Abstract
Interferon lambda 3 (IFNL3) and epidermal growth factor receptor (EGFR) single nucleotide polymorphisms (SNPs) may play a key role in the spontaneous clearance of hepatitis C virus (HCV) and treatment responses. The aim of this study was to evaluate the effect of IFNL3 SNPs and EGFR rs11506105 on treatment outcomes in patients with chronic HCV (CHC). IFNL3 SNPs and EGFR rs11506105 were genotyped by PCR-restriction fragment length polymorphism and PCR-sequencing, respectively, in 235 naïve patients with CHC infection. The frequency of rapid virologic response (RVR), complete early virologic response (cEVR) and sustained virologic response (SVR) were 52.3%, 76.2% and 64.7% respectively. The results of this study showed that RVR was associated with ALT (P=0.015), AST (P=0.020), IFNL3 rs12979860 (CC) (P=0.043), rs12980275 (AA) (P=1 × 10−4), and EGFR rs11506105 (AA) (P=0.010), and IFNL3 rs12979860 (CC) (P=0.048), rs12980275 (AA) (P=0.022), and EGFR rs11506105 (AA) (P=0.006) were correlated with cEVR. HCV genotype (P=0.007), IFNL3 rs12979860 (CC) (P=0.023), IFNL3 rs12980275 (AA) (P=1 × 10−4), EGFR rs11506105 (AA) (P=0.005), RVR (P=1 × 10−4), and cEVR (P=0.003) were significant predictors for SVR. These results, for the first time, revealed that beside IFNL3 SNPs, EGFR rs11506105 is strongly associated with RVR, cEVR and SVR. EGFR rs11506105 besides IFNL3 SNPs could predict treatment responses in CHC patients.
Similar content being viewed by others
Introduction
Despite the identification of new drugs against hepatitis C virus (HCV) or direct-acting antiviral agents, HCV infection is a major public health problem. The recent estimates of HCV disease worldwide have shown a 2.5% increased rate compared to 15 years ago, equivalent to >185 million infections.1 Chronic HCV (CHC) infection can cause liver failure, liver cirrhosis, hepatocellular cancer (HCC) and death, especially in human immunodeficiency virus-positive patients on highly active antiretroviral therapy.2
The prevalence of HCV infection in Iran is very low and is estimated to be around 0.5%. The cost of HCV treatment, comparable to low and middle-income countries is an important burden on health-care centers, and withholding treatment on those who achieved sustained virologic response (SVR) could be cost saving. Hepatitis C virus is classified into seven distinct genotypes (1–7), and HCV genotypes 1a and 3a are the most common types in Iran.3, 4, 5
Pegylated interferon alpha and ribavirin (pegIFN-α/RBV), sofosbuvir, ledipasvir and daclatasvir are now available in Iran.6 The standard treatment for CHC is a combination of pegIFN-α/RBV in a 24th or 48th course, which is associated with several side effects, including depression, hemolytic anemia, hepatic decompensation and thrombocytopenia. In HCV genotypes 2 and 3, the rate of SVR is 70–90% and nearly 50% for HCV genotype 1 and genotype 4. We require several studies that will assess genetic host factors in Iranian patients with CHC infection. It is possible that researchers may find a genetic variation similar to interferon lambda 3 (IFNL3) (formerly known as interleukin-28B) that predicts response to HCV therapy or spontaneous clearance. The effect of single nucleotide polymorphisms (SNPs) near IFNL3 on treatment outcomes in pegIFN-α/RBV therapy is very interesting.
