAT; and fibrosis, agranulocyte adhesion and diapedesis, and atherosclerosis signaling in liver.
Conclusions:
Taken altogether, the results provide robust evidence of major divergence in the transcriptomes, phenotypes and metabolic processes between obesity susceptibility and obesity resistance in the HFD-fed C57BL/6J mice.
Similar content being viewed by others
Introduction
Obesity is positively associated with insulin resistance, dyslipidemia and other chronic disorders, all of which characterize metabolic syndrome.1 Obesity is characterized by expansion of adipose tissue and fat droplet storage when energy intake exceeds energy expenditure. There is a direct relation between the percentage of body fat and dietary fat.2 Nonetheless, in rodents, the metabolic responses to a high-fat diet (HFD) can be different even under conditions of isocaloric feeding within a group of animals with the same genetic background; hence, some rodents can show a more obesity-prone (OP) or more obesity-resistant (OR) phenotype. Several recent studies on OP and OR rats revealed differences in metabolic, transcriptomic and proteomic profiles.3, 4, 5, 6, 7 Transcriptomic analysis that is based on the microarray technology showed that genes associated with the metabolism of energy, lipids and glucose are modulated by a HFD.8, 9 Recent advances in high-throughput transcriptomic methods (based on next-generation sequencing) now allow for the measurement of gene expression across a broader dynamic range, with increased specificity and sensitivity as compared with microarrays.10, 11 One of the most frequently analyzed genetic features, mRNA, can now be quantified on a genome-wide level by means of next-generation sequencing of mRNAs (mRNA-Seq) with greater specificity and sensitivity than previously possible. This is the first study to utilize mRNA-Seq to study the differences in transcriptomic profiles of adipose and hepatic tissues between OP and OR C57BL/6J mice fed an isoenergetic HFD.
First, we characterized the divergent changes in the metabolic profile and phenotype of HFD-fed mice that were either susceptible or resistant to diet-induced obesity, as compared with normal diet (ND)-fed mice. Second, using mRNA-Seq, we analyzed transcriptional responses and identified molecular pathways (using Ingenuity Pathway Analysis; IPA) that underlie susceptibility or resistance to diet-induced obesity in C57BL/6J mice.
Materials and methods
Animals
Male C57BL/6J mice (4-week-old; Jackson Laboratory, Bar Harbor, ME, USA) were fed lab chow diet for the first week for adaptation. After that, the mice were randomly assigned to 2 groups: ND (17.2 kcal% dietary fat; Harlan, Madison, WI, USA; n=10) and HFD (60 kcal% dietary fat; Harlan; n=20). After 6 weeks on the HFD, 20 mice were subdivided on the basis of body weight gain into an OP group (n=8; ⩾43 g) and OR group (n=7; ⩽39 g). Differences in metabolic and inflammatory phenotypes were then characterized. mRNA-Seq was used to identify divergent changes in the transcriptome in ND-fed, OP and OR mice. Body weight and blood glucose level were measured every 1 to 2 weeks. At the end of the diet period, all mice were anesthetized with isoflurane after a 12-h fast. Blood samples were collected in tubes containing EDTA from the inferior vena cava for quantification of glucose, plasma lipids and hormones. The adipose tissue depots were dissected, rinsed with physiological saline, weighed, immediately frozen in liquid nitrogen and stored at −70 °C until analysis. All experiments were performed in accordance with protocols for animal studies approved by the Ethics Committee of Kyungpook National University (approval No. KNU 2012-136).
Methods
Methods and any associated references are available in the online version of the paper.
Results
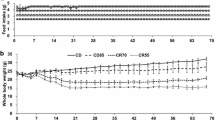
Body weight gain and food efficiency are augmented in OP mice and attenuated in OR mice
Figure 1a represents changes in body weight during the experimental period of 12 weeks, excluding the 1-week adaptation period. The initial body weight of the mice was approximately the same in all the three groups. Some mice were resistant to the HFD-induced body weight gain, whereas other mice showed a significant body weight gain as early as week 1 on the HFD (Figure 1a). We subsequently classified these mice as either OP (characterized by a greater body weight gain) or OR (characterized by a lesser body weight gain) as shown in Figure 1a. The overall body weight gain was significantly greater in the HFD-fed OP mice (0.30±0.011 g per day, P<0.001) than in the ND-fed mice (0.11±0.004 g per day) but lower in the HFD-fed OR mice (0.21±0.006 g per day, P<0.001). Energy intake was not different between OP and OR mice, and was greater in comparison with the ND-fed mice (Figure 1c). Hence, the food efficiency ratio was significantly lower in the OR group than in the OP group (P<0.05) although food efficiency ratio of these groups was significantly higher than that of the ND group (Figure 1d).
