Abstract
Monoculture cultivation patterns in agro-ecosystems only provide less varied soil organic matter (SOM) molecules of plant origin. Whether and how the natural fallow stage between cultivation seasons facilitates the restoration of SOM molecular diversity and mitigates the adverse impacts of constant cropping pattern is elusive. Here, we utilized FT-ICR-MS, UHPLC-MS/MS, and high-throughput sequencing to investigate the biological change processes in SOM molecular composition under cultivation and fallow status in a long-farmed paddy field. Our study showed that SOM molecular diversity increased by 45.70%–85.36% in fallow stage compared to rice cultivation season. SOM molecular diversity was positively correlated with bacterial diversity and root exudate molecular diversity, and negatively correlated with fungal diversity. Notably, root exudate molecular diversity accounted for 48.48% of the variation in SOM molecular diversity. The increased SOM molecular diversity in fallow stage was attributed more to the diverse plant-produced molecules than the microbe-consumed molecules. Plant species turnover resulted in the conversion of root exudate components to Organoheterocyclic compounds and Organic acids/derivatives from rice planting stage to fallow stage. Recruited microbes were dominated by Basidiomycita, Ascomycota, Acidobacteria, Chloroflexi and Proteobacteria, resulting in the transformation from carbohydrates, lipid-like SOM molecules to lipid-like and lignin-like SOM molecules. Both field and microcosm experiments confirmed that root exudates are the main source of SOM molecules, and are influenced by the soil microbial community. This study provides solid evidence that fallow status in agro-ecosystems provides explosion of biodiversity and counteracts the negative effects of long-term monoculture cultivation on SOM diversity.
Graphical Abstract

Highlights
• Natural fallow promotes soil organic matter molecular diversity in paddy fields.
• Plant diversity and bacterial diversity increase in fallow stage.
• Root exudates increase SOM molecular diversity by mediating microbial communities.
• Variations in the composition of biological communities drive transformations in SOM molecular composition.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Soil organic matter (SOM) is the largest reservoir of organic carbon (C) in terrestrial ecosystems, and its turnover plays a key role in the global elemental cycle and future climate change (Schmidt et al. 2011; Seneca et al. 2021). Increasing evidence suggests that the diversity and composition of SOM molecules have emerged as potentially critical controls on soil organic carbon persistence (Davenport et al. 2023; Jones et al. 2023; Chen et al. 2022). SOM is a complex mixture of plant- and microbial-derived polymers and their degradation products. The fate of SOM molecules is closely linked to the metabolism of complex biomes (Hu et al. 2022b, a; Zhou et al. 2002). There are two pathways for the formation of SOM molecules: one is the entry of plant litter or root exudate molecules into the soil, where they combine with minerals physically or chemically to form plant-derived SOM molecules; the other is the conversion of plant-derived carbon by microbial metabolism to microbial-derived SOM molecules (Liang et al. 2017). The molecular diversity and composition of plant-derived SOM molecules varies depending on the diversity and type of litter or root exudate molecules of different species (Davenport et al. 2023). Microorganisms play a key role in the conversion of plant-derived carbon into SOM molecules. Microorganisms act as “funnels” to utilize and decompose plant-derived carbon into low molecular weight molecules, thereby increasing the diversity of SOM molecules (Davenport et al. 2023; Liang et al. 2017). Microbial “recipes”, namely the selective utilization of carbon sources, determine the potential for conversion of plant-derived C to microbial-derived SOM molecules (Huang et al. 2023). In addition, the prevalence of cross-feeding mechanisms among microorganisms leads to the interconversion of microbial-derived SOM molecules, which enriches the molecular diversity of SOM (Hu et al. 2022a). Thus, different resource preferences of microbial species and complex cross-feeding mechanisms result in SOM composition and diversity that may vary with unique microbial communities (Liu et al. 2023a, b). Environmental stresses select habitat-specific plant communities, and plant diversity and plant input types act as “sources” of SOM to alter SOM molecular diversity and composition (Córdova et al. 2018). In addition, different types of plant communities selectively screen microbes with appropriate transporter proteins and metabolic pathways to use a given plant-derived C, thus reshaping the distribution patterns of SOM molecular composition and diversity (Baran et al. 2015). Therefore, plant communities, as the source of SOM molecules, and microbial communities, as intermediate mediators between plant-derived molecules and SOM molecules, have a profound impact on SOM molecule composition and diversity. However, the understanding of the pattern of SOM molecule formation is still stuck on the interactions between SOM molecules and microbial communities, and little is known about the upstream control of microbial communities and SOM molecule transformations by plant communities.
Agroecosystems are active components of the global C cycle, hosting a range of processes that degrade and generate SOM molecules (Li et al. 2018; Wu et al. 2021). Monoculture cropping patterns in agroecosystems diminish the multifunctionality of the ecosystems, as well as reduce the sources of SOM molecular diversity. This is one of the causes of carbon emissions from farmland and contributes to climate warming (Wen et al. 2024). Fallowing is a way to return farmland to its natural state with rapid plant colonization and increased biodiversity. As an effective strategy of maintaining ecosystem services and restoring soil nutrient conversion capacity, fallow mitigates the negative effects of cropping patterns and increases potential sources of SOM molecular diversity. However, there are knowledge gaps about the biological mechanisms of SOM molecular diversity succession in agroecosystems under both the cultivation and fallow states. Paddy ecosystems, as typical artificial wetlands, play a key role in organic carbon cycling. In paddy ecosystems, long-term cultivation can alter the diversity and composition of biological communities, leading to unique patterns of distribution of SOM molecular groups (Wurz et al. 2022). Natural fallow in winter returns the paddy fields to natural ecosystems, creating unique plant communities, increasing biodiversity and reshaping the interactivity between above-ground plants and below-ground microorganisms. Fallowing led to an increase in the contribution of plant-derived SOM molecules to SOM formation (Ma et al. 2024). In addition, fallow may attenuate or even reverse the feedback effects of biomes on SOM molecules under long-term cropping patterns. Thus, exploring how biomes mediate the successional pattern of SOM molecules remains challenging in a whole annual period.