IFNL3 rs12979860, rs8099917 and rs12980275 are associated with treatment outcomes among patients with HCV infection.7, 8, 9 The IFNL3 rs12979860 CC genotype, as compared to CT or TT genotypes, is powerfully associated with SVR after pegIFN-α/RBV combination therapy.7 The HCV patients with the IFNL3 rs8099917 TT genotype and IFNL3 rs12980275 AA genotype demonstrated a good chance of infection removal.10
On the cell surface, epidermal growth factor receptor (EGFR) is activated by binding its ligands, including epidermal growth factor and transforming growth factor-α, which leads to activation of the signaling pathway. One of the EGFR signaling roles is liver regeneration following acute and chronic liver, moreover in HCC and cirrhosis.1, 11 A study in Iran revealed that EGFR rs11506105 genotype is associated with spontaneous clearance in patients with HCV infections.1 So far, the relationship between EGFR rs11506105 and response to therapy has not been discussed in any study. In the current study, we were curious whether EGFR rs11506105 predicts treatment outcomes in patients with HCV infection.
The progression of direct-acting antiviral agents on HCV may reduce the importance of SNP genotyping in predicting treatment outcomes. direct-acting antiviral agent therapy for the HCV in several countries has not yet been approved; the combination of pegIFN-α/RBV is the standard treatment. Hence, IFNL3 and other SNP genotyping may also predict how HCV responds to pegIFN-α/RBV.6
The aim of the current study was to evaluate the effect of EGFR rs11506105 on the rapid virologic response (RVR), completely early virologic response (cEVR) and SVR to HCV genotypes 1a/2a/3a. In addition, we determined the impact of three different IFNL3 polymorphisms (rs12979860, rs8099917 and rs12980275) on treatment outcomes.
Results
Patient’s characteristics
A total of 235 subjects were included in this study. One hundred and twenty (51.1%) patients were infected with HCV subtype 1a, and 97 (41.3%) and 18 (7.6%) patients were infected with HCV subtype 3a and HCV subtype 2a, respectively. The mean age in all patients was 42.3±10.8 years. In all, 166 (70.6%) patients were male. Baseline demographic and virologic properties of the all patients are summarized in Table 1.
IFNL3 and EGFR frequency in patients with CHC infection
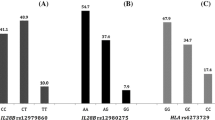
The distribution of IFNL3 rs12979860 (1A), rs8099917 (1B), rs12980275 (1C) and EGFR rs11506105 (1D) is depicted in Figure 1. The genotypes IFNL3 rs12979860 CT (45.5%), rs8099917 TT (68.1%), rs12980275 AA (54.9%) and EGFR rs11506105 AA (47.2%) were mostly revealed in patients with CHC infection. IFNL3 rs12979860, rs8099917, rs12980275 and EGFR rs11506105 in patients with CHC infection were in Hardy–Weinberg equilibrium. The LD of IFNL3 rs12979860 and rs12980275 was high (D′=0.850, r2=0.710), while between IFNL3 rs12979860 and rs8099917 was moderate (D′=0.460, r2=0.510).
IFNL3 SNPs and EGFR rs11506105 compared with HCV genotypes and HCV-RNA viral load
The association between EGFR rs11506105 and IFNL3 SNPs (rs12979860, rs12980275, rs8099917) and HCV-RNA viral load and HCV genotype in CHC patients was determined. Between HCV genotypes with IFNL3 rs8099917 (P=0.013); and EGFR rs11506105 (P=0.005) were a significant association. After stratification for HCV genotype (1a versus 2a/3a), again, there was association between IFNL3 rs8099917 (P=0.003) and EGFR rs11506105 (P=0.001) with HCV genotype (Supplementary Tables S1 and S2). In the current study, only the IFNL3 rs8099917 (P=0.048) was significantly associated with HCV-RNA viral load.