(a) Changes in body weight during 12 weeks. (b) Body weight gain per day. (c) Differences in energy intake. (d) Food efficiency ratio. (e) Muscle weight. (f) Liver weight. (g) Hematoxylin and eosin staining (H&E; upper panel) and Masson’s trichrome staining (MT; lower panel) of the liver. (h) Total weight of WAT. (i) H&E and MT staining of epididymal adipocytes (magnification × 200) from ND-fed C57BL/6J mice or HFD-fed OP or OR C57BL/6J mice. ND, AIN-93G; HFD, 60% kcal from fat. The data are presented as mean±s.e.m. abcMean values not sharing a common superscript were significantly different among the groups (P<0.05).
eWAT and liver were enlarged in OP mice, with visible immune-cell infiltration, but these effects were attenuated in OR mice
The significant reduction in muscle weight that we observed in the HFD-fed mice was reversed in the OR group (Figure 1e). The liver weight per 100 g of body weight was significantly greater in the OP group than in the ND group, but lower in the OR group than in the OP group (Figure 1f). Tissue morphology analysis revealed that the accumulation of hepatic lipid droplets decreased in the OR group compared with the OP group. Moreover, Masson’s trichrome staining of the liver revealed notable hepatic fibrosis around the vessels in the OP group (Figure 1g). WAT weight per 100 g of body weight was significantly greater in the HFD-fed mice than in the ND group; perirenal, subcutaneous, mesenteric, interscapular, visceral and total WAT deposits were smaller in the OR group than in the OP group (Figure 1h). In addition, epididymal-adipocyte size of OP mice was visibly larger than that of the ND-fed mice according to the hematoxylin and eosin staining staining, whereas the adipocyte size of OR mice was visibly smaller than that of OP mice. Masson’s trichrome staining of epididymal white adipose tissue (eWAT) revealed a normal fat pad with densely packed hexagonal adipocytes in ND-fed mice, without evidence of immune-cell infiltration. There was visible morphological evidence of infiltration by immune cells in OP mice in contrast to ND-fed mice. OR mice showed weak infiltration by immune cells according to Masson’s trichrome staining (Figure 1i).
Lipid profiles of plasma and liver improved in OR mice compared with OP mice
The total cholesterol level in plasma was significantly lower in the OR group than in the OP group (Figure 2a). The levels of hepatic fatty acids, triglycerides and cholesterol were significantly lower in the OR group than in the OP group (Figure 2b). Furthermore, the levels of plasma glutamic oxaloacetic transaminase and glutamic pyruvic transaminase were significantly higher in both the OP group and OR group than in the ND group, but were lower in the OR group than in the OP group (Figure 2c). Of the two enzymes involved in fatty-acid oxidation, the activity of β-oxidation was increased in both the OP and OR groups compared with the ND group, whereas the activity of CPT was increased only in the OR group (Figure 2d). The activity of the rate-limiting enzyme for cholesterol biosynthesis in the liver, HMGCR, was markedly reduced in the OR group compared with the OP group (Figure 2d).
(a) Plasma total cholesterol and triglyceride concentrations. (b) Hepatic lipid profiles. (c) Plasma glutamic oxaloacetic transaminase (GOT) and glutamic pyruvic transaminase (GPT) levels. (d) Hepatic activities of lipid-regulating enzymes. (e, f) Energy expenditure. (g) Oxygen consumption (VO2) in ND-fed C57BL/6J mice or HFD-fed OP and OR C57BL/6J mice. ND, AIN-93G; HFD, 60% kcal from fat. Data are presented as mean±s.e.m. abcMean values not sharing a common superscript were significantly different among the groups (P<0.05).
Energy expenditure is decreased in OP mice, but increased in OR mice
The energy expenditure decreased in the OP group, relative to the ND group, during both the light phase and dark phase, whereas energy expenditure of the OR group significantly increased as compared with that of the OP group during the dark phase (Figures 2e and f). Consistent with the energy expenditure, VO2 was significantly lower in the OP group than in the ND group during the dark phase, whereas OR mice showed higher VO2 than did the OP group during the dark phase (Figure 2g).
