Abstract
Anti-tumor cellular immunotherapies that implement a suicide gene system can limit potential undesirable effects. In a haplo-identical bone marrow transplant clinical trial, over 90% of iCaspase-9-expressing cells were eradicated after AP1903 exposure, and signs of graft-versus-host disease disappeared. Nevertheless, low numbers of genetically modified T cells survived this treatment. We studied genetically modified cell lines (GMCL) that carried a dual iCaspase-9/ΔCD19 DNA construct (ΔCD19=truncated CD19). With AP1903 exposure, a low percentage of cells (1.47±0.67%; n=5 replications) persisted in vitro. Repeated exposures to increasing AP1903 doses generated low (GMCLLR) and high AP1903-responders (GMCLHR), which expressed different levels of surface ΔCD19 and intracellular iCaspase-9. Compared with GMCLHR, GMCLLR exhibited higher methylation of 5′-long-terminal repeat (LTR) promoters, both in the number of sequences with at least one methylated CpG (16 vs 51.5%, respectively) and in the number of CpG islands (1.2 vs 8.9%, respectively). Four days of 5-azacytidine exposure reduced methylation and increased ΔCD19 and iCaspase-9 expression. Interestingly, LTR demethylation restored GMCLLR sensitivity to AP1903 by 24.3-fold (1.8 vs 43.8%) without affecting GMCLHR. We showed that 5′-LTR-methylation inhibited transgene expression and caused AP1903 hypo-responsiveness. Treating with a hypomethylating agent restored AP1903 sensitivity. This approach can be applied in further clinical trials to improve iCaspase-9 response if low response is detected.
Similar content being viewed by others
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) remains one of the most efficient therapeutic tools for treating some hematological disorders.1 Graft-versus-host disease (GvHD) remains a major source of morbidity and mortality after HSCT. GvHD results when mature immunocompetent donor T cells present in the graft recognize alloantigens expressed by the recipient. Depleting the T cells in a graft efficiently prevents GvHD, but it impairs desirable activities, such as graft-versus-leukemia (GvL), anti-viral immune response and graft rejection control. Alternatively, a conditional in vivo T-cell depletion, performed at the time of GvHD, can bypass these complications. This approach efficiently preserves beneficial (that is, GvL) and suppresses deleterious (that is, GvHD) graft effects.2, 3
Many approaches for modulating alloreactivity have been evaluated in several clinical trials in the context of allo-HSCT. One approach is to transfer a ‘suicide gene’ to cells, which sensitizes cells to a prodrug. The prodrug is not toxic to unmodified cells. Currently, several suicide gene systems are available.4, 5 Two of the most promising suicide gene systems, which have been validated in both pre-clinical models and clinical trials, are the Herpes Simplex virus-thymidine kinase/ganciclovir (HS-TK/GCV) system6, 7, 8, 9 and the chimeric, inducible caspase-9/AP1903 (iCasp9/AP1903) system.10, 11, 12
We and others have identified obstacles that limit in vitro production of genetically modified T cells (GMTCs).13, 14, 15, 16, 17, 18, 19 Some limitations have also been pointed out in the HS-TK/GCV/Neo system. For example, it takes 5–7 days to eliminate GMTCs; some non-human transgenes are immunogenic;20, 21 GCV may interfere with expression, in cases of CMV reactivation; the G418 selection displays toxicity; and the response may be incomplete, because only dividing GMTCs are affected or because the HS-TK gene may undergo alternative splicing.22, 23 These drawbacks may be circumvented with the iCasp9/ΔCD19/AP1903 system.24, 25, 26 This system is based on human-derived transgenes, which reduce immunogenicity; it is designed to eliminate GMTCs rapidly (within a few hours), including non-dividing cells; and it utilizes a non-toxic, synthetic chemical inductor of dimerization (CID), known as AP1903. This prodrug does not preclude the use of ganciclovir as a CMV prophylactic or treatment. The iCasp9-mediated suicide system has been widely applied in cell therapy approaches, including HSCT,12 induced pluripotent stem cell safeguarding,27 chimeric antigen receptor T-cell redirection28, 29 or mesenchymal stromal cell transfers.30 However, the most convincing demonstration of the power of this suicide tool was its use in T-cell allodepletion in a clinical trial, in the context of haplo-identical transplantation.10 In that study, T cells modified with iCasp9 induced GvHD, but within 30 min after AP1903 administration the major portion of cytotoxic donor cells (>90%) were eliminated, and concomitantly, the GvHD was eradicated. However, a small proportion of donor CD19+/CD3+ GMTCs remained in the peripheral blood of patients after treatment with AP1903, and these cells remained detectable for 9 months after infusion. When these GMTCs were recovered from blood samples with fluorescence-activated cell sorting, it was shown that, in vitro, these residual cells were sensitive to a second dose of AP1903. Moreover, in vitro reactivation of GMTCs with OKT3 and an anti-CD28 antibody, 22 days post-transduction, increased transgene expression, which led to restored sensitivity, and the killing efficiency improved.12 Nevertheless, this method would be risky in patients since infusion of an anti-CD28 mAb (TGN1412) into six healthy volunteers in a phase I clinical trial caused a cytokine release storm, followed by severe adverse effects, which limited the use of this approach.31
Another way to increase transgene expression is to inhibit DNA methylation.32 5-azacytidine (5-aza) is a fully compatible candidate, because it is known to trap DNA methyl transferase 1,33 and it is well-tolerated after HSCT.34 Moreover, it can preserve the GvL effect by upregulating the expression of tumor antigens in malignant cells, and it mitigates GvHD by expanding the number of regulatory T cells.33, 35
The aim of this work was to characterize cells that were genetically modified to express iCasp9, but exhibited low-AP1903 responses. We investigated the mechanism underlying this incomplete apoptosis sensitivity and we tested 5-aza solutions to improve the responses to AP1903 treatment. Our results might provide a basis for future clinic trials to test 5-aza associated with AP1903 as an improved suicide gene therapy for controlling allogeneic HSCT.