Ultrahigh-resolution Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) represents an advanced mass spectrometry technique (Koch et al. 2007; Nebbioso and Piccolo 2013; Qi et al. 2022). This method excels in the complete separation and analysis of complex mass peaks along with their corresponding molecular formulae, owing to its exceptional ultra-high resolution and accuracy (Wu et al. 2021; Kellerman et al. 2014; Hu et al. 2022b, a). Attributed to the advances in SOM extraction methods, we can analyze the composition of SOM molecules rather than just DOM molecules using FT-ICR-MS (Wu et al. 2022). The development of this technology has made it possible to resolve SOM and biome interactions at the molecular level, by using combined multi-omics analysis. In this study, we used FT-ICR-MS, UHPLC-MS/MS and high-throughput sequencing to explore patterns of synergistic variations between SOM molecules and biotic community under artificial management of agroecosystems.
Here, we conducted a year-long experiment in a paddy field that has been continuously cultivated for 33 years to investigate the effects of biome-mediated SOM molecular diversity and composition under different cropping stages. This study aimed to investigate (1) the dynamic patterns of the diversity and composition of SOM molecules and biomes in paddy soils under cultivation and fallow stages, and (2) the coupling relationship between SOM molecules and biological communities (including microbial community in the belowground and plant community in the aboveground) in molecular level. On this basis, we formulated the following scientific hypotheses: (1) biological community and SOM molecular diversity increase during natural fallow stage; (2) biological community diversity promotes SOM molecular diversity; and (3) the proportion of plant-derived SOM molecules (e.g. lignins) increases in SOM during fallow stage.
2 Materials and methods
2.1 Study site and experimental description
The soil used for this study was from the paddy field at the Yingtan National Agroecosystem Field Experiment Station (28°15′30″ N, 116°55′30″ E) of the Chinese Academy of Sciences in Yujiang Country, Jiangxi Province, China. Four fertilization treatments were set up in this study and sampled at three tillage stages. The fertilization treatments were set as: no fertilizer (CK), chemical fertilizer (NPK), organic fertilizer (OM) and chemical and organic fertilizer combined application (NPKOM). Specific fertilizer application rates were described in detail in the supplementary material (Table S1). Sampling was conducted at two states: cultivation state (late rice stage [LRS] and early rice stage [ERS]) and natural states (fallow stage, FS) in October 2022, July 2023 and March 2023, respectively (Fig. S1). These three time points were chosen because the aboveground plants are at maturity and the relationship between plants, microbes, and SOM molecules is less disturbed by artificial activity. The experiment consisted of 4 fertilization regimes and × 3 tillage stages with 3 replicates, each replicate was carried out in 5.5 × 5.5 m soil blocks. Three soil cores (6 cm in diameter) were randomly collected (a total of 36 soil samples) from each block. Fresh soil collected was divided into two sub-samples, one was immediately sent to the laboratory and stored at -40 °C, and the other was air-dried for the determination of soil properties.
2.2 DNA extraction, PCR, and Illumina sequencing
FastDNA SPIN Kit was used to extract bacterial DNA from 0.5 g of soil (MP Biomedicals, USA) following the manufacturer’s protocol. The prokaryotic 16S rRNA gene V4-V5 variable region was amplified with 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′) universal primers. Fungal richness was determined via full-length internal transcribed spacer (ITS) amplicon sequencing using the primers ITS1 (5′-GGAAGTAAAAGTCGTAACAAGG-3′)/ITS5F (5′-GCTGCGTTCTTCATCGATGC-3′). Bioinformatic processing was performed as explained above. Raw sequence data were processed within the QIIME2 environment (release 2021.8), denoising sequences with the available DADA2 pipeline. Representative sequences of the generated bacterial and fungal ASVs were aligned against the SILVA 138 database and the UNITE reference database using an open-reference Naïve Bayes feature classifier, respectively. Details are in the Supplemental Information.
2.3 SOM extraction and FT-ICR-MS analysis
SOM was extracted from soil samples (2 g) using ultrapure water at a soil: water ratio of 1:10 and then shaken for 2 h at room temperature in a horizontal shaker (repeated three times). The solution was centrifuged at 2800 g for 20 min and filtered through a 0.45 μm membrane filter. The supernatant was freeze-dried and stored at -40℃. The remaining soil was rinsed with 6 mL of methanol (HPLC grade; Merck, Germany) + 12 mL of CHCl3 (HPLC grade; Merck, Germany) and transferred to a PTFE tube, shaken for 1 h and then centrifuged at 2800 g for 10 min. Mix the SOM organic extract with the corresponding SOM water extract. The mixtures were dried under vacuum centrifugation, resuspended in 1 mL of methanol, vortexed for 10 s, and then centrifuged at 10,000 rcf for 5 min.