Changes in plasma glucose and insulin levels are augmented in OP mice, but somewhat attenuated in OR mice
Plasma glucose was significantly upregulated in both the OP and OR group in comparison with the ND group, but significantly lower in the OR group than in the OP group (Figure 3a). The plasma insulin level was higher in the OP group than in the ND group, but significantly lower in the OR group than in the OP group (Figure 3b). Consistent with the plasma insulin level, immunohistochemical staining of the pancreas revealed a visibly increased size of the islet boundary and greater numbers of insulin-positive β-cells in the OP group than in the ND or OR groups (Figure 3e). The ND and OR groups had a smaller hormone-stained area (meaning preservation of pancreatic cells) in comparison with OP mice (Figure 3e). Insulin resistance according to HOMA-IR was significantly augmented in the OP group compared with the ND group, but significantly attenuated in the OR group (Figure 3c). There were no significant differences in plasma glucagon levels among the groups (Figure 3d). The HFD-induced upregulation of hepatic glycogen was attenuated in the OR group (Figure 3f), whereas glucokinase activity significantly increased in the OR group compared with the OP group (Figure 3g).
(a) Plasma glucose concentration. (b) Plasma insulin concentration. (c) Insulin resistance according to the homeostasis model assessment for insulin resistance (HOMA-IR). (d) Plasma glucagon concentration. (e) Differences in immunohistochemical staining for pancreatic insulin. (f) Hepatic glycogen content. (g) Hepatic glucokinase activity. (h) Hepatic phosphoenolpyruvate carboxykinase (PEPCK) activity. (i) Hepatic glucose-6-phosphatase (G6Pase) activity. (j) Differences in plasma leptin, (k) resistin, (l) adiponectin, (m) tumor necrosis factor-α, (n) MCP-1, (o) PAI-1, (p) IL-6 and (q) IFN-γ among ND-fed C57BL/6J mice and HFD-fed OP and OR C57BL/6J mice. ND, AIN-93G; HFD, 60% kcal from fat. The data are presented as mean±s.e.m. abcMean values not sharing a common superscript were significantly different among the groups (P<0.05).
Levels of adipokines and proinflammatory cytokines are elevated in OP mice, and these changes are attenuated in OR mice
The plasma concentrations of leptin and resistin were elevated in the OP group in comparison with the ND group, but these changes were attenuated in the OR group (Figures 3j and k). There were no differences in plasma adiponectin levels among the groups (Figure 3l). We observed significantly higher plasma concentrations of MCP-1 (monocyte chemotactic protein 1), PAI-1 (plasminogen activator inhibitor 1), IL-6 (interleukin 6) and interferon-γ (IFN-γ) in the OP group than in the ND group, but the upregulation of MCP-1, PAI-1, IL-6 and IFN-γ was significantly attenuated in the OR group compared with the OP group. Furthermore, MCP-1, PAI-1, IL-6 and IFN-γ levels were not different between OR and ND-fed mice (Figures 3n–q).
Transcriptomic profiles of eWAT and hepatic tissue revealed common and divergent transcriptomic patterns between OP and OR mice
To identify the global transcriptomic profiles associated with obesity susceptibility and obesity resistance, we performed mRNA-Seq on eWAT and liver samples obtained from ND-fed, OP and OR mice, and systematically analyzed the results as follows. First, we identified differentially expressed genes (DEGs) between OP and ND-fed mice, between OR and ND-fed mice, and between OR and OP mice using the cutoff set to fold change⩾|2|and P-value <0.05. In eWAT, 3366 DEGs were identified between OP and ND-fed mice (770 upregulated and 2596 downregulated), 235 DEGs were identified between OR and ND-fed mice (151 upregulated and 84 downregulated) and 1871 DEGs were identified between OR and OP mice (1622 upregulated and 249 downregulated; Figure 4a). In the liver, 1561 DEGs were identified between OP and ND-fed mice (1119 upregulated and 442 downregulated), 620 DEGs were identified between OR and ND-fed mice (239 upregulated and 381 downregulated) and 1029 DEGs were identified between OR and OP mice (883 upregulated and 146 downregulated; Figure 4b). The Venn diagram indicates the number of genes regulated commonly or specifically in various groups. There were 168 DEGs in eWAT that were either upregulated or downregulated in both OP and OR mice (Figure 4c). These commonly regulated DEGs were associated with a broad range of gene ontologies including upregulation of leukocyte activation, immune responses, as well as downregulation of some defense-response transcripts in eWAT (Supplementary Table S2). In contrast, the commonly upregulated DEGs in the liver were associated with cholesterol and steroid metabolism, and with redox reactions (oxidation reduction), whereas the downregulated DEGs were not significantly enriched in any specific gene ontology (Figure 4d, Supplementary Table S3).