Results
AP1903 exposure led to incomplete eradication of iCasp9/ΔCD19+ GMTC
We examined specimens from four healthy donors that participated in a pre-clinical study, performed in the context of a future clinical trial (Side by CIDe). We prepared peripheral blood mononuclear cell (PBMC) samples to study the response to the AP1903 compound. PBMCs were transduced with a retroviral construct that conferred iCasp9/ΔCD19 expression. The mean transduction efficiency, analyzed with CD3/CD19 staining and flow cytometry, was 36.3±4.8% (n=4). Four days after transduction, we submitted the transduced cell product to CD19 immunomagnetic cell sorting. The percentage of CD3+/CD19+ cells in the final sorted product was 94.9 ±1.9% (n=4, data not shown).
To assess killing efficiency, we used Trucount tubes, which provided clear discrimination between dead cells and cell debris; this assay provided an accurate estimation of the % Cell death (Figure 1a). One day after AP1903 exposure, the mean killing efficiency observed for the four pre-clinical samples was 95.8±3.1% for the final product (immunomagnetic cell-sorted fraction) and 97.0±2.5%, for the isolated fraction of CD3+/CD19+ GMTCs (immunomagnetic cell-sorted fraction gated on CD3+/CD19+ cells) (Figure 1b). Thus, a mean of 3±2.5% of CD3+/CD19+ GMTCs remained alive after AP1903 exposure.
Exposure to AP1903 led to incomplete eradication of iCaspase-9/ΔCD19+ genetically modified T cells. (a) Dot plots show characteristics of GMTCs before (top row) and after (bottom row) treatment with AP1903. FSC/SSC and CD3/CD19 were used to characterize cells and cell phenotype. Annexin V/7-AAD staining was used to study cell death induced by AP1903 treatment. Alive, viable cell fraction; All ly, all lymphocytes. (b) Histogram shows percentage of cell death in four patient samples from a pre-clinical trial. Killing efficiency was normalized to that observed in control cells (untreated cells); light gray bars, final product; dark gray bars, isolated CD19+ GMTCs.
In vitro repeated AP1903 exposure of iCasp9/ΔCD19+ transduced T-cell lines led to low-responder cells
We generated AP1903-resistant cell lines by repeatedly exposing cells to increasing doses of AP1903, from 0.1 to 10 nm (clinical dose). As shown in Figure 2a, first, we found that CEM and Jurkat T-cell lines had different sensitivities to AP1903. With exposure to the first (and lowest) AP1903 dose, we noticed that 21% of CEM and 43% of Jurkat cells were sensitive to AP1903, and underwent apoptosis.
Genetically modified T-cell lines with low response to AP1903 were generated in vitro after repeated exposure to AP1903. Two different T-cell lines, CEM (triangles) and Jurkat (circles) were genetically modified (GMCLs) to express the suicide gene iCaspase-9 and the cell surface marker CD19. GMCLs were exposed to increasing doses of AP1903. (a) Kinetics of cell death after 24-h AP1903 exposure. Every 10 days, cells that survived the prior AP1903 exposure were again exposed to an increased dose of AP1903. The percentage of cells killed was determined with trypan blue staining. (b) Cell death evaluated in T-cell lines with annexin-V–7-AAD staining revealed two populations of cells with low (GMTLLR) and high (GMTLHR) responses to AP1903 (n=3 for each T-cell lines).
Second, we found that, interestingly, after successive exposures to AP1903, in the presence of the highest dose (10 nm), 99% of both cell lines remained alive (Figure 2b). This population of AP1903-resistent cells was used in this study. Thus, these data highlighted two cell populations with different AP1903 response levels, which we called high- and low-responder cells (GMTCHR and GMTCLR, respectively).
Low-AP1903 response was directly linked to the iCaspase-9 pathway
To determine whether the low response to AP1903 was directly linked to the iCaspase-9 pathway, we induced apoptosis by irradiating cells (70 Gy). A killing efficiency analysis demonstrated that the two cell lines were not different in their sensitivity to irradiation (Figure 3a). Moreover, western blot analyses indicated that a caspase cascade was activated by irradiation in the CEM cell lines. We detected caspase-9 and -3 cleavage fragments in GMCLHR treated with either AP1903 or irradiation. In contrast, we detected high levels of cleaved-caspase-9 and -3 in GMCLLR only after irradiation (Figure 3b). The cell surface expression levels of cell-death receptors, Fas and TRAIL (DR4 and DR5), were not different between GMCLHR and GMCLLR (Supplementary Data S1). These results indicated that the low response of GMCLLR to AP1903 exposure was directly linked to the iCasp9 pathway. The same results were obtained with Jurkat GMCLs (data not shown).
Low-AP1903 response was directly linked to the iCaspase-9 pathway. (a) Histogram shows percentage of CEM cell death at 24 h after irradiation (70 Gy) or AP1903 (10 nm) exposure. Cell death was evaluated in the indicated cell populations with annexin-V–7-AAD staining (n=3). (b) Western blot analysis of caspase pathway after irradiation (70 Gy) or AP1903 (10 nm) exposure. CEM cell lines were tested 24 h after treatment in the indicated cell populations. Actin hybridization was used as a protein-loading control (n=3).
