Abstract
MicroRNAs (miRNAs) are important regulators that play key roles in tumorigenesis and tumor progression. In this study, we investigate whether let-7b acts as a tumor suppressor to inhibit invasion and metastasis in gastric cancers. We analyzed the expression of let-7b in 60 pair-matched gastric neoplastic and adjacent non-neoplastic tissues by quantitative real-time polymerase chain reaction. Functional analysis of let-7b expression was assessed in vitro in gastric cancer cell lines with let-7b precursor and inhibitor. The roles of let-7b in tumorigenesis and tumor metastasis were analyzed using a stable let-7b expression plasmid in nude mice. A luciferase reporter assay was used to assess the effect of let-7b on inhibitor of growth family, member 1 (ING1) expression. Real-time PCR showed decreased levels of let-7b expression in metastatic gastric cancer tissues and cell lines that are potentially highly metastatic. Cell invasion and migration were significantly impaired in GC9811-P and SGC7901-M cell lines after transfection with let-7b mimics. Nude mice with xenograft models of gastric cancer confirmed that let-7b could inhibit gastric cancer metastasis in vivo after transfection by the lentivirus pGCsil-GFP- let-7b. Luciferase reporter assays demonstrated that let-7b directly binds to the 3′-UTR of ING1, and real-time PCR and western blotting further indicated that let-7b downregulated the expression of ING1 at the mRNA and protein levels. Our study demonstrates that overexpression of let-7b in gastric cancer can inhibit invasion and migration of gastric cancer cells through directly targeting the tumor metastasis-associated gene ING1. These findings help clarify the molecular mechanisms involved in gastric cancer metastasis and indicate that let-7b modulation may be a bona fide treatment of gastric cancer.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is the fourth most common cancer and is the second most common cause of death from cancer in the world.1, 2, 3 For most solid malignancies, metastasis is the predominant cause of cancer death.4, 5, 6 Elucidation of the molecular mechanisms that regulate the sequential steps of metastasis formation is critical for the reduction of cancer mortality. The regulatory mechanism involved in the development of GC is not well understood; the discovery of critical carcinogenic pathways and the identification of new therapeutic targets for GC are crucial for local and global public health.
MicroRNAs (miRNA) are a novel class of small non-coding RNAs that typically inhibit the translation and stability of messenger RNAs (mRNAs) by binding to the 3′-untranslated regions (3′-UTR) of their target mRNAs.5 MiRNAs have6, 7, 8 nucleotides and are found in all multi-cellular eukaryotic cells. MiRNAs have important roles in various biological and pathological processes, such as development, cell proliferation, differentiation, apoptosis, inflammation, stress response and migration.7, 8, 9 Increasing evidences have suggested that miRNAs are deregulated or upregulated in all types of cancers, acting either as tumor suppressors (for example, miR-34, miR-15/16, let-7, miR-200 family) or as oncogenes (for example, miR-155, miR-222/221, miR-17-5p, miR-21),1, 3, 8, 10 in which the miRNAs play key roles in important aspects of tumorigenesis, such as cancer initiation, differentiation, growth and progression,3, 11, 12, 13, 14, 15 mainly by interfering with the expression of target genes involved in cell cycle, apoptosis, cell migration and invasion, and angiogenesis.
The let-7 family is a conserved family of miRNAs. Let-7 was originally observed in the nematode Caenorhabditis elegans,10 and 14 members have been found to date.11 Recently, the expression levels of many let-7-family members were found to be reduced in a variety of cancers. For example, let-7 is downregulated in lung cancer, melanoma, and head and neck squamous carcinoma, whereas overexpression of let-7 can inhibit cancer cell growth.12, 13 Several oncogenes, such as RAS, MYC and HMGA2, are direct targets of let-7.14 Let-7b is one member of the let-7 family. Previous studies have shown that let-7b is upregulated in primary breast cancer and promotes angiogenesis;15 downregulation in PAH16 was caused by chronic hypoxia or monocrotaline in rats, and the level was decreased in plasma vesicles of non-small cell lung cancer patients.17 In addition, let-7b can affect cell proliferation by targeting Kallikrein-related peptidases, a family of serine proteases that have been shown to be dysregulated in several malignancies, including ovarian cancer.18
In this study, we investigated the biological effects and potential mechanisms of let-7b in gastric carcinoma. We examined the expression of let-7b in GC and found that let-7b was associated with differentiation and lymphatic metastasis of human GC. Exogenous expression of let-7b inhibits invasion, tumorigenesis and metastasis formation both in vitro and in vivo. Using bioinformatics analysis, we identified inhibitor of growth family, member 1 (ING1) as a putative let-7b target.