Transcriptomic modulation in eWAT and hepatic tissue of OP mice in response to the HFD
The OP phenotype was underpinned by a set of 3198 DEGs (in eWAT) that underwent changes in expression only in the OP group. The 689 upregulated DEGs in adipose tissue of OP mice include large groups of immune-response genes involved in leukocyte, lymphocyte and T-cell activation, in immune pathways, and in regulation of cytokine production (Supplementary Table S4). Conversely, the 2509 downregulated DEGs in eWAT of OP mice included transcripts involved in sperm motility as well as a number of transcripts associated with spermatogenesis (Supplementary Table S4). In the liver, the 949 upregulated DEGs of OP mice included the transcripts involved in muscle contraction, electron transport chain and inflammatory response, whereas 322 downregulated DEGs were associated with biological processes related to enzyme-linked receptor protein signaling pathway (Supplementary Table S5).
Transcriptomic modulation in eWAT and hepatic tissue of OR mice in response to the HFD
The OR phenotype was associated with 67 DEGs (in eWAT) that underwent changes in expression only in the OR group. Only three upregulated DEGs were observed in adipose tissue of OR mice (Foxa1, Syt8 and Ctse). In contrast, there were 64 downregulated DEGs in adipose tissue, which were associated with defense responses and a response to bacterial infection (Supplementary Table S6). In the liver, 70 upregulated DEGs and 260 downregulated DEGs of OR mice were not significantly enriched in any specific gene ontologies.
Comparison of transcriptomic modulation in eWAT and hepatic tissue between OR and OP mice
Between the eWAT transcriptomes of OR and OP mice, 249 downregulated DEGs were associated with the gene ontology terms ‘immune response,’ ‘cell activation,’ ‘defense response’ and ‘leukocyte activation’; 1622 upregulated DEGs were involved in ‘sexual reproduction,’ ‘male gamete generation’ and ‘spermatogenesis’ (Supplementary Table S7). In the liver, upregulated DEGs were not significantly enriched in any specific gene ontologies, whereas 146 downregulated DEGs were associated with gene ontologies involving downregulation of muscle contraction, extracellular structure organization and collagen fibril organization (Supplementary Table S8).
Differential molecular pathways in eWAT and hepatic tissue of OP and OR mice according to IPA
To identify the interactive molecular pathways associated with obesity susceptibility and obesity resistance, we performed IPA and obtained Figures 5 and 6. The top-ranked canonical pathways in eWAT and hepatic tissue are shown in Figures 5 and 6 as a comparison of OP and ND-fed mice. The OP phenotype in eWAT was associated with upregulation of diverse genes related to immune-response signaling, including leukocyte extravasation, phagocytosis in macrophages and monocytes, communication between cells of innate and adaptive immunity, TREM1 signaling and many genes involved in inflammatory signaling (Figure 5a). Particularly, the majority of genes in the leukocyte extravasation signaling pathway (the top canonical pathway in eWAT) were downregulated in the OR group compared with the OP group (Figure 5b). We also found that canonical pathways, such as atherosclerosis signaling, sperm motility mechanisms, cAMP-mediated signaling and AMP-activated protein kinase signaling, were altered in the eWAT transcriptome of OR mice compared to OP mice (Figures 5c and d). In the liver, the OP phenotype was associated with upregulation of oxidative phosphorylation (OXPHOS), mitochondrial dysfunction, hepatic fibrosis, and cholesterol biosynthesis (Figure 6a). In contrast, upregulation of OXPHOS-related genes in the OP group was attenuated in the OR group (Figure 6b). Furthermore, we found that the upregulation of the pathways associated with hepatic fibrosis, agranulocyte adhesion and diapedesis, atherosclerosis signaling and OXPHOS was attenuated in OR mice (Figures 6c and d).
(a) Significant canonical pathways in the eWAT of HFD-fed OP mice versus ND-fed mice. (b) A heat map of the genes involved in leukocyte extravasation signaling in eWAT. (c) Significant canonical pathways in the eWAT of OR versus OP mice. (d) Expression of genes related to significant canonical pathways in the eWAT of OR mice versus OP mice. The significant pathways were obtained by IPA.
(a) Significant canonical pathways in the liver of HFD-fed OR mice versus HFD-fed OP mice. (b) A heat map of the genes involved in oxidative phosphorylation in the liver. (c) Significant canonical pathways in the liver of OR mice versus OP mice. (d) Expression of genes related to significant canonical pathways in the liver of OR mice versus OP mice. The significant pathways were obtained by IPA.
Discussion
This study provides robust evidence of a distinct OP phenotype and a distinct OR phenotype in C57BL/6J mice fed an isocaloric HFD. The emergence of divergent phenotypes is evident in the HFD-fed mice despite the identical genetic background and environmental conditions.