GMCLs with a low-AP1903 response displayed the lowest ΔCD19 and iCaspase-9 expression
We first analyzed ΔCD19 expression on the cell surfaces of the GMCLHR and GMCLLR with flow cytometry. We showed that ΔCD19 expression was higher in GMCLHR compared with GMCLLR; for CEM cell lines, the ratios of fluorescence intensity (RFIs) were 147.7±30 vs 45.1±7.6 (n=2), respectively, and for Jurkat cell lines RFIs were 6.8±01 vs 2.7±0.3 (n=3), respectively (Figure 4a). Moreover, the fold changes in RFI between GMCLHR and GMCLLR were similar for CEM and Jurkat cell lines (3.3-fold and 2.5-fold, respectively).
Direct correlation between ΔCD19 and iCaspase-9 transgene expression and AP1903 killing efficiency. (a) Fluorescence-activated cell-sorting (FACS) histograms show ΔCD19 cell surface expression in T-cell lines (Jurkat and CEM) that were unmodified (WT), or genetically modified (GMCL) and showed high or low responses to AP1903 (GMCLHR and GMCLLR, respectively). Values on each histogram indicate the mean fluorescence intensity (MFI) detected from fluorescently labeled anti-ΔCD19 antibodies. (b) Western blot analysis shows caspase-9 processing in untransduced, GMCLHR and GMCLLR cells before (untreated) and after AP1903 (10 nm, 24 h) exposure. Actin was used as a protein-loading control (n=3). (c) Cell-sorting protocol for isolating three different CEM and Jurkat GMCL subpopulations, according to the MFI of ΔCD19 signals (MFI values are shown on each histogram). GMCLHigh, high ΔCD19 expression; GMCLLow, low ΔCD19 expression; GMCLMed, intermediate ΔCD19 expression. (d) The percent AP1903 killing efficiency is shown for the three different GMCL subpopulations, determined by annexin-V–7-AAD staining (n=2). (e) Correlation between ΔCD19 cell surface expression (RFI: Ratio Fluorescence Intensity of ΔCD19/CD4) and killing efficiency (%) for two different T-cell lines, Jurkat (diamonds) and CEM (circles). R2 is the correlation coefficient.
The pSFG.iCasp9-ΔCD19 retroviral vector was based on the viral 2A system, which provided stoichiometric expression of the iCaspase-9 and ΔCD19 proteins. The 2A-like sequence used in the construct is derived from Thosea Asigna insect virus. It is one of the most efficiency 2A-like system allowing >99% cleavage between a glycine and terminal proline residue.12, 36 Based on our previous results, we hypothesized that GMCLLR would express low levels of iCaspase-9. This hypothesis was confirmed in western blots, which showed that the pro-iCaspase-9 signal (47 kDa) was always higher in the GMCLHR than in the GMCLLR. The 82 kDa polyprotein iCasp9-2A-ΔCD19 has never been detected by western blot. Interestingly, treating with AP1903 led to full proteolytic processing of pro-iCaspase-9 to iCaspase-9 -cleaved forms (30–36 kDa) in the GMCLHR, but not in the GMCLLR (Figure 4b). Due to the multicistronic system (viral 2A peptide), ΔCD19 expression levels corresponded to iCaspase-9 expression levels; therefore, ΔCD19 expression reflected the sensitivity to AP1903-induced apoptosis.
To confirm the direct links between ΔCD19 cell surface expression, intracellular iCaspase-9 production and sensitivity to AP1903, we performed cell-sorting to identify three different ΔCD19-positive GMCL fractions, according to CD19 fluorescence intensity (Figure 4c). These CEM GMCL fractions showed increasing killing efficiencies of 54.8%, 61.6.0% and 85.3%, respectively, that corresponded to low (RFI=2.3±0.28), medium (RFI=4.3±0.28) and high (RFI=6.3±1.98) ΔCD19 expression (Figure 4d). Plotting the ΔCD19 RFIs against the corresponding killing efficiencies confirmed a direct correlation (R=0.9221) between killing efficiency and transgene expression levels (Figure 4e).
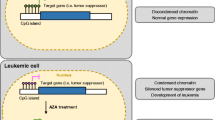
Retroviral LTR promoter methylation status was associated with the response to AP1903
In gene therapy, it is well-known that viral LTR promoter methylation leads to transcriptional repression, and thus, transgene silencing. This type of transcriptional repression can be alleviated by treating with a hypomethylating agent. We reasoned that treating our cells with a hypomethylating agent might restore transgene expression and sensitivity to AP1903-induced apoptosis. We determined that 5-aza may be a good candidate for hypomethylation, because, in allogeneic HSCT, it is used to mitigate GvHD without compromising GvL. To treat genetically modified cell lines (GMCLs) with 5-aza, we first defined the best treatment sequence to optimize the improvement in transgene expression. Thus, cell lines were incubated with 5-aza for 1–5 days, and on the last day AP1903 (10 nm) was added (Figure 5a). Based on the levels of transgene expression, killing efficiency and 5-aza toxicity, we determined that 4 days of 5-aza exposure provided the greatest improvement in GMCLLR suicidal activity, with<20% toxicity (Figure 5b).