Methods
Tissue collection
Primary gastric tumor tissues, adjacent non-tumor gastric tissues and distant metastatic gastric tissues were obtained from patients who underwent surgery at the first affiliated hospital of Bengbu Medical College in Bengbu (Anhui), China. All samples were clinically and pathologically shown to be correctly labeled. Patients offering samples for the study signed informed consent forms.
Cell culture
The human GC cell lines GC9811, GC9811-P, SGC7901-NM and SGC7901-M were conserved in our own laboratory and were cultured in RPMI1640 (HyClone, Rockville, MD, USA), supplemented with 10% fetal bovine serum (GIBCO, Waltham, MA, USA), 100 units per ml penicillin, and 0.1 mg ml−1 streptomycin at 37 °C in a humidified 5% carbon dioxide incubator.
Western blot
Cells were harvested, washed twice in phosphate-buffered saline and lysed in lysis buffer (protease inhibitors were added immediately before use) for 30 min on ice. Lysate was centrifuged at 10 000 r.p.m. and the supernatants were collected and stored at −70°C in aliquots. All procedures were carried out on ice. Protein concentration was determined using BCA assay kit (Tianlai Biotech, SpectraMax Plus384; Molecular Devices, Sunnyvale, CA, USA).
miRNA real-time PCR
Cells (1 × 102–1 × 107) cells were harvested, washed in phosphate-buffered saline once and stored on ice; complete cell lysate was prepared by addition of 600 μl lysis binding buffer and vertex; 60 μl miRNA homogenate addictive was added to the cell lysate and mixed thoroughly by inverting several times; sample was stored on ice for 10 min, followed by addition of equal volume (600 μl) of phenol: chloroform (1:1) solution; sample was mixed by inverting for 30–60 s, and then centrifuged at 12 000 g for 5 min; the supernatant was transferred to a new tube and the volume was estimated; one-third volume of 100% ethanol was added and mixed; the mixture was loaded to the column at room temperature and centrifuged at 10 000 g for 15 s; the flow-though was collected and the volume was then estimated; two-third volume of 100% ethanol was added and mixed; the mixture was loaded to column at room temperature and centrifuged at 10 000 g for 15 s; the flow-through was discarded; 700 μl miRNA wash solution was added to the column, followed by centrifugation at 10 000 round/min for 10 s; the flow-through was discarded; 500 μl miRNA wash solution was added to the column, followed by centrifugation at 10 000 round/min for 10 s; the flow-through was discarded; the column was transferred to a new tube and 100 μl preheated elution solution (95 degree) was added at room temperature; RNA was collected by centrifugation at 12 000 g for 30 s.
Wound healing assay
As reported previously,6, 11 the gastric cells transiently transfected with miR-31 precursor or negative control miRNA precursor were seeded in a 100-mm Petri dish. A wound was made by scratching on the Petri dish bottom, followed by another 48 h growth.
Immunoblotting
For immunoblotting, the human GC cells were collected 72 h after transfection with miR-31 precursor or negative control miRNA precursor. Cells were lysed using 1 × RIPA buffer (Upstate Biotechnology, Lake Placid, NY, USA) containing a protease inhibitor cocktail (Sigma, St Louis, MO, USA). After cell lysis, 45 mg of protein was loaded on a 10% SDS gel followed by transfer to PVDF membrane. Antibodies against SGPP2 (Abcam, Cambridge, MA, USA), Smad4 (Cell Signaling Technologies, Danvers, MA, USA) and Actin (Sigma) were used. Secondary antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The detected signals were visualized by an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China), as recommended by the manufacturer.