Correlation between IFNL3 and EGFR SNPs and treatment responses
Table 2 shows the relationships between treatment outcomes and demographic factors. RVR, cEVR and SVR rates were significantly higher among patients with the favorable genotypes of IFNL3 SNPs (rs12979860 CC, rs12980275 AA and rs8099917 TT), compared to patients with unfavorable genotypes of IFNL3 (rs12979860 CT/TT, rs12980275 AG/GG and rs8099917 TG/GG genotypes) (Figures 2 and 3). In EGFR rs11506105 SNPs, the frequency of RVR, cEVR and SVR was remarkably higher among patients with the AA genotype compared to AG and GG alleles (Figure 3). Also, ALT (P=0.017) and HCV genotypes (P=0.016) were significant predictors for SVR. Of the 235 patients, 79 subjects had favorable genotypes (IFNL3 rs12979860 CC, rs8099917 TT, rs12980275 AA and EGFR rs11506105 AA). Patients with co-expression of the favorable genotypes demonstrated a better response to pegIFN-α/RBV combination therapy compared to unfavorable genotypes. The rates of RVR, cEVR and SVR among 79 patients with all favorable alleles were 68 (86.1%), 63 (79.7%) and 72 (91.1%), respectively. The AUC-–receiver operator characteristic was 0.93 for RVR, 0.82 for cEVR and 0.96 for SVR, but the AUC was 0.71 by adding ALT and AST in RVR and was 0.82 by adding HCV genotypes in SVR, suggesting that genetic factors of host and viral factors were generally important to resolution of virus during treatment (Figure 4).
Factors related to RVR, cEVR and SVR responses
By multivariate logistic regression analysis, we investigated the factors related to RVR, cEVR and SVR (Table 3 and supplementary Table S3). RVR was associated with ALT (odds ratio (OR) 2.50, 95% CI 1.66–4.01, P=0.015), AST (OR 0.10, 95% CI 0.06–0.89, P=0.020), IFNL3 rs12979860 (CC) (OR 1.74, 95% CI 1.02–4.13, P=0.043), IFNL3 rs12980275 (AA) (OR 5.93, 95% CI 2.94–11.98, P=1 × 10−4), EGFR rs11506105 (AA) (OR 2.09, 95% CI 1.19–3.67, P=0.010), and IFNL3 rs12979860 (CC) (OR 1.71, 95% CI 1.08–3.32, P=0.048), IFNL3 rs12980275 (AA) (OR 0.35, 95% CI 0.14–0.86, P=0.022), and EGFR rs11506105 (AA) (OR 2.36, 95% CI 1.28–4.24, P=0.006) were correlated with cEVR. Multivariate logistic regression analysis showed that HCV genotype (OR 0.25, 95% CI 0.08–0.82, P=0.007), IFNL3 rs12979860 (CC) (OR 0.32, 95% CI 0.12–0.86, P=0.023), IFNL3 rs12980275 (AA) (OR 2.37, 95% CI 1.23–4.03, P=1 × 10−4), EGFR rs11506105 (AA) (OR 2.61, 95% CI 1.83–5.74, P=0.005), RVR (OR 1.53, 95% CI 1.16–3.14, P=1 × 10−4), and cEVR (OR 3.94, 95% CI 2.45–8.25, P=0.003) were significant predictors for SVR.