Methylation status of the retroviral LTR promoter was associated with the level of response to AP1903. (a) Schematic timeline of 5-azacytadine (5-aza) treatments. 5-aza (1 μm) was applied for 1–5 days (D-1 to D-5) and AP1903 (10 nm) was applied on the last day (D-1) before analysis (D0). (b) The % of cell death in the presence of 5-aza alone (black bars) or 5-aza combined with AP1903 (5-aza+AP1903, gray bars) in genetically modified CEM cells that showed high or low responses to AP1903 (HR and LR, respectively). (c) Schematic representation of the PCR target region (primers indicated with arrows) and the CpG positions (vertical lines). (d) Graphic representation of methylation patterns in genetically modified CEM cell lines (CEM GMCL) that showed high or low responses to AP1903 (GMCLHR and GMCLLR, respectively). Each row corresponds to one DNA sequence, and each column represents one CpG site, numbered from 1 to 16, starting from the 5′ end of the LTR PCR product: box colors represent residues that were methylated (black), unmethylated (gray) or unknown (white). (e) Methylation levels (%) were calculated over all individually analyzed sequences (left axis, white bars) or based on the number of CpG sites (right axis, gray bars). The percent cell death in each population, in the presence or absence of 5-aza is also indicated (black bars, left axis). Numbers of individual DNA sequences analyzed: CEM GMCLHR (n=25), CEM GMCLHR5-aza (n=35), CEM GMCLLR (n=33), CEM GMCLLR5-aza (n=28).
We analyzed the methylation status in the majority (16 out 20) of CpG islands located in the 5′-LTR promoters in DNA isolated from CEM GMCLHR or -GMCLLR, after treating with or without 5-aza (Figure 5c). Figure 5d shows the results of sequencing individual cloned PCR products obtained from bisulfite DNA. The squares indicate the 16 CpG islands, and black squares correspond to methylated sequences. It was clear that the methylation status of GMCLLR was higher than that of GMCLHR, whether we considered the number of sequences that carried at least one methylated CpG island (16 vs 51.5%) or the number of methylated CpG islands among all CpG sites (1.2 vs 8.9%) (Figures 5d and e). In GMCLLR, 5-aza treatment reduced the percent methylation from 51.5 to 25% (2.06-fold reduction) of the number of sequences, and from 8.9 to 3.57% (2.49-fold reduction) of CpG sites (Figures 5d and e). Functionally, the reduction in methylation of the DNA in GMCLLR after treatment with 5-aza was correlated to the increase in cell death (from 1.8 to 43.8%=24.3-fold increase; Figures 5d and e). Interestingly, in GMCLHR, we did not identify any significant effect of 5-aza treatment, on either the methylation level (methylation without vs with 5-aza: 16 vs 14.2% of sequences and 1.2 vs 1.07% of CpG islands) or cell death (death without vs with 5-aza: 88.1 vs 92.8%), consistent with an absence of 5-aza toxicity.
Based on these data, we concluded that the methylation status was directly correlated with the sensitivity to AP1903-induced cell suicide.
5-aza exposure increased GMCLLR transgene expression and AP1903 sensitivity
We then subjected GMCLHR and GMCLLR to 5-aza treatment for 4 days, followed by AP1903 exposure on the last day. Flow cytometry analysis of ΔCD19 transgene expression did not show any effect on GMCLHR, but we noted a 4.5-fold increase (n=5) on GMCLLR (Figures 6a and b). A western blot analysis showed that 5-aza increased both the pro- and cleaved forms of iCaspase-9 in GMCLHR and GMCLLR. Addition of AP1903 led to an increase in pro-iCaspase-9 cleavage in GMCLLR (Figure 6c). Finally, we demonstrated that 5-aza added to AP1903 treatment induced better killing efficiency (22.1-fold compared with AP1903 alone) in GMCLLR (Figure 6d). To conclude, we showed that pretreatment with 5-aza reduced DNA methylation, which enhanced the expression of the suicide transgene and increased AP1903-induced apoptosis.
5-azacytidine exposure increased transgene expression and improved AP1903 GMCL sensitivity. (a) Fluorescence-activated cell-sorting (FACS) histograms show cell surface ΔCD19 expression in T-cell lines (Jurkat and CEM) that were genetically modified (GMCL) and showed high or low responses to AP1903 (GMCLHR and GMCLLR, respectively). Cells were pretreated for 4 days in the absence (ϕ) or presence of 5-azacytadine (1 μm 5-aza). Values on each histogram indicate the mean fluorescence intensity (MFI) detected from fluorescently labeled anti-ΔCD19 antibodies. (b) Histogram represents the ratio of fluorescence intensity of ΔCD19/CD4 (ΔCD19 RFI) for each cell line (GMCLHR and GMCLLR), after no treatment (untreated) or 4 days of 5-aza pretreatment (n=2 for CEM cell lines and n=3 for Jurkat cell lines). (c) Western blot analysis shows caspase-9 processing and actin expression (as a loading control) in Jurkat and CEM GMCLHR and GMCLLR cells. Cells were untreated (−) or treated (+) with 1 μm 5-aza (4 days), 10 nm AP1903 (1day) or both. (d) Killing efficiency in the absence (untreated) or presence of 5-aza alone or 5-aza+AP1903 treatment in CEM and Jurkat wild-type (WT), GMCLHR and GMCLLR cells (n=2 for CEM cell lines and n=3 for Jurkat cell lines).
Discussion
It was previously demonstrated that, in HSCT applications, GvHD, immune reconstitution, and viral and bacterial infections could be modulated early after HSCT with the administration of allogeneic T cells engineered to express a gene that conferred susceptibility to apoptosis (suicide gene). Two suicide gene systems, the HS-TK/GCV system37 and, more recently, the iCaspase-9/AP1903 system, have been validated in clinical trials.11 Previous studies have shown that the iCaspase-9/AP1903 system was safe and powerful. In four young patients that had developed GvHD, a single dose of dimerizing drug (AP1903) eliminated over 90% of the GMTC within 30 min of administration, and the GvHD had resolved.10 However, although the GvHD disappeared, a small number of non-clinically alloreactive GMTCs remained that had not responded to AP1903. In suicide gene systems, control over adverse events requires, as much as possible, the depletion of GMTCs. In our clinical approach (Side by CIDe), only allo-GMTCs will be infused into patients; therefore, unlike the clinical trial of Di Stasi et al.,10 the donor alloreactive CD25+ T cells cannot be depleted with the anti-CD25 immunotoxin, RFT5-SMPT-dgA. Therefore, this study focused on mechanisms of AP1903 resistance.