Tumor-bearing (xenografts) study
As reported recently,8, 15 1.5 × 105 murine gastric adenocarcinoma cells re-suspended in 150 ul phosphate-buffered saline were injected subcutaneously into the flank of the normal C57B/l6 mice at age about 8 weeks (five mice per group). Both gastric cell line and the mice were in C57BL/6 background and no rejection occurred. The animals were maintained in a pathogen-free barrier facility and closely monitored by animal facility staff. The grown tumors (xenografts) were measured every 3 days starting 23 days post inoculation of cells using caliper as length × width × width/2 (mm3). miR-31 precursor or negative miRNA (6.26 ug; GenePharma, Shanghai, China) mixed with 1.6 ul transfection reagent Lipofectamine 2000 (Invitrogen, Rockville, MD, USA) in 50 ul phosphate-buffered saline were injected into the tumors every 3 days, for a total of three times. Thirty-two days after inoculation, the animals were killed and the xenografts were isolated, the weight (gram) and volume (mm3) of the xenografts were determined. All procedures were conducted according to the Animal Care and Use guideline approved by Xinxiang Medical University Animal Care Committee.
Statistical analysis
Student’s t-test (two-tailed), one-way analysis of variance and Mann-Whitney test were employed to analyze the in vitro and in vivo data using SPSS 12.0 software (Chicago, IL, USA). P-value<0.05 was defined as statistically significant.
Results
The level of let-7b expression was frequently decreased in human GC metastatic cell lines and tissues
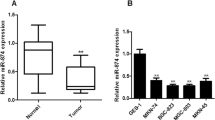
To determine whether let-7b expression is associated with GC, we examined and compared let-7b expression in primary GC tissues and in pair-matched adjacent non-tumor tissues using quantitative RT-PCR. A decrease of let-7b expression was found in 70.18% of the 57 patients with GC with a median change by about 1.83-fold (P<0.05, Figure 1a). We then examined the mRNA expression levels of let-7b in two pairs of human GC cell lines, GC9811, GC9811-P and SGC7901-NM, SGC7901-M. As shown in Figures 1b and A and B, expression of let-7b was lower in the GC cells, GC9811-P and SGC7901-M, with high metastatic potential, whereas let-7b was more highly expressed in the GC cells, GC9811 and SGC7901-NM, with low metastatic potential. To further validate the role of let-7b in GC cells metastasis, we compared the expression levels of let-7b in fresh human GC tissue specimens with metastases from eight individuals, to normal gastric tissues (NG), primary GC tissues and distant metastatic GC tissues (MC), respectively, by real-time PCR. Intriguingly, there was remarkable downregulation of let-7b in MC and GC, compared with let-7b expression in NG (Figure 1b). We observed that either the loss of or decreased expression of let-7b in tumors and their metastatic tissues resulted in enhanced metastasis in GC tissues. The results all suggested that the expression of let-7b was negatively correlated very closely with decreased GC metastases and might play a vital role in pathological processes. We have thus provided clinical and cellular conceptual evidence for a metastatic switch in cancer development that suggests a key role for the expression of let-7b in cancer development.
Expression of let-7b was downregulated in gastric cancer metastatic cell lines and tissues. (a) Level of let-7b expression in gastric cancer tissues (n=60) was higher than that in non-tumor tissues (n=60) (A). The values (ΔCt) are relative to those of U6 small RNA (B). (b) The mean fold changes of let-7b gene expression in GC9811 and GC9811-P (A). The mean fold changes of let-7b gene expression in SGC7901-NM and SGC7901-M (B). Student’s t-test, n=3. (C) The relative expression of let-7b to 5S. *P<0.01. One-way ANOVA, n=8. Real-time PCR show that the relative expression of let-7b in eight adjacent non-cancerous gastric tissue samples, and in gastric cancer cell lines GC9811, SGC7901-NM is higher than that in their primary gastric cancer and distant gastric metastasis specimens and the high potential metastasis gastric cancer cell GC9811-P and SGC7901-M. Each sample was analyzed in triplicate and normalized to 5S. Fold change was calculated by 2-ΔΔCt.