Deuterated octadecanoic acid was added to the sample as an internal standard at a dose of 15 μL per mL of sample (5 × 10–7 mol/L). The ESI FT-ICR MS (Bruker, Billerica, MA, USA) was equipped with a 9.4 T actively shielded superconducting magnet interfaced with negative ion mode electrospray ionization. Each sample was injected into the ESI source using a syringe pump at a rate of 180 μL/h. The polarization voltage was 4.0 kV. Capillary column introduction and exit voltages were 4.5 kV and 320 V, respectively. Ions accumulate in the hexapole for 0.001 s before transfer to the ICR cell. The m/z range was 150–800 Da. A 4 M word size was selected for time domain signal acquisition. The signal-to-noise ratio and dynamic range were enhanced by accumulating 128-fold domain FT-ICR transients. Details of data analysis are included in the Supplementary Information.
2.4 Root exudates extraction and UHPLC-MS/MS Analysis
The abundance of plants in the plots was counted before the collection of root exudate. Plants from each plot were mixed into one sample for root exudate collection based on the species abundance ratio. Harvest the entire plant's roots and promptly rinse them with flowing deionized water to eliminate soil residues. Submerge the complete root system in a plastic container containing 20–50 mL of deionized water, with the specific volume depending on the root system's size. Cover the container with aluminum foil to induce darkness around the roots. Maintain the roots in water under controlled climatic conditions, mirroring those of plant growth, for 24 h. Subsequently, collect the root exudate solution, freeze-dry it into a lyophilized powder, accurately measure the weight, and store it in a 1.5 mL centrifuge tube.
To compare the effects of litter leachate and root exudates on the formation of SOM molecules, we collected and measured the diversity of litter leachate. For the extraction of litter leachate, we collected and mixed plant leaves from the fallow stage of each block according to their biomass-specific gravity. The litter from each block was mixed and put into 0.02 mm mesh sizes litter decomposition bags and buried in the ground. After three months, the soil was rinsed off and the litter leachate was extracted according to the root exudate extraction method described above. Details of the test methods are in the Supplementary Information.
2.5 Microcosm experiment
In order to avoid the influence of various factors in the field on the experimental results, we established a microcosmic experiment to verify the contribution of biodiversity to the diversity of SOM molecules. The root exudates diversity validation test was set up with five treatments, and each treatment was set up with two different plant species, with specific information in the Supplementary Information (Table S2). We extracted microbial suspensions from fresh soil and obtained microbial suspensions with 1, 10–1, 10–2, 10–4 and 0 by gradient dilution. Four root exudates mixtures were added before the start of incubation, and then the microbial suspension was added to sterilized soil for three months in a constant temperature incubator at 37 °C. SOM molecular diversity, microbial diversity, and root exudates diversity of the soil were determined at the end of the incubation. Details are in the Supplementary Information.
2.6 Statistical analyses
The diversity index was calculated in the “vegan” package (R Core Team. 2018). SOM molecular, root exudate structure and microbial community composition were assessed using Principal Coordinate Analysis. Redundancy analysis and hierarchical partitioning analysis were used to characterize the effect of biodiversity on the SOM molecular structure. We performed Procrustes and mantel tests to investigate the association between microorganisms and SOM molecules. SparCC was applied to construct a dichotomous network of root exudate, microbes-SOM molecules. When the correlation threshold in network was |ρ|> 0.3 and p < 0.05, we considered that there was a potential interaction between root exudate molecules, microorganisms and SOM molecules. In addition, we selected the microorganisms and root exudate molecules that were included in the network and performed the SparCC network analysis between the two to explore the interactions between the root exudate molecules and the microorganisms. Then, we added these links to the microbe, root exudate-SOM molecule network. The association between plant communities and root exudates was characterized by co-occurrence networks. We found potential associations between plant communities and root exudate molecules by a “pairwise” approach. With the “psych” and “reshape2” packages, we retained links with r > 0.5 and p < 0.05 to represent the relationship between plant production of root exudate molecules. Detailed information is in the Supplementary Information.
3 Results
3.1 Variations of the diversity and composition of biological communities and SOM molecules in different growing stages
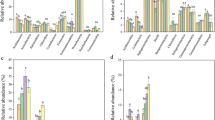
The richness index was used to characterize changes in diversity of biotic communities and SOM molecules across fertilization regimes and cropping stages. In fallow stage, the diversity of SOM molecules, root exudate molecules and bacteria increased by 45.70%–85.36%, 171.67%–177.38% and 13.78%–19.14%, respectively, as compared to rice cultivation stages, whereas the fungal diversity decreased by 35.58%–39.40% (Figs. 1 and S2). Fertilization practices were also found to significantly increase the diversity of SOM molecules (F = 3.45, p < 0.05), plants (F = 8.81, p < 0.001), bacteria (F = 5.63, p < 0.01) and fungi (F = 21.13, p < 0.001). The cropping stage had a greater effect on the SOM molecular diversity than fertilization regime (Fig. S6 and Table S3), indicating the important impacts of aboveground plant community. Plant communities mediate the process of SOM molecular diversity formation through root exudates and litter leachates. Thus, we further compared the molecular diversity of root exudates and litter leachates during the fallow period (Fig. S2). It was found that the molecular diversity of litter leachates was lower than that of root exudates and was limitedly affected by fertilization regime (F = 1.61, p > 0.05). Plant root exudates received a significant effect of the cropping stage (F = 343.06, p < 0.001) and were not sensitive to the fertilization regime (F = 1.74, p > 0.05).