First, we confirmed that the small percentage of GMTCs that showed a partial response to AP1903 comprised cells with low-ΔCD19 expression. Thus, due to the viral 2A-like cleavage peptide system, these cells also showed low-iCaspase-9 expression. We assessed transgene expression with either flow cytometry to detect ΔCD19 or with western blotting to detect iCaspase-9 cleavage. In both cases, transgene expression was directly correlated to killing efficiency. Although iCaspase-9 and ΔCD19 proteins are separately expressed after 2A peptide self-cleavage, both proteins are translated from the same mRNA. Transcription of iCaspase-9/ΔCD19 mRNA is driven by a single LTR promoter. In gene therapy, it was shown that reduction of transcriptional activity in host cells can be due to silencing retroviral elements involving different defense mechanisms. This can be mediated by endogenous factors that bind to the LTR promoter or by methylation of CpG islands to inhibit transgene expression.38 Our results showed that both iCaspase-9 and ΔCD19 were simultaneously downregulated in GMCLs, which indicated that the LTR was a good target for studying methylation status. Thus, we hypothesized, and confirmed, that the weak transgene expression in GMCLLR was associated with a higher level of methylation in the LTR promoter than that found in GMCLHR. This finding was consistent with that of Straathof et al.,11 who also demonstrated, with an iCaspase-9-2A-GFP retroviral vector, that low-GFP expression in GMTCs was associated with low-AP1903 responses. In the present work, we provide one explanation for reductions in transgene expression. However, low-response phenomenon is a low-frequency event.
For clinical purposes, cells that express a non-immunogenic membrane-bound marker (ΔCD20, ΔCD34, ΔCD19 or ΔNGFR) can be selected with immunomagnetic cell sorting to identify cells with high-transgene expression. However, some GMTCs with low-transgene expression may be selected with immunomagnetic cell sorting, and these may cause adverse events, because they can escape AP1903 toxicity. Thus, an additional treatment may be useful for restoring AP1903 sensitivity to GMTCs that express iCaspase-9. It was shown that a second in vitro activation (OKT3 / CD28) of iCaspase-9 in GMTCs12 and a second in vitro exposure of GMTCs to the dimerizer, after circulating in vivo,10 could eliminate residual GMTCs. The former treatment is not relevant for clinical purposes, because it will have an important impact on unmanipulated T cells derived from the bone marrow graft. Moreover, the use of in vivo anti-CD28 mAbs can lead to serious adverse events (cytokine storm), as previously described.31
The hypomethylating agent, 5-aza, provides several beneficial effects. It is compatible with HSCT settings, because it is well-tolerated after transplantation.34 Also, it upregulates HLA-DR expression, and it upregulates the epigenetically silenced tumor antigens; consequently, it induces a CD8+ T-cell response against tumor antigens, and thus, contributes to the GvL effect.33, 39 5-aza also inhibits DNA methyl transferase activity, which increases iCaspase-9 transgene expression and increases AP1903 killing efficiency. Finally, it increases regulatory T-cell expansion, and consequently limits GvHD.33, 35
The present work confirmed that, among the GMTCs produced with our clinical-grade supernatant, a small portion that expressed the iCaspase-9 suicide gene displayed a low response to the AP1903 dimerizer. This study was the first to show that the low sensitivity to AP1903 was linked to a high level of methylation in the LTR promoter. This epigenetic phenomenon could be reversed by treating with the 5-aza hypomethylating agent. As reported by others, 5-aza treatment has already been tested to induce demethylation of gene in the treatment of tumor-bearing mice.40 Plumb et al.,40 demonstrated that 5-aza could increase gene expression without toxicity (weight loss or other sign). Moreover, 5-aza could also be used to enhance other molecules such lintuzumab in pre-clinical models of acute myeloid leukemia.41 However, this strategy remains to be tested in future clinical trials, and the best sequence of treatment remains to be defined. Moreover, in the present study, we did not identify a mechanism that conferred a specific resistance to AP1903; thus, the suicide system iCasp9/AP1903 represents an optimal system for safe, effective gene therapy. The monitoring of circulating GMTCs of our phase I-IIB clinical trial ‘Side by CIDe’, a biomedical research based on GMTCs expressing iCaspase-9 and ΔCD19 in context of hematopoietic stem cell allo-graft, will give us complementary information. It will allow us to better characterize the low-responder cells after in vivo injection of AP1903.
In conclusion, this study addressed an interesting issue in HSCT, and our results provided a means to improve and ensure GvHD control in cases of persistent, alloreactive GMTCs. This approach should be evaluated in future clinical trials.
Material and methods
Peripheral blood cells, cell lines and retrovirus
Human T-cell lines, CEM (ATCC, Molsheim, France; CCL-119) and Jurkat (DSMZ, Braunshweig, Germany; ACC282) were stored in an internal master cell bank for non-ambiguous identification. Cell lines were cultured in RPMI1640 (Lonza, Paris, France) supplemented with 10% heat-inactivated, endotoxin-free fetal calf serum (Invitrogen, Cergy-Pontoise, France).
Peripheral blood cell samples of healthy donors were collected at the French Blood Center (Besançon, France) after obtaining written informed consent.