C57BL/6J is the strain used most widely in preclinical experimental models of obesity and of the related complications, but various studies have shown interindividual variability in weight gain and in physiological responses to an HFD.12, 13, 14 Acute and chronic overeating experiments in young men have also revealed individual differences in the responses to overeating.15, 16, 17, 18 In humans, however, it is difficult to identify the relative contribution of specific genetic and environmental factors to those responses to dietary changes. In the present study, divergence in the body weight gain, in response to the HFD, is apparent as early as week 1, with a significantly greater body weight gain in mice classified as OP compared with OR mice. Several research groups have attempted to identify the physiological factors that determine whether a mouse is more susceptible or less susceptible to diet-induced obesity. These researchers, however, have mainly focused on identification of differences in whole-body physiological or metabolic adaptations between different mouse strains rather than within the same mouse strain.19
Obesity susceptibility and resistance result in distinct body weight gain, tissue morphology and metabolism. Energy intake appears not to differ between our OP and OR mice; therefore, the food efficiency ratio is significantly lower in OR mice, indicating that these mice are less capable of energy storage. This result can be explained by increased energy expenditure and VO2 in OR mice compared with OP mice during the dark phase.
A major proportion of the excess body weight gain per day in OP mice was due to expansion of WAT, including the epididymal, perirenal, mesenteric and interscapular depots. Epididymal adipocytes were enlarged and lipid laden in OP mice, and there was evidence of infiltration by collagen fibrils between the adipocytes. In contrast, OR mice had smaller adipocytes, with only traces of infiltration by collagen fibrils.
Exposure to an HFD induces metabolic alterations including dyslipidemia and hepatic steatosis, and we observed differences in metabolic profiles between OP and OR mice. The total cholesterol level in plasma was attenuated in the OR group compared with the OP group. The levels of hepatic lipids including fatty acids, triglycerides and cholesterol were significantly lower in the OR group than in the OP group, with simultaneous alterations in CPT and HMGCR activities. In addition, plasma glutamic oxaloacetic transaminase and glutamic pyruvic transaminase levels (hepatic toxicity markers) were significantly lower in the OR group than in the OP group. Over time, OP mice developed aberrations in plasma glucose and insulin levels with enlargement of pancreatic β-cells. Generally, the state of insulin resistance, such as that observed during obesity and type 2 diabetes mellitus, is closely related to the downregulation of hepatic glycolytic enzymes and decreased glucose utilization.20 In the present study, the activity of hepatic glucokinase in OR mice significantly increased, whereas the hepatic glycogen level decreased compared with those in OP mice. This finding implies enhanced hepatic glucose utilization in OR mice. Generally, previous evidence has proved that the hepatic gluconeogenic enzymes such as PEPCK and G6Pase are upregulated in obesity, insulin resistance and type 2 diabetes.21, 22, 23, 24, 25 However, the expression/activation of PEPCK and G6Pase is conflicting in hepatic gluconeogenesis of DIO models. Do et al.26 reported that activities of PEPCK and G6Pase are consistently suppressed in the HFD-fed mice and mRNA expression of PEPCK is consistently downregulated during the development of DIO. In this study, both mRNA expressions (data not shown) and activities of PEPCK and G6Pase were significantly lower in DIO mice (both OP and OR mice) compared with ND mice, in agreement with previous results.26, 27, 28 Moreover, a recent report suggests that increased mRNA expression of PEPCK and G6Pase does not account for the hepatic insulin resistance and increased gluconeogenesis in type 2 diabetic subjects.29
Chronic low-grade inflammation of adipose tissue and skeletal muscle as well as systemic inflammation are the hallmarks of a HFD and obesity.17, 30 The excess lipid accumulation that was observed here in eWAT of OP mice was associated with the upregulation of inflammation markers including MCP-1, PAI-1, IL-6 and IFN-γ. In addition, leptin and resistin levels were higher in OP mice compared with ND-fed mice, but this change was attenuated in OR mice.
Proinflammatory cytokines have been shown to participate in insulin resistance.31 The downregulation of inflammation markers in OR mice suggests that they may be more capable of coping with the excess fat intake, by rapidly clearing metabolic byproducts and via metabolic homeostasis including glucose homeostasis, as opposed to OP mice.