The dominant plant species changed across tillage stages. Rice (Oryza sativa) was the dominant species in the cropping phase, while the weed (Alopecurus aequalis) was the dominant species in the fallow phase (Fig. S3). SOM molecules, plant root exudate molecular types and microbial taxa (phylum) varied with cropping stages (p < 0.001), but were not sensitive to fertilization regimes (p > 0.05; Fig. 2). During the fallow period, for SOM molecules, the relative proportions of lignin increased and carbohydrates decreased (Fig. 2a); for root exudate molecules, the relative proportions of organic acids and derivatives, organoheterocyclic compounds, phenylpropanoids and polyketides increased while the lipid-like molecules decreased (Fig. 2b); and the relative abundance of Ascomycota, Chloroflexi and Proteobacteria increased as compared with rice cropping stages (Fig. 2c, d). In addition, root exudates and litter molecules differed significantly in composition during the fallow period. Root exudates and litter molecules were not sensitive to changes in fertilization regimes (p > 0.05; Figs. S5 and S7). In general, we found that the cropping stage increased the diversity and altered the composition of SOM molecules and biomes. Therefore, in the subsequent analysis, we mainly focused on the synergistic patterns of change in the diversity of biomes and SOM molecules across different cultivation stages.
3.2 The relationship between biodiversity and SOM molecular diversity
We further analyzed the influence of the diversity of bacteria, fungi, plants and their root exudate molecules on the formation of the SOM molecular diversity. The results showed that SOM molecular diversity was positively correlated with bacterial diversity (r = 0.62, p < 0.001), root exudate molecular diversity (r = 0.91, p < 0.001), and plant diversity (r = 0.31, p = 0.06), whereas it was significantly negatively correlated with fungal diversity (r = -0.82, p < 0.001; Fig. 3). Then, we investigated the impacts of the diversity of biomes and their metabolites on SOM molecular structure. Mantel and Procrustes analysis revealed that plant (r = 0.74, M2 = 0.31, p < 0.001), root exudate molecules (r = 0.71, M2 = 0.38, p < 0.001), bacterial (r = 0.68, M2 = 0.60, p < 0.001), and fungal (r = 0.54, M2 = 0.56, p < 0.001) community composition significantly influenced SOM molecules composition, and plant and root exudate molecules had greater effects on SOM than microbes (Fig. S8). The results of RDA and hierarchical partitioning showed that root exudate molecules had the greatest effect (48.48%) on the composition and diversity of SOM (Fig. 4).
Associations between root exudate molecules, bacteria, fungi, and SOM molecules were characterized by constructing co-occurrence networks (Fig. S9). The number of nodes but not the number of links increased in fallow stage compared to the rice cultivation stages (Table S4). Specifically, the links between root exudate molecules and SOM molecules increased, but the links between fungi and SOM molecules decreased in the fallow stage. The decreased N/P (negative links/positive links) in the network implied that the network of SOM molecules became more unstable during the fallow phase. Meanwhile, the percentage of lignin in SOM molecules increased while lipids and carbohydrates decreased during the fallow stage in the network (Table S5).
3.3 Biological factors driving changes in SOM molecular diversity and composition
Changes in SOM molecule diversity (richness index) were closely related to dynamics between newly produced molecules and consumed molecules. Unique SOM molecules and root exudate molecules from rice cultivation and fallow stages were used to characterize the relationship between newly formed molecules and old consumed molecules. The results showed that 378 SOM molecules and 1258 root exudate molecules were consumed, while 2642 SOM molecules and 1318 root exudate molecules were newly generated from late rice cultivation stage to fallow stage (Fig. S10). The consumed SOM molecules were dominated by lipids and carbohydrates molecules, while the newly-formed SOM molecules were dominated by lipids, lignin, and proteins/amino sugars molecules (Figs. S12 and 5a, b). The consumed root exudate molecules were dominated by lipid-like molecules, while the imported new root exudate molecules were dominated by organoheterocyclic compounds, organic acids/derivatives molecules (Fig. S13). From fallow stage to early rice cultivation stage, we found that 1413 SOM molecules and 1314 root exudate molecules were consumed, and 551 SOM molecules and 1253 root exudate molecules were newly formed (Fig. S11). The consumed SOM molecules were dominated by lipids, lignin, and proteins/amino sugars, while the new SOM molecules formed were dominated by lipids and carbohydrates (Figs. S14 and 5c, d). The consumed root exudate molecules were dominated by organoheterocyclic compounds, organic acids and derivatives, while the imported new root exudates were dominated by lipid-like molecules. During the fallow stage, root exudates dominated by organoheterocyclic compounds and organic acids derivatives recruited Ascomycota, Chloroflexi and Proteobacteria, and metabolized to form a consortium of SOM molecules dominated by lipids, lignin and proteins/amino sugars (Fig. 5b and c). During the cultivation stages, root exudate molecules dominated by lipid-like molecules recruited Basidiomycita, Rozellomycota, Proteobacteria, and formed a consortium dominated by lipids and carbohydrates molecules (Fig. 5a and d).
Network plot showing the association between newly produced (b, d) and consumed (a, c) biotic factors with SOM molecules in neighboring phenological stages. Nodes shape represent different groups. Different colors represent different classifications. A connection stands for a strong (Spearman's ρ > 0.30) and significant (p < 0.05) correlation
To further validate the unique SOM molecular consortium and biological community formation processes under different tillage states, we constructed temporal SpaCC bipartite network. From the cultivation stage to the fallow stage, nodes (541) and links (345) were higher in the new SOM molecule formation network than in the old SOM molecule consumption network (nodes: 251, links: 149). Bacteria with produced and consumed SOM molecules had the highest number of links, 148 and 81, respectively (Table S6); from the fallow stage to the cultivation stage, nodes (752) and links (700) were higher than in the new SOM molecule formation network (nodes: 448, links: 405). Root exudate molecules with produced and consumed SOM molecules had the most links, 128 and 343, respectively (Table S7). In addition, we compared the community structure of consumed and newly produced molecules in the network. The compositions of newly produced root exudate molecules, bacterial community and SOM molecules from late cultivation stage to the fallow stage were not significantly different from those consumed from the fallow stage to early cultivation stage. This verifies that the fallow stage recruited a different biological community from the tillage stages and thus formed a unique consortium of SOM molecules (Fig. S15). Overall, more kinds of root exudate molecules were generated in fallow stage, and more kinds of SOM molecules retained in soil as the increasing number of SOM molecules in fallow stage exceeded the consuming number in the rice growing seasons.