Let-7b inhibited the invasive and metastatic abilities of GC cells in vitro
Because let-7b acts as an agitator in invasion and metastatic processes, we investigated the effects of decreased let-7b on less metastatic cell lines and increased let-7b on more metastatic cell lines. To accomplish this, short interfering RNA oligonucleotides of let-7b were constructed and introduced into GC9811 cells and SGC7901 -NM cells for metastasis assays in vitro, as GC9811-let-7b-short interfering RNA cells, SGC7901-NM-let-7b-short interfering RNA cells and control cells. Concurrently, mimic-let-7b and negative control oligonucleotides were also constructed and introduced into GC9811-P cells and SGC7901-M cells for metastasis assays in vitro, as GC9811-P-let-7b-mimic cells, SGC7901-N-has let-7b mimic cells and control cells. Depletion of let-7b significantly impaired the ability of GC9811 cells to migrate and invade through the matrigel-coated membranes or the non-matrigel-coated membranes towards serum-containing medium in a modified Boyden chamber assay (Figure 2a). Increased expression of let-7b significantly suppressed the ability of GC9811-P cells to migrate and invade through matrigel-coated membranes or non-matrigel-coated membranes towards serum-containing medium in a modified Boyden chamber assay (Figure 2a), when compared with the control cells. Similar results were found in SGC7901-NM cells (Figure 2b) and SGC7901-M cells (Figure 2b), whereas let-7b knockout in GC cell lines resulted in higher invasion and migration rates. Furthermore, wound healing assay shows that let-7b inhibits the cell healing (Figure 2c).
Effect of let-7b on tumor cell invasion and metastasis of GC9811 and GC9811-P Cells. (a) Representative fields of invasive cells on the membrane (A). (magnification of 6200). Average invasive cell number per field (B). The invasive cell number of GC9811 or GC9811-P cells transfected with let-7b-inhibitors is drastically increased than that transfected with negative control. *P<0.05, Student’s t-test, n=10. (b) The migration cell number of GC9811 or GC9811-P cells transfected with let-7b-mimics is drastically decreased than that transfected with negative control. *P<0.05, Student’s t-test, n=10. (c) Wound healing assay shows that let-7b inhibits the cell healing.
Ectopic expression of let-7b inhibits tumorigenesis and metastasis in vivo
We then tested whether the ectopic expression of let-7b could promote tumor growth in vivo. We constructed a let-7b expression vector and selected SGC-7901 cells stably transfected with let-7b. Subsequently, let-7b-transfected SGC-7901 cells and control-transfected SGC-7901 cells were injected subcutaneously into nude mice, and tumor formation was monitored. After 30 days, the animals were killed, and the tumor volume was measured. Tumor growth was significantly inhibited in the mice injected with let-7b-transfected SGC-7901 cells compared with control-transfected SGC-7901 cells (Figures 3a and b). The average tumor volume of mice inoculated with let-7b-transfected SGC-7901 cells at day 30 was 23.81±1.95 cm3, which was significantly smaller than that of mice inoculated with control-transfected SGC-7901 cells (29.78±2.36 cm3, P<0.05, Figures 3a and b). Thus, let-7b can inhibit tumorigenesis in vivo. We found that ectopic expression of let-7b inhibited migration and invasion of GC cells in vitro; therefore, we further tested whether the ectopic expression of let-7b could affect tumor metastasis in vivo. SGC-7901 cells stably transfected with let-7b or control vector were injected into 4-week-old male nude mice through the tail vein. One month after the injection, the mice were killed. The average number of hepatic metastatic nodes per mouse was much smaller in mice injected with SGC-7901 cells stably transfected with let-7b vector than that in the control group (3±0.68 vs 12.32±0.21, P<0.01) (Figure 3c). In addition, immunohistochemical analysis confirmed that the ING1 protein was downregulated in let-7b-transfected SGC-7901 cells compared with control-transfected SGC-7901 cells (Figure 3d). Thus, these results indicate that let-7b has the ability to inhibit metastasis of GC cells in vivo, which is consistent with the data obtained from the in vitro migration and invasion assays.