Discussion
In the current study, we evaluated the host factors (IFNL3 SNPs and EGFR rs11506105) influencing the RVR, cEVR and SVR responses in CHC patients treated with pegIFN-α/RBV. Epidermal growth factor receptor is a receptor of tyrosine kinase that controls key processes such as survival, cell proliferation, tumorigenesis, differentiation during development and tissue homeostasis.12 It was recently shown that EGFR participates in the virus entry into the cell, a complex multi-stage performance involving interactions of HCV envelope glycoproteins (E1 and E2) with multiple cellular cofactors, for instance cluster of differentiation 81 (CD81), glycosaminoglycans, ephrinA2 receptor (EphA2), claudin-1 (CLDN1), occludin, low-density lipoprotein receptor (LDLR) and scavenger receptor class B type I (SR-B1).13, 14
EGFR cannot directly react with the viral particle, but it increases the formation of the CD81–CLDN1 complex that is needed for HCV entry into the cell.13 Carapito et al. reported that EGFR rs11506105, by an unknown mechanism, affects the action of EGFR on CLDN1 and/or CD81 trafficking that are obligatory to form receptor complexes for efficient HCV entry into the cell.1
EGFR is overexpressed in the liver of about 50% of patients with CHC and is often accompanied by transforming growth factor-α in cirrhotic patients compared to non-cirrhotic patients.15 However, the levels of epidermal growth factor and transforming growth factor-α were lower in CHC patients compared to HCC patients.16 Moreover, EGFR plasma concentrations are higher in patients with HCC caused by hepatitis B virus and HCV, advancing a proposal that plasma EGFR could be considered as a marker for HCC, particularly when carcinogenesis is influenced by viral hepatitis virus infection.17
Only one study investigated the relationship between EGFR rs11506105 and HCV viral spontaneous clearance, and no study about the relationship between EGFR rs11506105 and treatment responses has been done. Carapito et al. showed that EGFR rs11506105 is strong independent predictive factor of HCV viral spontaneous clearance.1
In our study, for the first time, we examined the relationship between EGFR rs11506105 and RVR, cEVR and SVR responses, as predictors after pegIFN-α/RBV in CHC patients. The results of the present study showed that the EGFR rs11506105 genotype is strongly correlated with RVR, cEVR and SVR in CHC patients. As a result, EGFR rs11506105 genotyping can be an appropriate factor before pegIFN-α/RBV combination therapy.
In the other part of the current study, we investigated the correlation between HCV viral loads with EGFR and IFNL3 SNPs. Only IFNL3 rs8099917 genotype was associated with HCV-RNA viral load. In addition, IFNL3 rs8099917 and EGFR rs11506105 genotypes were correlated with all HCV genotypes. Inconsistent with other reports, there was no significant association between IFNL3 rs12980275 and IFNL3 rs12979860 genotypes with HCV genotypes.4, 16
Several studies have been shown that IFNL3 SNPs are one of the most powerful factors that contribute to RVR, EVR and SVR responses in patients treated with pegIFN-α/RVB combination therapy.4, 16, 18, 19
In the current study, several factors such as liver enzymes (ALT and AST), IFNL3 rs12979860 (CC), IFNL3 rs12980275 (AA) were associated with RVR. Patients with IFNL3 rs12979860 CC, and IFNL3 rs12980275 AA genotypes more likely achieved RVR compared to patients with unfavorable genotypes. This finding is in agreement with Sedighimehr et al.16 who exhibited a high rate of RVR in IFNL3 rs12980275 AA. Furthermore, several studies have shown that IFNL3 rs12979860 is correlated with RVR. The frequency of the CC genotype for achieving RVR is higher than CT and TT genotypes,16, 19, 20, 21 but Lagging et al. demonstrated IFNL3 rs12979860 was not associated with RVR, which is inconsistent with our study.22
Several studies have suggested that IFNL3 rs12979860 and rs12980275 genotypes are correlated with the cEVR.16, 23 The present study showed that the frequency of cEVR was higher in patients with IFNL3 rs12979860 CC and rs12980275 AA genotypes.
In our study, beside the EGFR rs11506105 genotype, HCV genotypes, IFNL3 rs12979860 (CC), rs12980275 (AA), RVR and cEVR were independent factors for prediction of SVR response.