Plant community succession resulted in changes in root exudate molecular composition across tillage stages. Lipid-like molecules in root exudate were produced mainly by Oryza sativa, Setaria viridis and Carex heterostachya at rice cultivation stage (Fig. 6a and d); whereas organoheterocyclic compounds and organic acids/derivatives molecules were produced mainly by Hemistepta lyrata, Lapsanastrum apogonoides, Cyperus iria and Rotala indica at fallow stage. Organoheterocyclic compounds and organic acids/derivatives molecules were consumed by various microbial taxa from fallow stage to early rice stage (Fig. 6b and c). The study showed that Basidiomycita, Ascomycota, Acidobacteria, Chloroflexi and Proteobacteria were observed to be involved. Differently, Rozellomycota, Desulfobacterota, Acidobacteria, Chloroflexi, Proteobacteria and Ascomycota exhibited a preference for lipid-like molecules at rice cultivation stage. Our study showed that root exudate molecules had a greater influence than microbes on the formation of SOM molecular diversity and compositional patterns. However, the field experiment was coupled with different cultivation stages and fertilization practices, and influenced by many environment factors. To avoid the interference of these factors, we arranged microcosmic experiments to verify the contribution of root exudate molecular diversity and microbial diversity to the formation of SOM molecular diversity. Microcosm experiment demonstrated that SOM molecular diversity had significant positive correlations with root exudate molecular diversity and microbial diversity (Fig. S16). This positive feedback was strong at low thresholds and weakened above thresholds (1770.60–1775.03). In addition, the independent effect of root exudate molecular diversity on SOM molecular diversity was higher than that of microbial diversity.
4 Discussion
4.1 SOM molecular diversity increased during the fallow stage
The process of SOM molecular diversity formation and the biological community are closely related; however, their coupling relationship is uncharacterized. Our study provides insight into the relationship between biomes and SOM molecule formation in agroecosystems, and provides theoretical support for weed management in fallow state. The results supported our hypothesis (1) that SOM molecules, bacteria, plants and their root exudate molecular diversity significantly increased during the fallow period; however, fungal diversity decreased (Fig. 1). Fertilizer application increased SOM molecules and biodiversity, but the enhancement effect was lower compared to the cropping period. Although exogenous inputs increased biodiversity, their effects on root exudate molecules were not significant. Root exudate molecules were the primary influential factor on SOM molecules, thus, the effect of exogenous inputs on the diversity of SOM molecules was limited (Figs. 4 and S8). The paddy fields returned to a natural ecosystem with no anthropogenic disturbance in the fallow stage, and aboveground weeds rapidly colonized and encroached on the niche through the chemosensory effects of root exudate molecules (Korenblum et al. 2022). On the one hand, competition among plants promotes diversity (Descombes et al. 2020); on the other hand, competition also leads to niche differentiation, which results in the coexistence of more species and mediates an increase in root exudate molecular diversity (Pastore et al. 2021). It is well known that such chemical sequences in the rhizosphere recruit microbial communities with a preference for this (Han et al. 2020; Feng et al. 2023). Bacteria preferred an accessible resource (root exudate molecules) over fungi, facilitating rapid colonization of the bacterial community. Competitive interactions between microorganisms also inhibited fungal growth, thus reducing fungal diversity. In addition, fungal diversity increased in high-humidity habitats, which explained why fungal diversity was higher during rice cultivation relative to dry and cold fallow stage (Zhang et al. 2021).
4.2 Increased SOM molecules are mainly derived from (weed) root exudates during the fallow stage
Changes in SOM molecular diversity (richness index) result from a shift in the dynamic balances between the production of new molecules and the consumption of old molecules (Figs. S10 and S11). We constructed a temporal network to characterize the potential biological factors underlying the changes in SOM molecular diversity and composition during the succession from late to fallow to early rice stage. Elevated SOM molecular diversity during the fallow period was attributed to a faster rate of newly produced molecules than consumed molecules. Anthropogenic disturbances such as fertilizers and pesticides are reduced and weeds colonize rapidly at fallow stage. Different plant species produce differential root exudate molecules, and these metabolic molecules recruit microorganisms with different preferences (McLaughlin et al. 2023; Zhalnina et al. 2018; Preece and Peñuelas 2020). Thus, increased above-ground plant diversity usually leads to an explosion of bacterial diversity in the below-ground (Oelmann et al. 2021). More newly produced root exudate molecules and recruited bacteria mediated the increase in SOM molecules during the fallow stage compared to the rice cropping stages. Although fungi were closely associated with SOM molecule formation, the link between root exudate molecules and bacteria with SOM molecules exceeded 80%, which diluted the effect of fungi on SOM molecules (Table S6 and S7).