Let-7b inhibits tumorigenesis and metastasis in vivo. (a) Photographs of tumors in nude mice resulting from injection of let-7b expression vector (let-7b mimic) or control plasmid (control) stably transfected GC9811-P cells. (b) Tumorigenesis curves. Rapid tumor growth was observed in the GC9811-P group (data expressed as mean±s.d.; P<0.05). (c) Photographs of the number of liver metastatic nodes in nude mice injected with let-7b mimic or control vector stably transfected GC9811-P cells. The arrows indicate liver metastatic nodes. (d) Histological examination found that the expression of ING1 was markedly reduced in the let-7b-transfection group compared with the control groups. aP<0.05, compared with miR-control transfectants. Scale bars: 200 μm.
Let-7b downregulates ING1 expression by directly targeting its 3′-UTR
To further explore the mechanism by which let-7b suppresses GC invasion and metastasis, we analyzed potential downstream tumor metastasis-related target genes in silico. We focused on let-7b target genes, especially those genes that were migration- and/or invasion-related, and found that ING1, involved in the promotion of cancer cell migration or invasion, might be the target gene of let-7b. In an effort to determine whether ING1 is regulated by let-7b through direct binding to its 3′-UTR, we constructed full-length wild-type and mutant fragments of ING1 mRNA 3′-UTR, and inserted them into the region immediately downstream of a luciferase reporter gene (Figure 4a). Subsequently, let-7b mimic oligos were co-transfected with different luciferase 3′-UTR constructs into GC9811-P cells. We found that let-7b decreased the relative luciferase activity in the wild-type 3′-UTR of ING1 (Figure 4b). However, luciferase activity did not drop sharply in the UTRs with mutant binding sites, when compared with the mut-type counterparts (Figure 4c). These data support that ING1 is direct target of let-7b. RT-PCR showed that the expression of ING1 in GC9811 cells and SGC7901-NM cells transfected with let-7b-mimics is downregulated compared with those cells transfected with control constructs (Figure 4d). Western blotting results showed that the expression of ING1 in GC9811 and SGC7901-NM cells transfected with let-7b-mimics is downregulated compared with those transfected with negative control (Figure 4e). Furthermore, Real-time PCR shows that mRNA relative expression of ING1 in GC tissues and distant metastatic tissue are downregulated compared with their normal adjacent non-cancerous specimens (Figure 5a). Immunohistochemistry showed that the expression of ING1 in GC lymph node metastasis is upregulated compared with its primary GC tissue (Figure 5c), and it is upregulated in GC tissue compared with its matched adjacent normal stomach tissue (Figure 5b). These data demonstrate that let-7b can downregulate the mRNA expression of ING1 and can repress protein translation of it.
Let-7b directly targets ING1. The sequence of the let-7b-binding site within the ING1 3′-UTR and mutant (mut) ING1 3′UTR. Luciferase activity of the ING1 3′-UTR (a) or mut ING1 3′-UTR (b) reporter gene in GC9811 cells infected with the let-7b, let-7b-inhibitor, blank or empty mimic oligo. The assays showed that luciferase activities in the group were significantly decreased compared with those of the mutant and negative control groups, *P<0.05. (c) The expression of ING1 was analyzed by qRT-PCR. ING1 was decreased in GC9811 cells and SGC7901 cells transfected with let-7b-mimics compared with GC9811 cells and SGC7901 cells transfected with let-7b-negative control. (d) Let-7b suppresses the endogenous protein levels of ING1, as detected by western blotting. Stable transduced SGC7901-NM and GC9811 cells ectopically expressing let-7b were used for western blot analysis. Effect of let-7b on ING1 in GC9811 cells (right). Effect of let-7b on ING1 in SGC7901-NM cells (left).
The expression of ING1 in gastric tumor specimens. (a) Real-time PCR show that the relative expression of ING1 in eight distant gastric metastasis specimens is higher than that in their primary gastric cancer and adjacent non-cancerous gastric tissue samples. Each sample was analyzed in triplicate and normalized to 18S. (b) Expression of ING1 in primary gastric cancer (Ca) and its adjacent normal stomach tissue (N) by immunohistochemistry. (c) Expression of ING1 in primary gastric cancer (Ca) and its matched lymph node metastasis tissue (M) by immunohistochemistry.
Discussion
Invasion and metastasis, two of the most important hallmarks of cancer, are the leading factors of malignant cancer that lead to lethality, especially for GC.