Important virus-related factors in SVR include HCV-RNA viral load and HCV genotypes. In the current study and consistent with other studies, SVR was associated with HCV genotypes but not HCV-RNA viral load.19, 24 We found that patients who achieved SVR were IFNL3 rs12979860 CC (59.2%) genotype. Sedighimehr et al. and Sarrazin et al. demonstrated that 52.0 and 45.9% of CHC patients with the IFNL3 rs12979860 CC genotype achieve SVR, respectively.16, 25 Consistent with other studies, we also showed that IFNL3 rs12980275 AA was a significant predictor for SVR in patients with different HCV genotypes. IFNL3 rs12979860 and rs12980275 may hence be used as a prognostic factor for the RVR, cEVR and SVR.16, 26
Interestingly, in this report there was no correlation between IFNL3 rs8099917 and treatment responses. Several studies have been shown that IFNL3 rs8099917 is a powerful predictor factor for RVR, cEVR and SVR.23, 27
However, notwithstanding several association reports, the molecular mechanisms of IFNL3 SNPs to viral responses of HCV remain predominantly unknown and yet represent a major challenge in the future for understanding virus–host interactions.
Nevertheless, a few studies have mentioned functional roles of IFNL3 SNPs. For example, in the CpG region upstream of IFNL3, a TT/-G polymorphism has been revealed to increase the expression of both IFN-γ-inducible protein 10 (IP-10) and IFNL3.28
We showed that RVR and cEVR were significantly associated with SVR. Monitoring HCV RNA viral load at 4th (RVR) and 12th (cEVR) are part of the routine appraisement of the response rate to pegIFN-α/RBV combination therapy, and their presence is considered as an important factor to achieve SVR.19 Sedighimehr et al and Yu et al. showed that the HCV-RNA viral load at 4th and 12th are independent predictive factors for SVR.16, 29
Co-expression of the favorable genotypes indicated a better chance to achieve SVR as compared to the other genotypes. Another time, these findings confirmed the significant impact that IFNL3 SNPs, beside EGFR rs11506105, have on treatment outcome. A large number of studies have been reported that IFNL3 favorable genotypes predict a better chance of achieving SVR.16, 26
In view of the fact that the treatment of HCV is evolving quickly, the clinical use of IFNL3 genotype as an outcome predictor in various settings is still open to discussion. Patients with HCV-1 first-generation protease inhibitors in combination with pegIFN-α/RBV (triple therapy) were approved by Food and Drug Administration in 2011, and the response rate was raised to around 70%. It seems that, in contrary to patients with dual PEG-IFNα/RBV therapy, the role of IFNL3 genotypes for outcome prediction is decreased in patients treated with triple therapy. However, triple therapy enhances the costs of treatment and adverse effects, which are usually the cause of treatment discontinuation.30
In this study, IFNL3 SNPs have been suggested to select HCV-1 patients with high possibility of response to pegIFN-α/RBV therapy. Other similar studies have demonstrated that the application of IFNL3 genotype to guide the utilization of triple therapy is cost effective.30, 31 The pegIFN-α/RBV therapy may still be as a choice, and the IFNL3 genotype can be a predictive factor for response in this setting. According to IFN-free treatment regimens, the IFNL3 genotype can still indicate a possible predictor to the duration of tailored treatments in previous non-response to combined treatment patients and also in patients with cirrhosis and human immunodeficiency virus co-infection who are classified in the poor responder groups. Nevertheless, further studies are essential to prove these issues.30, 32
The limitations of this study were the absence of patients with spontaneous clearance, data related to cirrhosis and fibrosis, and lack of data on the impact of the relationship between ethnic with IFNL3 SNPs.
In conclusion, besides reaffirming the role of IFNL3 in RVR, cEVR and SVR, the current study revealed for the first time the relationship between EGFR rs11506105 and HCV treatment outcomes, and we demonstrated that EGFR rs11506105 is strongly associated with RVR, cEVR and SVR. These findings bring valuable data in the context of the development of new therapeutic approaches for HCV-infected patients.
Patients and methods
Study population
This is a cross-sectional study that included 235 naïve patients with CHC infection who were treated with the pegIFN-α/RBV combinations as initial antiviral treatments at Pasteur Institute of Iran (PII) from 20 April 2014 to 15 January 2016. The study was carried out according to the Declaration of Helsinki and relevant local regulations and was approved by the Ethical Committee of PII. Written informed consent was obtained from all patients. The inclusion criteria were HCV RNA and HCV-Ab positive, treatment-naïve patients with HCV genotype 1, 2 and 3, and age more than 18 years old. The exclusion criteria included co-infection with other hepatitis and human immunodeficiency virus, use of immunosuppressive drugs, other previous antiviral treatments for HCV infection, liver transplantation, liver cirrhosis and HCC.