The composition of the SOM molecular consortium and biological communities also underwent succession at different cultivation stages. In the temporal network, there was a high degree of consistency in the production of new SOM molecules from the late rice stage to the fallow stage and the consumption of old SOM molecules from the fallow stage to the early rice stage (Figs. 5 and S15). Similarly, the same pattern was observed in root exudate molecules and bacterial communities. The SOM molecules were dominated by lipid and lignin molecules in the fallow stage, whereas lipids and carbohydrates molecules dominated in the cultivation stages (Figs. 5 and 6). Organoheterocyclic compounds and Organic acids/derivatives dominated the root exudates molecules in the fallow stage, whereas lipid-like molecules dominated in the cultivation stages. This suggests that the diversity and composition of SOM molecules are predictable and regulated by biological factors that evolve with the tillage stage. In addition, more than 80% of the new SOM molecules produced (in the fallow stage) were retained in the soil during the succession from the fallow stage to the early rice stage. More than 90% of the newly recruited microorganisms (in the fallow stage) were present in the rice cultivation stage, however, the newly produced root exudate molecules were almost absent. On the one hand, root exudates turn over quickly in the soil and are difficult to retain for long periods (Panchal et al. 2022); on the other hand, these SOM molecules that are able to remain in the soil for long periods are closely associated with microbial turnover (Domeignoz-Horta et al. 2021). This is because microbial preferences for SOM molecules promote changes in SOM molecular composition.
The results also confirmed our hypothesis (2) that increased diversity of bacterial and root exudate molecules drives the dispersal pattern of SOM molecules. Root exudates had the greatest impact on the SOM molecular diversity and composition (Fig. 5). Root exudate molecules affect SOM molecules through two pathways: new SOM molecules are formed by physical mineralization and chemical decomposition after directly entering the soil, and the formation of microbial-derived C assimilated by microorganisms, thus accumulating in the soil (Liang et al. 2017). Previous studies have suggested that microorganisms consume plant-derived organic C to sustain catabolism and anabolism, converting SOC to respiratory CO2 (Liu et al. 2023a, b). In this process, microorganisms reduce SOM molecular diversity. For example, microbial diversity generally decreases along the soil profile, and the decrease in SOM molecular diversity along the profile may be related not only to assimilation but also to changes in microbial diversity (Gao et al. 2023; Davenport et al. 2023). The assimilation of “microbial funnels” reduces SOM molecular diversity, however, when the number of “microbial funnels”, namely, the microbial diversity, increases, the SOM molecular diversity may also show a divergent pattern. Thus, changes in SOM molecular diversity mediated by microbial assimilation and diversity are dynamic processes. When microbial diversity induces more metabolites than substrates for anabolic catabolism, SOM molecular diversity shows a divergent pattern. Aboveground plant communities reshape microbial communities and SOM molecular consortium patterns through root exudates and litter leachates. However, the contribution of root exudates and litter leachates molecular diversity to SOM molecular diversity has not been reported. In the present study, we found that the molecular diversity of litter leachates was significantly lower than that of root exudates and had no significant effect on SOM molecular diversity by comparing the metabolic molecular diversity of root exudates and litter leachates. This suggests that root exudate is a more important factor of plant-derived SOM molecules than litter.
Changes in above-ground plant diversity and composition were most readily observed at different stages of cultivation. The composition of plants and their root exudate molecules in the rice cultivation stages (early and late rice stages) was similar as no dispersion pattern was exhibited in the PCOA1 axis. In contrast, the composition and structure of dominant plants and their root exudate molecules changed significantly in the fallow stage compared to the rice cultivation stages. Selection effect theory suggests that dominant species that colonize a habitat are more capable of influencing the structure and function of an ecosystem. Rice (Oryza sativa) was the dominant species during the cultivation stage, and its root exudates were dominated by lipids that recruited Rozellomycota, Desulfobacterota, Acidobacteria, Chloroflexi, Proteobacteria and Ascomycota during the rice cultivation stages (Figs. 6 and S13). Through physicochemical processes and microbial metabolism, they were converted into carbohydrates, lipid-like SOM molecules sequestered in the soil; aboveground plant communities underwent succession with Hemistepta lyrata, Lapsanastrum apogonoides, Cyperus iria and Rotala indica becoming the dominant species in the fallow period. Plant species turnover resulted in the conversion of root exudate components to Organoheterocyclic compounds and Organic acids/derivatives, and reshaped the microbial community. Recruited microbes were dominated by Basidiomycita, Ascomycota, Acidobacteria, Chloroflexi and Proteobacteria. Synergistic changes in plants and microbes resulted in the transformation of SOM molecules towards lipid and lignin molecules. In summary, changes in the diversity and composition of SOM molecules were co-regulated by aboveground plant communities and belowground microbial communities. Succession of dominant species in the plant community led to changes in the type of root exudates and reshaped the microbial community. In brief, dynamic changes in biological factors determine the fate of SOM molecular diversity and composition.
4.3 Limitations
We did not discuss the effect of season on the formation of SOM molecules, and the effect of season on the formation of SOM molecules may be limited. Because changes in the diversity and composition of SOM molecules were not observed to differ between the early and late rice stages. The FTICR-MS can only measure part of the SOM due to the limitation of the quality window. We can only qualitatively determine the origin of SOM molecules but not quantitatively. Future studies should combine FT-ICRMS and biomarker methods to elucidate the origin of SOM molecules from both quantitative and qualitative perspectives. In addition, the development of causal inference networks can help to understand the transformation process of SOM molecules rather than through simple correlation networks.
5 Conclusions
Continuing technological developments have pushed us to explore SOM formation and transformation at a deeper level. We elucidated the interactions between SOM molecules and biomes by the combined multi-omics analyses. Agricultural fallow maintains weed diversity and mitigates the negative effects of long-term tillage on SOM molecular diversity. Biological factors, especially root exudate molecules, promote SOM molecular diversity. Plant community succession leads to the conversion of root exudate components to predominantly Organoheterocyclic compounds and Organic acids/derivatives during fallow stage. Unique microbial communities (Chloroflexi and Proteobacteria) are recruited, which in turn cause the transformation from carbohydrates, lipid-like SOM molecules to lipid-like and lignin-like SOM molecules. Thus, changes in above-ground plant and below-ground microbial communities determine the fate of SOM molecules. Our study provides a framework for future insights into SOM succession at the molecular level in agricultural soils.