The long-term survival of patients with GC after curative resection is confounded by a high recurrence rate, which is mainly due to the spread of lymphatic metastasis.4, 19 Therefore, the identification of metastatic factors and an understanding of the underlying molecular pathways involved in the progression of metastasis are critical issues. Recent studies have shown that miRNAs play a fundamental role in the invasion and metastasis of GC,15, 16, 18 thereby opening a novel avenue to investigate the molecular mechanism of GC progression and to develop potential therapeutics against GC.
In the present study, we demonstrated that let-7b expression in metastatic GC cell lines was downregulated when compared with let-7b expression in non-metastatic GC cells. Ectopic expression and short interfering RNA knockdown of let-7b confirmed its invasion-suppressing activity in vitro and in vivo. Moreover, we show that ING1 is a direct target for let-7b and this let-7b-mediated suppression of ING1 is dependent on its 3′-UTR. Therefore, these results highlight the significance of let-7b as a tumor suppressor in cell invasion and metastasis by targeting ING1 in GC.
MiRNAs are a class of small, non-coding RNAs that can regulate gene expression by either inducing the degradation of target mRNAs or by impairing the translation of their target mRNAs. MiRNAs can also upregulate gene expression by targeting the UTR of their target genes. Many studies have revealed that aberrantly expressed miRNAs participate in tumorigenesis in temporal and spatial manners.20 For example, miR-10b,21 miR-373 and miR-520c13, 14, 15, 16, 17, 18, 19, 20, 21 stimulated cancer cell migration and invasion in breast cancer. Additionally, miR-155 can promote tumor invasion and metastasis in breast cancer by downregulating its target, Rho A22 and promote tumor invasion and metastasis in pancreas duct carcinoma by repressing the expression of TP53INP1.23 The miRNA-200 family (miRNA-200a, miRNA-200b, miRNA-200c, miRNA-141 and miRNA-429) can inhibit tumor invasion and metastasis by regulating the epithelial to mesenchymal transition.24 Although many studies have been carried out to investigate the mechanism of miRNA and tumor metastasis, the mechanism was not clear. Most importantly, few studies have been carried out on the mechanisms of GC metastasis regulation by miRNAs.
The let-7b gene is located at 9q22.3, and is involved in a variety of physiological and pathological processes, including angiogenesis,25 immunocyte differentiation,26 replicative senescence,27 growth arrest,28 pulmonary arterial hypertension29 and carcinogenesis.30 Let-7b is downregulated in several malignancies. A highly characterized example is renal cell carcinoma, in which let-7b downregulation led to a significant decrease in kallikrein expression.30 Kallikreins can also be targeted by let-7b in ovarian cancer.31 In papillary thyroid cancer, reduced expression of let-7b might be an essential molecular event in RET/PTC malignant transformation.32 In the case of primary breast cancer, however, let-7b was upregulated when compared with normal adjacent tumor tissues.33 In our study, we demonstrated that let-7b is also downregulated in MC tissues and GC cell lines with high metastatic potential, and we further explored the mechanism by which let-7b reduced tumor invasion and metastasis.
To explore the molecular mechanism underlying let-7b function, we searched for its direct target genes using bioinformatics analysis of miRNA-mRNA 3′-UTR matching. Among the putative targets for let-7b, ING1 was detected by the TargetScan program. This gene encodes a tumor suppressor protein that can induce cell growth arrest and apoptosis; the encoded protein is a nuclear protein that physically interacts with the tumor suppressor protein TP53 and is a component of the p53 signaling pathway.29 The bioinformatics analysis revealed that the conserved binding sites on ING1 that can be recognized by let-7b are located in the 3′-UTR. To test this assumption, we investigated whether let-7b affects ING1 protein levels. We found that let-7b leads to a significant decrease in ING1 protein levels, suggesting that ING1 is a functional target of let-7b. Lastly, results from our dual-luciferase reporter assays suggest that ING1 is a functional downstream target of let-7b.
In conclusion, our study demonstrates that let-7b can suppress the invasion and metastasis of GC by directly binding the 3′-UTR of ING1, its target.34, 35 Though there is still much to learn about the role of let-7b in GC tumorigenesis, let-7b provides us with a new potential target for GC treatment.