Each patient with HCV subtype 1a and 2a/3a were treated by RBV (800–1200 mg per day) plus Peg-IFNα-2a (180 μg per week) for 48 and 24 weeks, respectively.16
Interpretation of treatment response
Quantitative HCV RNA was assessed at baseline and at 4, 12, 24 and 48 weeks. Undetectable HCV RNA in serum after 4 and 12 weeks after the initiation of combination therapy was defined RVR and cEVR, respectively. SVR was defined as complete elimination of HCV RNA in serum 6 months after the cessation of antiviral treatment.16
Laboratory analysis
HCV genotyping and HCV RNA viral load were carried out by the AmpliSens HCV-1/2/3-FEP PCR kit (InterLabService Ltd., Moscow, Russia) and the Amplicor Monitor HCV 2.0 (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) respectively, according to the manufacturer’s instructions. HCV viral load was determined in all patients at baseline (0), 4th (RVR), 12th (cEVR), end-of-treatment (24th and 48th treatment sessions, depending on HCV genotype) and 24th post-treatment (SVR). Serum alkaline phosphatase (ALK), alanine aminotransferase (ALT), aspartate aminotransferase (AST) were analyzed using routine laboratory methods.
Single nucleotide polymorphisms genotyping
Human DNA for SNPs detection was extracted from 235 peripheral blood mononuclear cells by Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. IFNL3 SNPs (rs12979860, rs8099917 and rs12980275) were genotyped using PCR restriction fragment length polymorphism assay, as previously described.5, 33 The EGFR rs11506105 was determined by using direct PCR sequencing. For direct sequencing, the EGFR rs11506105 region was amplified by primers F-5′-TTGAATGTGGTTTCGTTGGA-3′ and R-5′-GTTGTGGCAGCAGTCACT-3′. The product size was 207 bp. The GeneJET PCR Purification Kit (Thermo Fisher Scientific) was used for PCR products purification. Afterwards, PCR products were sequenced by an ABI Automated Sequencer (Applied Biosystems, Foster City, CA, USA). The MEGA version 6.0 software was used to analyze raw sequencing data.34
Statistical analysis
The Hardy–Weinberg disequilibrium test was carried out as reported.35 Haploview version 4.2 software was used for determined linkage disequilibrium. Continuously distributed variables were described by reporting their mean. Median and inter-quartile ranges were reported for ALT, AST and ALK levels because the distributions were highly positively skewed. The statistical significance of the differences in mean levels of normally distributed variables was examined using the t-test. The Mann–Whitney U for skewed variables. Chi-square test was performed to test univariate statistical association between categorical variables at baseline. Multivariate logistic regression analysis was used for risk factors associated with RVR, cEVR and SVR. Following Jewell, Hosmer-Lemeshow guideline was used for variable selection in multivariate logistic modeling. In addition, heterogeneity in baseline was adjusted in SNP association analysis including potential confounding variables of ALT, AST, ALK, IFNL3 rs12979860, IFNL3 rs12980275, IFNL3 rs8099917, EGFR rs11506105 that differed between groups at baseline. All P-values were two-tailed, with P<0.05 considered statistically significant. Receiver operator characteristic curve was constructed to estimate the level of IFNL3 SNPs and EGFR rs11506105 in relation to RVR, cEVR and SVR. Analyses were performed using IBM SPSS for windows version 22.0 statistical software (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp.).