Retroviral supernatant was produced at the Cell & Gene Therapy Center (Houston, TX, USA) from a PG13 packaging cell line that had been transfected with the SFG.iCasp9-2A-ΔCD19.vector.10
Retroviral transduction of iCaspase-9/ΔCD19 gene to produce GMCs
GMTCs (primary cells or cell lines) that expressed iCaspase-9/ΔCD19 were generated by retroviral transduction in a six-well-retronectin (1.2 μg cm−2 in PBS overnight at 4 °C, Takara, Shiga, Japan) -coated plate. PBMCs were activated with CD3/CD28 beads for 3 days prior to transduction. Cells were then spin-transduced (1.5 h, 2000 g, 10 °C) with the retroviral particles. Transduction efficiency was determined by performing flow cytometric analysis to identify ΔCD19 cell surface marker expression. Four days after transduction, ΔCD19-positive cells were magnetically labeled with CD19 microbeads (CD19 microbeads, Miltenyi Biotec, Bergisch Gladbach, Germany) and loaded onto a MACS Column (Miltenyi Biotec), according to the manufacturer’s protocol. The isolated ΔCD19-expressing cells were expanded and cryopreserved.
All GMCs used in this study met the predefined release criteria for clinical trials (Side by CIDe). Cultures comprised >92% CD3+/CD19+ cells, and AP1903 exposure induced over 90% growth inhibition.
Induction of apoptosis with AP1903±5-aza and generation of low-responder cell lines
Suicide gene functionality in GMCs was assessed by adding to the culture medium the synthetic molecule AP1903 (Bellicum Pharmaceuticals, Houston, TX, USA) at 10 nm final concentration. During cell production, the functionality of iCaspase-9 transgene was verified by adding AP1903 on the day following CD19 immunomagnetic cell sorting. Cell death was evaluated 24 h later with flow cytometry.
To generate AP1903-low-responder cell lines (GMCLLR), viable CEM and Jurkat cells that expressed iCaspase-9+/ΔCD19+ were treated with serially increasing doses of AP1903 (from 0.1 to 10 nm). In some cases, a single dose of 5-aza (1 μm, Sigma-Aldrich, St Louis, MO, USA) was added to GMCLs from 1 to 5 days before, or concurrently with, AP1903 treatment.
Immunophenotyping and cell-death analysis
Cell-surface phenotype was investigated with the following monoclonal antibodies: anti-CD3, anti-CD4, (Becton Dickinson, Franklin Lakes, NJ, USA) and anti-CD19 (Miltenyi Biotec). Antibody binding was detected on a Canto II flow cytometer with Diva software (Becton Dickinson). Appropriately matched isotype controls were included in all immunophenotyping analyses.
Cell death was first assessed visually by counting dead cells that could take up trypan blue. For more precise cell-death analysis, cells were stained with annexin-V and 7-aminoactinomycin (7-AAD) (Beckman Coulter, Fullerton, CA, USA) after 24 h of AP1903 treatment, and analyzed with flow cytometry. Fluorescence analysis was gated on CD3+/CD19+ positive cells. Cells were considered viable when they were negative for both annexin-V and 7-AAD.
The absolute cell-death quantification was determined with Trucount tubes (Becton Dickinson) after acquiring 5000 or 10000 fluorescent beads with flow cytometry. Thus, we evaluated cell killing as follows: %Dead cells=[1−(absolute number of viable cells in AP1903-treated cells/absolute number of viable cells in untreated cells)] × 100.
Western blotting
Cells were lysed, cooled by incubating on ice, by sonication in RIPA buffer supplemented with a protease inhibitor cocktail (complete Mini EDTA-free; Roche, Bâle, Switzerland). Lysate protein content was quantified with a BCA protein assay. Then, equivalent amounts of cell extract (20 μg proteins) were fractioned with SDS–PAGE, and separated proteins were electrotransferred onto Polyfluorure de vinylidene membranes. Membranes were probed overnight with primary antibodies (diluted at 1:1000) that bound to human caspases 3, 8, 9 and 10 (#9662, #4790 S, #9502 S, #9752 S, Cell Signaling, Danvers, MA, USA). We added antibodies (diluted 1:106) that recognized β-actin (clone AC15, #A5441, Sigma-Aldrich) as an internal loading control. For immunodetection, we added the following secondary antibodies (diluted at 1:24 000): horseradish peroxidase-conjugated goat anti-rabbit IgG (#111-035-144, Jackson, West Grove, PA, USA) or sheep anti-mouse IgG (#515-035-062, Jackson). Then, we added enhanced chemiluminescence detection reagents, and chemiluminescence was detected with a camera and Bio-1D software (Vilber-Lourmat, Collegien, France).
Methylation status analysis
Conversion of unmethylated DNA cytosine residues to uracil with bisulfite was performed with the MethylEdge Bisulfite Conversion System (Promega, Madison, WI, USA), according to the manufacturer’s protocol. Converted DNA was subjected to PCR with primers that specifically recognized the 5′-LTR promoter region, designed with Methyl primer express v1.0 software (Life technologies, Courtaboeuf, France). The region targeted by the primers included most of the CpG islands (16 of 20). The primer sequences were: 5LTRbi-Fw 5′-AAGGATTTGAAATGATTTTGTG-3′; 5LTRbi-Rv 5′-CTAAACAATCRAACAAACACAA-3′. The PCR products were gel-purified and cloned into the pCR2.1 TOPO vector with the TOPO TA Cloning Kit (Life technologies). DNA was purified from selected bacterial colonies with the NucleoSpin plasmid Kit (Macherey-Nagel, Düren, Germany). For each culture condition, at least 20 individual bacterial colonies were Sanger-sequenced. Then, sequences were aligned to the unmethylated LTR sequence for analysis with the BISMA software.42 To evaluate the methylation status, we analyzed the methylation level (%) observed over all individual sequences and the number of CpG sites that were found to be informative.