References
Chen J, Chen LJ, Zhou HC, Yang RB, Lu Y, Xia YL et al. Prognostic value of matrix metalloproteinase-9 in gastric cancer: a meta-analysis. Hepatogastroenterology 2014; 61: 518–524.
Yang Y, Bai ZG, Yin J, Wu GC, Zhang ZT . Role of c-Src activity in the regulation of gastric cancer cell migration. Oncol Rep 2014; 32: 45–49.
Li Y, Tan BB, Zhao Q, Fan LQ, Wang D, Liu Y . ZNF139 promotes tumor metastasis by increasing migration and invasion in human gastric cancer cells. Neoplasma 2014; 61: 291–298.
Yu SY, Li Y, Fan LQ, Zhao Q, Tan BB, Liu Y . Impact of Annexin A3 expression in gastric cancer cells. Neoplasma 2014; 61: 257–264.
Oo HZ, Sentani K, Sakamoto N, Anami K, Naito Y, Uraoka N et al. Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol Rep 2014; 32: 403–410.
Li XS, Xu Q, Fu XY, Luo WS . Heat shock protein 22 overexpression is associated with the progression and prognosis in gastric cancer. J Cancer Res Clin Oncol 2014; 140: 1305–1313.
Guan SS, Chang J, Cheng CC, Luo TY, Ho AS, Wang CC et al. Afatinib and its encapsulated polymeric micelles inhibits HER2-overexpressed colorectal tumor cell growth in vitro and in vivo. Oncotarget 2014; 5: 4868–4880.
Luo BH, Xiong F, Wang JP, Li JH, Zhong M, Liu QL et al. Epidermal growth factor-like domain-containing protein 7 (EGFL7) enhances EGF receptor-AKT signaling, epithelial-mesenchymal transition, and metastasis of gastric cancer cells. PLoS One 2014; 9: e99922.
Vaiopoulos AG, Kostakis ID, Gkioka E, Athanasoula KC, Pikoulis E, Papalambros A et al. Detection of circulating tumor cells in colorectal and gastric cancer using a multiplex PCR assay. Anticancer Res 2014; 34: 3083–3092.
Choi MR, An CH, Chung YJ, Choi YJ, Yoo NJ, Lee SH . Mutational and expressional analysis of ERBB3 gene in common solid cancers. APMIS 2014; 122: 1207–1212.
Musiani D, Konda JD, Pavan S, Torchiaro E, Sassi F, Noghero A et al. Heat-shock protein 27 (HSP27, HSPB1) is up-regulated by MET kinase inhibitors and confers resistance to MET-targeted therapy. FASEB J e-pub ahead of print 5 June 2014.
Xu L, Qu X, Li H, Li C, Liu J, Zheng H et al. Src/caveolin-1-regulated EGFR activation antagonizes TRAIL-induced apoptosis ingastric cancer cells. Oncol Rep 2014; 32: 318–324.
Aprile G, Giampieri R, Bonotto M, Bittoni A, Ongaro E, Cardellino GG et al. The challenge of targeted therapies for gastric cancer patients: the beginning of a long journey. Expert Opin Investig Drugs 2014; 23: 925–942.
Kurokawa Y, Matsuura N, Kawabata R, Nishikawa K, Ebisui C, Yokoyama Y et al. Prognostic Impact of Major Receptor Tyrosine Kinase Expression in Gastric Cancer. Ann Surg Oncol 2014; 21: 584–590.
Nielsen TO, Friis-Hansen L, Poulsen SS, Federspiel B, Sorensen BS . Expression of the EGF family in gastric cancer: downregulation of HER4 and its activating ligand NRG4. PLoS One 2014; 9: e94606.
Pertino MW, Lopez C, Theoduloz C, Schmeda-Hirschmann G . 1,2,3-triazole-substituted oleanolic Acid derivatives: synthesis and antiproliferative activity. Molecules 2013; 18: 7661–7667.
Rasul A, Khan M, Yu B, Ali M, Bo YJ, Yang H et al. a sesquiterpene lactone, induces apoptosis in SGC-7901 cells via mitochondrial and phosphatidylinositol 3-kinase/Akt signaling pathways. Arch Pharm Res 2013; 36: 1262–1269.