Availability of data and materials
Data will be made available on request.
References
Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, Da Rocha UN, Chakraborty R, Bowen BP, Karaoz U, Cadillo-Quiroz H, Garcia-Pichel F, Northen TR (2015) Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun 6:8289. https://doi.org/10.1038/ncomms9289
Chen S, Feng X, Lin Q, Liu C, Cheng K, Zhang X, Bian R, Liu X, Wang Y, Drosos M, Zheng J, Li L, Pan G (2022) Pool complexity and molecular diversity shaped topsoil organic matter accumulation following decadal forest restoration in a karst terrain. Soil Biol Biochem 166:108553. https://doi.org/10.1016/j.soilbio.2022.108553
Córdova SC, Olk DC, Dietzel RN, Mueller KE, Archontouilis SV, Castellano MJ (2018) Plant litter quality affects the accumulation rate, composition, and stability of mineral-associated soil organic matter. Soil Biol Biochem 125:115–124. https://doi.org/10.1016/j.soilbio.2018.07.010
Davenport R, Bowen BP, Lynch LM, Kosina SM, Shabtai I, Northen TR, Lehmann J (2023) Decomposition decreases molecular diversity and ecosystem similarity of soil organic matter. Proc Natl Acad Sci 120(25):e2303335120. https://doi.org/10.1073/pnas.230333512
Descombes P, Pitteloud C, Glauser G, Defossez E, Kergunteuil A, Allard PM, Rasmann S, Pellissier L (2020) Novel trophic interactions under climate change promote alpine plant coexistence. Science 370(6523):1469–1473. https://doi.org/10.1126/science.abd7015
Domeignoz-Horta LA, Shinfuku M, Junier P, Poirier S, Verrecchia E, Sebag D, DeAngelis KM (2021) Direct evidence for the role of microbial community composition in the formation of soil organic matter composition and persistence. ISME Commun 1:64. https://doi.org/10.1038/s43705-021-00071-7
Gao G, Li G, Liu M, Li P, Liu J, Ma S, Li D, Petropoulos E, Wu M, Li Z (2023) Changes in soil stoichiometry, soil organic carbon mineralization and bacterial community assembly processes across soil profiles. Sci Total Environ 903:166408. https://doi.org/10.1016/j.scitotenv.2023.166408
Han Q, Ma Q, Chen Y, Tian B, Xu L, Bai Y, Chen W, Li X (2020) Variation in Rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J 14(8):1915–1928. https://doi.org/10.1038/s41396-020-0648-9
Hu A, Choi M, Tanentzap AJ, Liu J, Jang KS, Lennon JT, Wang J (2022a) Ecological networks of dissolved organic matter and microorganisms under global change. Nat Commun 13(1):3600. https://doi.org/10.1038/s41467-022-31251-1
Hu A, Meng F, Tanentzap AJ, Jang KS, Wang J (2022b) Dark matter enhances interactions within both microbes and dissolved organic matter under global change. Environ Sci Technol 57(1):761–769. https://doi.org/10.1021/acs.est.2c05052
Huang W, Kuzyakov Y, Niu S, Luo Y, Sun B, Zhang J, Liang Y (2023) Drivers of microbially and plant-derived carbon in topsoil and subsoil. Global Change Biol 29(22):6188–6200. https://doi.org/10.1111/gcb.16951
Jones AR, Dalal RC, Gupta VV, Schmidt S, Allen DE, Jacobsen GE, Bird M, Grandy AS, Sanderman J (2023) Molecular complexity and diversity of persistent soil organic matter. Soil Biol Biochem 184:109061. https://doi.org/10.1016/j.soilbio.2023.109061
Kellerman AM, Dittmar T, Kothawala DN, Tranvik LJ (2014) Chemodiversity of dissolved organic matter in lakes driven by climate and hydrology. Nat Commun 5(1):3804. https://doi.org/10.1038/ncomms4804
Koch BP, Dittmar T, Witt M, Kattner G (2007) Fundamentals of molecular formula assignment to ultrahigh resolution mass data of natural organic matter. Anal Chem 79(4):1758–1763. https://doi.org/10.1021/ac061949s
Korenblum E, Massalha H, Aharoni A (2022) Plant–microbe interactions in the rhizosphere via a circular metabolic economy. Plant Cell 34(9):3168–3182. https://doi.org/10.1093/plcell/koac163
Li X, Chen Q, He C, Shi Q, Chen S, Reid B, Zhu Y, Sun G (2018) Organic carbon amendments affect the chemodiversity of soil dissolved organic matter and its associations with soil microbial communities. Environ Sci Technol 53(1):50–59. https://doi.org/10.1021/acs.est.8b04673
Liang C, Schimel JP, Jastrow JD (2017) The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol 2(8):1–6. https://doi.org/10.1038/nmicrobiol.2017.105
Liu S, Hao Z, Gao L, Fan LH, Yang F, Arash Z, Li M (2023) Spatial variation and relationship between soil dissolved organic matter and bacterial community in urban greenspaces. Carbon Res 2:13. https://doi.org/10.1007/s44246-023-00047-3
Liu M, Wei Y, Lian L, Wei B, Bi Y, Liu N, Yang G, Zhang Y (2023) Macrofungi promote SOC decomposition and weaken sequestration by modulating soil microbial function in temperate steppe. Sci Total Environ 899:165556. https://doi.org/10.1016/j.scitotenv.2023.165556
Ma L, Wang X, Fang Y, Vancov T, Jin X, Gao Q et al (2024) No-tillage farming for two decades increases plant- and microbial-biomolecules in the topsoil rather than soil profile in temperate agroecosystem. Soil Till Res 241:106108. https://doi.org/10.1016/j.still.2024.106108