References
Appelbaum FR . Haematopoietic cell transplantation as immunotherapy. Nature 2001; 411: 385–389.
Bonini C, Ferrari G, Verzeletti S, Servida P, Zappone E, Ruggieri L et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science 1997; 276: 1719–1724.
Tiberghien P, Cahn JY, Brion A, Deconinck E, Racadot E, Herve P et al. Use of donor T-lymphocytes expressing herpes-simplex thymidine kinase in allogeneic bone marrow transplantation: a phase I-II study. Hum Gene Ther 1997; 8: 615–624.
Marin V, Cribioli E, Philip B, Tettamanti S, Pizzitola I, Biondi A et al. Comparison of different suicide-gene strategies for the safety improvement of genetically manipulated T cells. Hum Gene Ther Methods 2012; 23: 376–386.
Introna M, Barbui AM, Bambacioni F, Casati C, Gaipa G, Borleri G et al. Genetic modification of human T cells with CD20: a strategy to purify and lyse transduced cells with anti-CD20 antibodies. Hum Gene Ther 2000; 11: 611–620.
Bonini C, Grez M, Traversari C, Ciceri F, Marktel S, Ferrari G et al. Safety of retroviral gene marking with a truncated NGF receptor. Nat Med 2003; 9: 367–369.
Ciceri F, Bonini C, Marktel S, Zappone E, Servida P, Bernardi M et al. Antitumor effects of HSV-TK-engineered donor lymphocytes after allogeneic stem-cell transplantation. Blood 2007; 109: 4698–4707.
Ciceri F, Bonini C, Stanghellini MT, Bondanza A, Traversari C, Salomoni M et al. Infusion of suicide-gene-engineered donor lymphocytes after family haploidentical haemopoietic stem-cell transplantation for leukaemia (the TK007 trial): a non-randomised phase I-II study. Lancet Oncol 2009; 10: 489–500.
Tiberghien P, Ferrand C, Lioure B, Milpied N, Angonin R, Deconinck E et al. Administration of herpes simplex-thymidine kinase-expressing donor T cells with a T-cell-depleted allogeneic marrow graft. Blood 2001; 97: 63–72.
Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med 2011; 365: 1673–1683.
Straathof KC, Pule MA, Yotnda P, Dotti G, Vanin EF, Brenner MK et al. An inducible caspase 9 safety switch for T-cell therapy. Blood 2005; 105: 4247–4254.
Tey SK, Dotti G, Rooney CM, Heslop HE, Brenner MK . Inducible caspase 9 suicide gene to improve the safety of allodepleted T cells after haploidentical stem cell transplantation. Biol Blood Marrow Transplant 2007; 13: 913–924.
Coito S, Sauce D, Duperrier A, Certoux JM, Bonyhadi M, Collette A et al. Retrovirus-mediated gene transfer in human primary T lymphocytes induces an activation- and transduction/selection-dependent TCR-B variable chain repertoire skewing of gene-modified cells. Stem Cells Dev 2004; 13: 71–81.
Sauce D, Tonnelier N, Duperrier A, Petracca B, de Carvalho Bittencourt M, Saadi M et al. Influence of ex vivo expansion and retrovirus-mediated gene transfer on primary T lymphocyte phenotype and functions. J Hematother Stem Cell Res 2002; 11: 929–940.
Sauce D, Bodinier M, Garin M, Petracca B, Tonnelier N, Duperrier A et al. Retrovirus-mediated gene transfer in primary T lymphocytes impairs their anti-Epstein-Barr virus potential through both culture-dependent and selection process-dependent mechanisms. Blood 2002; 99: 1165–1173.
Sauce D, Mercier P, Battini JL, Ferrand C, Certoux JM, Manel N et al. Preferential retroviral-mediated transduction of EBV- and CMV-specific T cells after polyclonal T-cell activation. Gene Ther 2004; 11: 1019–1022.
Ferrand C, Robinet E, Contassot E, Certoux JM, Lim A, Herve P et al. Retrovirus-mediated gene transfer in primary T lymphocytes: influence of the transduction/selection process and of ex vivo expansion on the T cell receptor beta chain hypervariable region repertoire. Hum Gene Ther 2000; 11: 1151–1164.
Deschamps M, Robinet E, Certoux JM, Mercier P, Sauce D, De Vos J et al. Transcriptome of retrovirally transduced CD8+ lymphocytes: influence of cell activation, transgene integration, and selection process. Mol Immunol 2008; 45: 1112–1125.
Sauce D, Rufer N, Mercier P, Bodinier M, Remy-Martin JP, Duperrier A et al. Retrovirus-mediated gene transfer in polyclonal T cells results in lower apoptosis and enhanced ex vivo cell expansion of CMV-reactive CD8 T cells as compared with EBV-reactive CD8 T cells. Blood 2003; 102: 1241–1248.
Mercier-Letondal P, Deschamps M, Sauce D, Certoux JM, Milpied N, Lioure B et al. Early immune response against retrovirally transduced herpes simplex virus thymidine kinase-expressing gene-modified T cells coinfused with a T cell-depleted marrow graft: an altered immune response? Hum Gene Ther 2008; 19: 937–950.
Traversari C, Marktel S, Magnani Z, Mangia P, Russo V, Ciceri F et al. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies. Blood 2007; 109: 4708–4715.
Garin MI, Garrett E, Tiberghien P, Apperley JF, Chalmers D, Melo JV et al. Molecular mechanism for ganciclovir resistance in human T lymphocytes transduced with retroviral vectors carrying the herpes simplex virus thymidine kinase gene. Blood 2001; 97: 122–129.