Hong KJ, Wu DC, Cheng KH, Chen LT, Hung WC . RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation. J Cell Physiol 2014; 229: 191–201.
Liu QS, Zhang J, Liu M, Dong WG . Lentiviral-mediated miRNA against liver-intestine cadherin suppresses tumor growth and invasiveness of human gastric cancer. Cancer Sci 2010; 101: 1807–1812.
Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R et al. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol 2014; 16: 374–379.
Guo LH, Li H, Wang F, Yu J, He JS . The tumor suppressor roles of miR-433 and miR-127 in gastric cancer. Int J Mol Sci 2013; 14: 14171–14184.
Fang Y, Shen H, Li H, Cao Y, Qin R, Long L et al. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim Biophys Sin (Shanghai) 2013; 45: 963–972.
Wu XJ, Mi YY, Yang H, Hu AK, Li C, Li XD et al. Association of the hsa-mir-499 (rs3746444) polymorphisms with gastric cancer risk in the Chinese population. Onkologie 2013; 36: 573–576.
Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H et al. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer 2013; 109: 2323–2330.
Yoon JH, Choi YJ, Choi WS, Nam SW, Lee JY, Park WS . Functional analysis of the NH2-terminal hydrophobic region and BRICHOS domain of GKN1. Biochem Biophys Res Commun 2013; 440: 689–695.
Iwaya T, Fukagawa T, Suzuki Y, Takahashi Y, Sawada G, Ishibashi M et al. Contrasting expression patterns of histone mRNA and microRNA 760 in patients with gastric cancer. Clin Cancer Res 2013; 19: 6438–6449.
Won KJ, Im JY, Yun CO, Chung KS, Kim YJ, Lee JS et al. Human Noxin is an anti-apoptotic protein in response to DNA damage of A549 non-small cell lung carcinoma. Int J Cancer 2014; 134: 2595–2604.
Nakadate Y, Kodera Y, Kitamura Y, Tachibana T, Tamura T, Koizumi F . Silencing of poly(ADP-ribose) glycohydrolase sensitizes lung cancer cells to radiation through the abrogation of DNA damage checkpoint. Biochem Biophys Res Commun 2013; 441: 793–798.
Shen J, Niu W, Zhou M, Zhang H, Ma J, Wang L et al. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One 2014; 9: e104510.
McLean MH, El-Omar EM . Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 2014; 11: 664–674.
Shin VY, Chu KM . MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 2014; 20: 10432–10439. Review.
Zhao H, Wang Y, Yang L, Jiang R, Li W . MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol Cell Biochem 2014; 385: 207–213.
Cao W, Yang W, Fan R, Li H, Jiang J, Geng M et al. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol 2014; 35: 1287–1295.
Kim YM, Kim IH, Nam TJ . Inhibition of AGS human gastric cancer cell invasion and proliferation by Capsosiphon fulvescens glycoprotein. Mol Med Rep 2013; 8: 11–16.
Tong F, Cao P, Yin Y, Xia S, Lai R, Liu S . MicroRNAs in gastric cancer: from benchtop to bedside. Dig Dis Sci 2014; 59: 24–30.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Han, X., Chen, Y., Yao, N. et al. MicroRNA let-7b suppresses human gastric cancer malignancy by targeting ING1. Cancer Gene Ther 22, 122–129 (2015). https://doi.org/10.1038/cgt.2014.75
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2014.75
- Springer Nature America, Inc.
This article is cited by
-
let-7b and let-7c microRNAs promote histone H2B ubiquitylation and inhibit cell migration by targeting multiple components of the H2B deubiquitylation machinery
Oncogene (2017)
-
MUFFINN: cancer gene discovery via network analysis of somatic mutation data
Genome Biology (2016)
-
Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2
Journal of Physiology and Biochemistry (2016)
-
RETRACTED ARTICLE: MicroRNA-216b is Down-Regulated in Human Gastric Adenocarcinoma and Inhibits Proliferation and Cell Cycle Progression by Targeting Oncogene HDAC8
Targeted Oncology (2016)