To characterize a biological process or a disease state, a variety of omics platforms have been utilized in experimental settings.32 In the present study, we performed mRNA-Seq on adipose and hepatic tissues to elucidate the molecular mechanisms behind the divergent phenotypic responses to an HFD as well as the pathological consequences. The mRNA-Seq data revealed several adaptive mechanisms that may explain why OR mice are protected from the pathological changes that occur in OP mice in response to the HFD. The transcriptomic modulation in OP mice is most likely the main driver of the pathological changes and metabolic complications associated with the HFD. Many groups of genes that are associated with enlargement of eWAT and immune-cell infiltration (such as genes controlling immune responses, cell activation and the inflammatory response) were highly upregulated in the eWAT of OP mice, but these changes were attenuated (or there was no change) in the eWAT of OR mice. On the other hand, many groups of genes that are associated with spermatogenesis, male gamete generation, sperm motility and glycolysis were downregulated only in the eWAT of OP mice in response to the HFD. Inflammatory abnormalities are known to occur prior to the development of metabolic disorders in various mouse strains with varying susceptibility to diet-induced obesity and diabetes.33 According to IPA, the OP phenotype is linked to upregulation of diverse signaling pathways associated with immune responses, including the pathways related to leukocyte extravasation, phagocytosis in macrophages and monocytes, communication between cells of innate and adaptive immunity, and many types of inflammatory signaling. In contrast, most of leukocyte extravasation-related genes were downregulated in OR mice. In addition, in OR mice, the upregulation of genes related to atherosclerosis signaling was attenuated, whereas expression of the genes linked to cAMP-mediated signaling was enhanced. Diet-induced obesity in rodents leads to a decrease in sperm motility and in the percentage of sperm with normal morphology.34, 35 In the present study, many groups of genes that are associated with sperm motility and spermatogenesis were strongly upregulated in the eWAT of OR mice compared with OP mice.
OR mice also showed significant alterations in the expression of genes involved in the AMP-activated protein kinase signaling pathway (in comparison with OP mice), which is known to contribute to cellular energy metabolism and whole-body energy.36 This result is supported by the increased energy expenditure in OR mice.
In the molecular pathways in the liver, according to IPA, the OP phenotype is associated with upregulation of mitochondrial dysfunction, hepatic fibrosis, cholesterol biosynthesis and OXPHOS (compared with ND-fed mice). In the liver of OP mice, the upregulation of OXPHOS transcripts including electron transport genes points to concerted enhancement of carbohydrate metabolism relative to the ND-fed mice, but these changes were attenuated in the OR group. OXPHOS is the metabolic pathway important for generation of energy, but also leads to ROS production, which can cause oxidative damage. Alterations in OXPHOS gene expression in various tissues have been linked to positive as well as negative effects in other studies. OXPHOS-related genes are upregulated in the liver of patients with type 2 diabetes.37 On the other hand, the expression and activity of components of OXPHOS are downregulated in human WAT, in direct correlation with the degree of obesity38 and expression of OXPHOS-related genes is markedly downregulated in the WAT of db/db mice and mice with diet-induced obesity.39 Furthermore, in the present study, the upregulation of pathways associated with hepatic fibrosis, agranulocyte adhesion and diapedesis, atherosclerosis signaling and hepatic OXPHOS was attenuated in the OR mice. Taken together, our findings suggest that the susceptibility or resistance to diet-induced obesity can be determined by distinct transcriptional programs because transcriptomic profiling revealed phenotypic differences between OR and OP mice in response to the HFD.
There are limitations to this study. First, C57BL/6J mice were chosen as a model of diet-induced obesity, because on a HFD they become obese and develop insulin resistance, and thus share many similarities with the humans who have these disorders. On the other hand, the drawback of using a single inbred strain is low-genetic heterogeneity. Nevertheless, we were able to identify significant differences in the transcriptome and phenotype between our OP and OR mice. Second, we performed mRNA-Seq on eWAT and hepatic tissue after 12 weeks when the OP phenotype and OR phenotype are well established. Therefore, the transcriptomic differences that were observed here may be partly owing to either the underlying phenotype or the inherent consequences of the HFD. OR mice appear to show both positive and negative adaptations to the HFD; therefore, sometime in their lifespan, they may go on to develop some of the pathological and metabolic complications of obesity, albeit at a lower frequency than the OP mice.
In conclusion, our findings reveal major divergence in the transcriptomes, metabolic processes and phenotypes between obesity susceptibility and obesity resistance in HFD-fed C57BL/6J mice.
References
Eckel RH, Grundy SM, Zimmet PZ . The metabolic syndrome. Lancet 2005; 365: 1415–1428.
Hill JO, Melanson EL, Wyatt HT . Dietary fat intake and regulation of energy balance: implications for obesity. J Nutr 2000; 130: 284S–288S.
Li H, Xie Z, Lin J, Song H, Wang Q, Wang K et al. Transcriptomic and metabonomic profiling of obesity-prone and obesity-resistant rats under high fat diet. J Proteome Res 2008; 7: 4775–4783.