References
Carapito R, Poustchi H, Kwemou M, Untrau M, Sharifi A, Merat S et al. Polymorphisms in EGFR and IL28B are associated with spontaneous clearance in an HCV-infected iranian population. Genes immun 2015; 16: 514–518.
Lavanchy D . The global burden of hepatitis C. Liver Int 2009; 29: 74–81.
Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 2014; 59: 318–327.
Fateh A, Aghasadeghi MR, Keyvani H, Mollaie HR, Yari S, Hadizade Tasbiti A et al. High resolution melting curve assay for detecting rs12979860 IL28B polymorphisms involved in response of Iranian patients to chronic hepatitis C treatment. Asian Pac J Cancer Prev 2015; 16: 1873–1880.
Fateh A, Aghasadeghi M, Siadat SD, Vaziri F, Sadeghi F, Fateh R et al. Comparison of three different methods for detection of IL28 rs12979860 polymorphisms as a predictor of treatment outcome in patients with hepatitis C virus. Osong Public Health Res Perspect 2016; 7: 83–89.
Alavian SM, Hajarizadeh B, Lankarani KB, Sharafi H, Daryani NE, Merat S et al. Recommendations for the clinical management of hepatitis C in Iran: a consensus-based national guideline. Hepat Month 2016; 16 (8): e40959.
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’hUigin C et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461: 798–801.
Dayyeh BKA, Gupta N, Sherman KE, de Bakker PI, Chung RT, Team ACTGAS. IL28B alleles exert an additive dose effect when applied to HCV-HIV coinfected persons undergoing peginterferon and ribavirin therapy. PLoS One 2011; 6: e25753.
Rao H, Wei L, Lopez‐Talavera JC, Shang J, Chen H, Li J et al. Distribution and clinical correlates of viral and host genotypes in Chinese patients with chronic hepatitis C virus infection. J Gastroenterol Gepatol 2014; 29: 545–553.
Zheng H, Li M, Chi B, Wu X-x, Wang J, Liu D-W . IL28B rs12980275 variant as a predictor of sustained virologic response to pegylated-interferon and ribavirin in chronic hepatitis C patients: a systematic review and meta-analysis. Clin Res Hepatol Gastroenterol 2015; 39: 576–583.
Komposch K, Sibilia M . EGFR Signaling in Liver Diseases. Int J Mol Sci 2015; 17: 30.
Schneider MR, Wolf E . The epidermal growth factor receptor ligands at a glance. J Cell Physiol 2009; 218: 460–466.
Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 2011; 17: 589–595.
Scheel TK, Rice CM . Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 2013; 19: 837–849.
Badawy AAG, El‐Hindawi A, Hammam O, Moussa M, Gabal S, Said N . Impact of epidermal growth factor receptor and transforming growth factor-α on hepatitis C virus‐induced hepatocarcinogenesis. Apmis 2015; 123: 823–831.
Sedighimehr P, Irani S, Sakhaee F, Vaziri F, Aghasadeghi M, Sadat SM et al. IL28B rs12980275 and HLA rs4273729 genotypes as a powerful predictor factor for rapid, early, and sustained virologic response in patients with chronic hepatitis C. Arch Virol 2016: 1–9.
Divella R, Daniele A, Gadaleta C, Tufaro A, Venneri MT, Paradiso A et al. Circulating transforming growth factor-β and epidermal growth factor receptor as related to virus infection in liver carcinogenesis. Anticancer Res 2012; 32: 141–145.
Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009; 461: 399–401.
Khubaib B, Saleem S, Idrees M, Afzal S, Wasim M . The genotype CC of IL‐28B SNP rs12979860 is significantly associated with a sustained virological response in chronic HCV‐infected Pakistani patients. J Digest Dis 2015; 16: 293–298.
Dong ZX, Zhou HJ, Xiang XG, Guo SM, Zhuang Y, Zhao GD et al. IL28B genetic variations are associated with treatment response of patients with chronic hepatitis C in a Chinese Han population. J Digest Dis 2015; 16: 90–97.