McLaughlin S, Zhalnina K, Kosina S, Northen TR, Sasse J (2023) The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat Commun 14(1):1649. https://doi.org/10.1038/s41467-023-37164-x
Nebbioso A, Piccolo A (2013) Molecular characterization of dissolved organic matter (DOM): a critical review. Anal Bioanal Chem 405:109–124. https://doi.org/10.1007/s00216-012-6363-2
Oelmann Y, Lange M, Leimer S, Roscher C, Aburto F, Alt F, Bange N, Berner D, Boch S, Boeddinghaus RS, Buscot F, Dassen S, De Deyn G, Eisenhauer N, Gleixner G, Goldmann K, Hölzel N, Jochum M, Kandeler E, Wilcke W (2021) Above- and belowground biodiversity jointly tighten the P cycle in agricultural grasslands. Nat Commun 12(1):4431. https://doi.org/10.1038/s41467-021-24714-4
Panchal P, Preece C, Peñuelas J, Giri J (2022) Soil carbon sequestration by root exudates. Trends Plant Sci 27(8):749–757. https://doi.org/10.1016/j.tplants.2022.04.009
Pastore AI, Barabás G, Bimler MD, Mayfield MM, Miller TE (2021) The evolution of niche overlap and competitive differences. Nat Ecol Evol 5(3):330–337. https://doi.org/10.1038/s41559-020-01383-y
Preece C, Peñuelas J (2020) A return to the wild: Root exudates and food security. Trends Plant Sci 25(1):14–21. https://doi.org/10.1016/j.tplants.2019.09.010
Qi Y, Xie Q, Wang JJ, Liu CQ, Fu PQ (2022) Deciphering dissolved organic matter by Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS): from bulk to fractions and individuals. Carbon Res 1:3. https://doi.org/10.1007/s44246-022-00002-8
R Core Team. R. (2018) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at https://www.R-project.org/
Schmidt MW, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, Trumbore SE (2011) Persistence of soil organic matter as an ecosystem property. Nature 478(7367):49–56. https://doi.org/10.1038/nature10386
Séneca J, Söllinger A, Herbold CW, Pjevac P, Prommer J, Verbruggen E, Sigurdsson BD, Peñuelas J, Janssens IA, Urich T, Tveit AT, Richter A (2021) Increased microbial expression of organic nitrogen cycling genes in long-term warmed grassland soils. ISME Commun 1(1). https://doi.org/10.1038/s43705-021-00073-5
Wen S, Hu A, Jiang S, Han L, Jang K, Tanentzap AJ, Zhong J, Wang J (2024) Temperature sensitivity of organic carbon decomposition in lake sediments is mediated by Chemodiversity. Global Change Biol 30(2):e17158. https://doi.org/10.1111/gcb.17158
Wu M, Li P, Li G, Liu K, Gao G, Ma S, Qiu C, Li Z (2022) Using potential molecular transformation to understand the molecular trade-offs in soil dissolved organic matter. Environ Sci Technol 56(16):11827–11834. https://doi.org/10.1021/acs.est.2c01137
Wu M, Li P, Li G, Petropoulos E, Feng Y, Li Z (2021) The chemodiversity of paddy soil dissolved organic matter is shaped and homogenized by bacterial communities that are orchestrated by geographic distance and Fertilizations. Soil Biol Biochem 161:108374. https://doi.org/10.1016/j.soilbio.2021.108374
Wurz A, Tscharntke T, Martin DA, Osen K, Rakotomalala AA, Raveloaritiana E, Andrianisaina F, Dröge S, Fulgence TR, Soazafy MR, Andriafanomezantsoa R, Andrianarimisa A, Babarezoto FS, Barkmann J, Hänke H, Hölscher D, Kreft H, Rakouth B, Guerrero-Ramírez NR, Grass I (2022) Win-win opportunities combining high yields with high multi-taxa biodiversity in tropical agroforestry. Nat Commun 13(1):4127. https://doi.org/10.1038/s41467-022-30866-8
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, Firestone MK, Northen TR, Brodie EL (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in Rhizosphere microbial community assembly. Nat Microbiol 3(4):470–480. https://doi.org/10.1038/s41564-018-0129-3
Zhang Z, Zhang J, Jiao S (2021) Fungi show broader environmental thresholds in wet than dry agricultural soils with distinct biogeographic patterns. Sci Total Environ 750:141761. https://doi.org/10.1016/j.scitotenv.2020.141761
Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O’Neill RV, Tiedje JM (2002) Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68(1):326–334. https://doi.org/10.1128/AEM.68.1.326-334.2002
Funding
This work was supported by the National Natural Science Foundation of China (42077021), Independent Deployment Project of Institute of Soil Science, Chinese Academy of Sciences (ISSAS2403), and Strategic Priority Research Program of the Chinese Academy of Sciences (XDA0440202).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Guozhen Gao. The first draft of the manuscript was written by Guozhen Gao. The manuscript was reviewed and edited by Jian Cui, Ming Liu, Pengfa Li, Meng Wu and Zhongpei Li. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Handling Editor: Fengchang Wu.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, G., Li, P., Liu, M. et al. Diverse biological communities promote SOM molecular diversity and compositional transformations during natural fallow stage in paddy fields. Carbon Res. 3, 65 (2024). https://doi.org/10.1007/s44246-024-00149-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44246-024-00149-6