Joo JI, Oh TS, Kim DH, Choi DK, Wang X, Choi JW et al. Differential expression of adipose tissue proteins between obesity-susceptible and-resistant rats fed a high-fat diet. Proteomics 2011; 11: 1429–1448.
Choi JW, Liu H, Choi DK, Oh TS, Mukherjee R, Yun JW . Profiling of gender-specific rat plasma proteins associated with susceptibility or resistance to diet-induced obesity. J Proteomics 2012; 75: 1386–1400.
Kim DH, Choi JW, Joo JI, Wang X, Choi DK, Oh TS et al. Changes in expression of skeletal muscle proteins between obesity-prone and obesity-resistant rats induced by a high-fat diet. J Proteome Res 2011; 10: 1281–1292.
Wang X, Choi JW, Joo JI, Kim DH, Oh TS, Choi DK et al. Differential expression of liver proteins between obesity-prone and obesity-resistant rats in response to a high-fat diet. Br J Nutr 2011; 106: 612–626.
López IP, Marti A, Milagro FI, Zulet Md Mde L, Moreno-Aliaga MJ, Martinez JA et al. DNA microarray analysis of genes differentially expressed in diet-induced (cafeteria) obese rats. Obes Res 2003; 11: 188–194.
Joo JI, Yun JW . Gene expression profiling of adipose tissues in obesity susceptible and resistant rats under a high fat diet. Cell Physiol Biochem 2011; 27: 327–340.
Malone JH, Oliver B . Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol 2011; 9: 34.
Guarnaccia M, Gentile G, Alessi E, Schneider C, Petralia S, Cavallaro S . Is this the real time for genomics? Genomics 2014; 103: 177–182.
Yang Y, Smith DL Jr, Keating KD, Allison DB, Nagy TR . Variations in body weight, food intake and body composition after long-term high-fat diet feeding in C57BL/6J mice. Obesity 2014; 22: 2147–2155.
Zhang LN, Morgan DG, Clapham JC, Speakman JR . Factors predicting nongenetic variability in body weight gain induced by a high-fat diet in inbred C57BL/6J mice. Obesity 2012; 20: 1179–1188.
Johnston SL, Souter DM, Tolkamp BJ, Gordon IJ, Illius AW, Kyriazakis I et al. Intake compensates for resting metabolic rate variation in female C57BL/6J mice fed high-fat diets. Obesity 2007; 15: 600–606.
Boon MR, Bakker LE, Haks MC, Quinten E, Schaart G, Van Beek L et al. Short-term high-fat diet increases macrophage markers in skeletal muscle accompanied by impaired insulin signalling in healthy male subjects. Clin Sci 2015; 128: 143–151.
Brøns C, Jensen CB, Storgaard H, Hiscock NJ, White A, Appel JS et al. Impact of short-term high-fat feeding on glucose and insulin metabolism in young healthy men. J Physiol 2009; 587: 2387–2397.
Johannsen DL, Tchoukalova Y, Tam CS, Covington JD, Xie W, Schwarz JM et al. Effect of 8 weeks of overfeeding on ectopic fat deposition and insulin sensitivity: testing the ‘adipose tissue expandability’ hypothesis. Diabetes Care 2014; 37: 2789–2797.
Alligier M, Gabert L, Meugnier E, Lambert-Porcheron S, Chanseaume E, Pilleul F et al. Visceral fat accumulation during lipid overfeeding is related to subcutaneous adipose tissue characteristics in healthy men. J Clin Endocrinol Metab 2013; 98: 802–810.
Poussin C, Ibberson M, Hall D, Ding J, Soto J, Abel ED et al. Oxidative phosphorylation flexibility in the liver of mice resistant to high-fat diet-induced hepatic steatosis. Diabetes 2011; 60: 2216–2224.
Jiang M, Zhang Y, Liu M, Lan MS, Fei J, Fan W et al. Hypermethylation of hepatic glucokinase and L-type pyruvate kinase promoters in high-fat diet-induced obese rats. Endocrinology 2011; 152: 1284–1289.
O'Brien RM, Granner DK . PEPCK gene as model of inhibitory effects of insulin on gene transcription. Diabetes Care 1990; 13: 327–339.
Magnusson I, Rothman DL, Katz LD, Shulman RG, Shulman GI . Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest 1992; 90: 1323–1327.
Sutherland C, O'Brien RM, Granner DK . New connections in the regulation of PEPCK gene expression by insulin. Phil Trans R Soc Lond B 1996; 351: 191–199.