Rosso C, Abate ML, Ciancio A, Strona S, Caviglia GP, Olivero A et al. IL28B polymorphism genotyping as predictor of rapid virologic response during interferon plus ribavirin treatment in hepatitis C virus genotype 1 patients. World J Gastroenterol 2014; 20: 13146.
Lagging M, Askarieh G, Negro F, Bibert S, Söderholm J, Westin J et al. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One 2011; 6: e17232.
Aziz H, Raza A, Ali K, Khattak JZK, Irfan J, Gill ML . Polymorphism of the IL28B gene (rs8099917, rs12979860) and virological response of Pakistani hepatitis C virus genotype 3 patients to pegylated interferon therapy. Int J Infect Dis 2015; 30: 91–97.
Ling Q, Chen J, Zhou H, Zhong J, Chen Y, Ye Q et al. Baseline factors associated with treatment response in patients infected with hepatitis C virus 1b by stratification of IL28B polymorphisms. Arch Virol 2015; 160: 1105–1112.
Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C et al. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol 2011; 54: 415–421.
Shaikh N, Waryah AM, Devrajani BR, Rajput MI, Hayat AS, Shaikh S . IL28B rs12980275 polymorphism shows association with response to treatment in Pakistani patients with chronic hepatitis C. J Med Virol 2015; 87: 814–820.
Akkız H, Akgöllü E, Bekar A, Yıldırım S, Sandıkçı M, Ülger Y et al. Relationship between IL28B gene rs8099917 polymorphism and SVR in Turkish patients with hepatitis C virus genotype 1. Clin Res Hepatol Gastroenterol 2015; 39: 711–717.
Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 2013; 210: 1109–1116.
Yu JW, Wang GQ, Sun LJ, Li XG, Li SC . Predictive value of rapid virological response and early virological response on sustained virological response in HCV patients treated with pegylated interferon α‐2a and ribavirin. J Gastroenterol Hepatol 2007; 22: 832–836.
Liang TJ, Ghany MG . Therapy of hepatitis C—back to the future. N Engl J Med 2014; 370: 2043–2047.
Liu S, Cipriano LE, Holodniy M, Owens DK, Goldhaber-Fiebert JD . New protease inhibitors for the treatment of chronic hepatitis C: a cost-effectiveness analysis. Ann Int Med 2012; 156: 279–290.
Cariani E, Roli L, Missale G, Villa E, Ferrari C, Trenti T . Interleukin 28B polymorphisms as predictors of sustained virological response in chronic hepatitis C: systematic review and meta-analysis. Pharmacogenom J 2016; 16: 18–29.
Venegas Santos ME, Villanueva Arancibia RA, González Lagos KV, Brahm Barril JR . IL28B polymorphisms associated with therapy responsee in Chilean chronic hepatitis C patients. World J Gastroenterol 2011; 17: 3636–3639.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evolut 2013; 30: 2725–2729.
Rodriguez S, Gaunt TR, Day IN . Hardy–Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 2009; 169: 505–514.
Acknowledgements
We would like to thank all of the patients who participated in the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Asnavandi, M., Zargar, M., Vaziri, F. et al. EGFR rs11506105 and IFNL3 SNPs but not rs8099917 are strongly associated with treatment responses in Iranian patients with chronic hepatitis C. Genes Immun 18, 144–151 (2017). https://doi.org/10.1038/gene.2017.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2017.12
- Springer Nature Limited
This article is cited by
-
The association between interferon lambda 3 and 4 gene single-nucleotide polymorphisms and the recovery of COVID-19 patients
Virology Journal (2021)
-
Interferon-λ rs12979860 genotype association with liver fibrosis in chronic hepatitis C (CHC) patients in the Pakistani population
Archives of Virology (2021)
-
First detection of human hepegivirus-1 (HHpgV-1) in Iranian patients with hemophilia
Scientific Reports (2018)