Deschamps M, Mercier-Lethondal P, Certoux JM, Henry C, Lioure B, Pagneux C et al. Deletions within the HSV-tk transgene in long-lasting circulating gene-modified T cells infused with a hematopoietic graft. Blood 2007; 110: 3842–3852.
Iuliucci JD, Oliver SD, Morley S, Ward C, Ward J, Dalgarno D et al. Intravenous safety and pharmacokinetics of a novel dimerizer drug, AP1903, in healthy volunteers. J Clin Pharmacol 2001; 41: 870–879.
Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR . Controlling signal transduction with synthetic ligands. Science 1993; 262: 1019–1024.
Fan L, Freeman KW, Khan T, Pham E, Spencer DM . Improved artificial death switches based on caspases and FADD. Hum Gene Ther 1999; 10: 2273–2285.
Zhong B, Watts KL, Gori JL, Wohlfahrt ME, Enssle J, Adair JE et al. Safeguarding nonhuman primate iPS cells with suicide genes. Mol Ther 2011; 19: 1667–1675.
Casucci MF, Nicolis di Robilant L, Dual B . Transgenesis of T cells with a novel CD44v6-specific chimeric antigen receptor and a suicide gene for safe and effective targeting of chemoresistance in hematopoietic tumors. Blood 2011; 2: 378–382.
Budde LE, Berger C, Lin Y, Wang J, Lin X, Frayo SE et al. Combining a CD20 Chimeric Antigen Receptor and an Inducible Caspase 9 Suicide Switch to Improve the Efficacy and Safety of T Cell Adoptive Immunotherapy for Lymphoma. PLoS One 2013; 8: e82742.
Ramos CA, Asgari Z, Liu E, Yvon E, Heslop HE, Rooney CM et al. An inducible caspase 9 suicide gene to improve the safety of mesenchymal stromal cell therapies. Stem Cells 2010; 28: 1107–1115.
Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med 2006; 355: 1018–1028.
Di Ianni M, Terenzi A, Perruccio K, Ciurnelli R, Lucheroni F, Benedetti R et al. 5-Azacytidine prevents transgene methylation in vivo. Gene Ther 1999; 6: 703–707.
Choi J, Ritchey J, Prior JL, Holt M, Shannon WD, Deych E et al. In vivo administration of hypomethylating agents mitigate graft-versus-host disease without sacrificing graft-versus-leukemia. Blood 2010; 116: 129–139.
de Lima M, Giralt S, Thall PF, de Padua Silva L, Jones RB, Komanduri K et al. Maintenance therapy with low-dose azacitidine after allogeneic hematopoietic stem cell transplantation for recurrent acute myelogenous leukemia or myelodysplastic syndrome: a dose and schedule finding study. Cancer 2010; 116: 5420–5431.
Goodyear OC, Dennis M, Jilani NY, Loke J, Siddique S, Ryan G et al. Azacitidine augments expansion of regulatory T cells after allogeneic stem cell transplantation in patients with acute myeloid leukemia (AML). Blood 2012; 119: 3361–3369.
Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY et al. High cleavage efficiency of a 2 A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 2011; 6: e18556.
Bonini C, Bondanza A, Perna SK, Kaneko S, Traversari C, Ciceri F et al. The suicide gene therapy challenge: how to improve a successful gene therapy approach. Mol Ther 2007; 15: 1248–1252.
Herbst F, Ball CR, Tuorto F, Nowrouzi A, Wang W, Zavidij O et al. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol Ther 2012; 20: 1014–1021.
Jabbour E, Giralt S, Kantarjian H, Garcia-Manero G, Jagasia M, Kebriaei P et al. Low-dose azacitidine after allogeneic stem cell transplantation for acute leukemia. Cancer 2009; 115: 1899–1905.
Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R . Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res 2000; 60: 6039–6044.
Sutherland MK, Yu C, Anderson M, Zeng W, van Rooijen N, Sievers EL et al. 5-azacytidine enhances the anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. MAbs 2010; 2: 440–448.
Rohde C, Zhang Y, Reinhardt R, Jeltsch A . BISMA—fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 2010; 11: 230.
Acknowledgements
We thank Pr Malcolm Brenner (Center for Cell and Gene Therapy, Baylor College of Medicine, Houston, USA) for providing research grade of packaging cell line and retroviral supernatant. We also thank Pr David Spencer (Bellicum Pharmaceuticals, Houston, USA) for providing research grade AP1903 Chemical Inducer Dimerizer and for revising this manuscript. This study was supported by the ‘Programme Hospitalier de Recherche Clinique’ (PHRC #950898), the ‘Ligue Nationale contre le Cancer’ (2013), Etablissement Français du sang (APR 2013) and Région de Franche-Comté (Grant 2013); it was co-funded, in part, by the 2013/A02 tender of the Association Capucine and the Société Française de Greffe de Moelle—Thérapie Cellulaire (SFGM-TC).
Author contributions
EB-R executed all experiments, including cell cultures, flow cytometry and western blotting, and she wrote the original draft of the manuscript. CG, II and J-MC assisted with retroviral transductions, GMTC production and AP1903 resistance. A-MM, PT and CB contributed to improving the manuscript and provided final approval. ED and FL provided relevant information of clinical interest and information on future clinical trials. CF and MD initiated and designed the study, participated in every step of the study, managed the whole project and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Gene Therapy website
Supplementary information
Rights and permissions
About this article
Cite this article
Bôle-Richard, E., Gamonet, C., Certoux, JM. et al. Exposure to hypomethylating agent, 5-azacytidine, may improve iCasp9 suicide gene therapy for treating GvHD in allografts. Gene Ther 23, 664–672 (2016). https://doi.org/10.1038/gt.2016.39
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2016.39
- Springer Nature Limited