Clore JN, Stillman J, Sugerman H . Glucose-6-phosphatase flux in vitro is increased in type 2 diabetes. Diabetes 2000; 49: 969–974.
Takamura T, Misu H, Matsuzawa-Nagata N, Sakurai M, Ota T, Shimizu A et al. Obesity upregulates genes involved in oxidative phosphorylation in livers of diabetic patients. Obesity 2008; 16: 2601–2609.
Do GM, Oh HY, Kwon EY, Cho YY, Shin SK, Park HJ et al. Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol Nutr Food Res 2011; 55: S173–S185.
Pang J, Rhodes DH, Pini M, Akasheh RT, Castellanos KJ, Cabay RJ et al. Increased adiposity, dysregulated glucose metabolism and systemic inflammation in Galectin-3 KO mice. PLoS One 2013; 8: e57915.
Rossmeisl M, Rim JS, Koza RA, Kozak LP . Variation in type 2 diabetes-related traits in mouse strains susceptible to diet-induced obesity. Diabetes 2003; 52: 1958–1966.
Samuel VT, Beddow SA, Iwasaki T, Zhang XM, Chu X, Still CD et al. Fasting hyperglycemia is not associated with increased expression of PEPCK or G6Pc in patients with Type 2 diabetes. Proc Natl Acad Sci USA 2009; 106: 12121–12126.
Kranendonk ME, van Herwaarden JA, Stupkova T, Wd Jager, Vink A, Moll FL et al. Inflammatory characteristics of distinct abdominal adipose tissue depots relate differently to metabolic risk factors for cardiovascular disease. Atherosclerosis 2015; 239: 419–427.
Maury E, Brichard SM . Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol 2010; 314: 1–16.
Hyduke DR, Lewis NE, Palsson BØ . Analysis of omics data with genome-scale models of metabolism. Mol Biosyst 2013; 9: 167–174.
Mori MA, Liu M, Bezy O, Almind K, Shapiro H, Kasif S et al. A systems biology approach identifies inflammatory abnormalities between mouse strains prior to development of metabolic disease. Diabetes 2010; 59: 2960–2971.
Fernandez CD, Bellentani FF, Fernandes GS, Perobelli JE, Favareto AP, Nascimento AF et al. Diet-induced obesity in rats leads to a decrease in sperm motility. Reprod Biol Endocrinol 2011; 9: 32.
Palmer NO1, Bakos HW, Fullston T, Lane M . Impact of obesity on male fertility, sperm function and molecular composition. Spermatogenesis 2012; 2: 253–263.
Long YC, Zierath JR . AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest 2006; 116: 1776–1783.
Misu H, Takamura T, Matsuzawa N, Shimizu A, Ota T, Sakurai M et al. Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia 2007; 50: 268–277.
Dahlman I, Forsgren M, Sjögren A, Nordström EA, Kaaman M, Näslund E et al. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesityand possibly involving tumor necrosis factor-α. Diabetes 2006; 55: 1792–1799.
Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M et al. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 2007; 56: 1751–1760.
Acknowledgements
This work was supported by the Bio-Synergy Research Project (NRF-2012M3A9C4048818) and the Science Research Center Project (NRF-2015R1A5A6001906) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
J-YC performed the experiments, analyzed the data and wrote/edited the manuscript. RAM analyzed the data and wrote the manuscript. E-YK, YJK and YH performed the experiments and reviewed the manuscript. JHYP, KWL, S-JK, JK and JWY designed the study and reviewed the manuscript. M-SC supervised this work and had full access to all the data and therefore takes full responsibility for the integrity of the results and accuracy of the data analysis.
Supplementary Information accompanies this paper on International Journal of Obesity website
Supplementary information
Rights and permissions
About this article
Cite this article
Choi, JY., McGregor, R., Kwon, EY. et al. The metabolic response to a high-fat diet reveals obesity-prone and -resistant phenotypes in mice with distinct mRNA-seq transcriptome profiles. Int J Obes 40, 1452–1460 (2016). https://doi.org/10.1038/ijo.2016.70
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2016.70
- Springer Nature Limited
This article is cited by
-
A combination of rebaudioside A and neohesperidin dihydrochalcone suppressed weight gain by regulating visceral fat and hepatic lipid metabolism in ob/ob mice
Food Science and Biotechnology (2024)
-
Obesity, not a high fat, high sucrose diet alone, induced glucose intolerance and cardiac dysfunction during pregnancy and postpartum
Scientific Reports (2021)
-
Combination of a high-fat diet with sweetened condensed milk exacerbates inflammation and insulin resistance induced by each separately in mice
Scientific Reports (2017)