Abstract
Corynebacterium glutamicum is widely used in the production of amino acids. C. glutamicum possesses seven sigma factors, among which SigD is responsible for the transcription of genes involved in the synthesis of mycolic acid (MA) and its derivatives, the unique cell envelope of C. glutamicum. To understand the influence of MA synthesis on amino acid production and membrane phenotype of C. glutamicum, the expression of sigD gene and some mycolyltransferase genes, i.e., cmt1, cop1 and cmt2, were regulated by several growth-regulated promoters in this study. Except for 2 mutant strains of Pcg3096-sigD and Pcg1633-cop1, the growth and 4-hydroxyisoleucine (4-HIL) titer of most modified strains did not change significantly. But the 4-HIL titer of PodhI-sigD strain increased by 20.73% (142.45 ± 3.69 mM) compared to that of control strain (117.99 ± 0.34 mM). After it was cultivated in bioreactor, 4-HIL titer reached 372.56 mM. This may be caused by the increase of MA content, and 17% decrease of cell hydrophobicity and 12% increase of membrane permeability were observed at the exponential phase. In conclusion, we proved that rearrangements in regulation of sigD expression contributed to the improved fermentation performance of C. glutamicum and promoted 4-HIL production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes is one of the four major non-communicable diseases worldwide. It is usually treated with oral medicines and insulin injection. In recent decades, fenugreek has been shown to have preventive and palliative effects on type I and type II diabetes, because the 4-hydroxyisoleucine (4-HIL) in fenugreek showed unique activity in stimulating insulin secretion as well as ameliorating insulin resistance [1]. 4-HIL was originally extracted from fenugreek seeds. The anti-diabetic activity of 4-HIL depends on glucose concentration, so the side effects caused by other chemicals for treating diabetes, such as hypoglycemia and gastrointestinal discomfort can be avoided [2]. 4-HIL can also enhance insulin sensitivity, thereby can be used for the treatment of type II diabetes [3]. Considering the limited amount of 4-HIL that can be extracted from plants, fermentation methods have been adopted to produce 4-HIL [4]. L-isoleucine dioxygenase (IDO) was found in Bacillus thuringiensis that catalyzes the C-4 hydroxylation of L-isoleucine (Ile) to form 4-HIL [5].
Corynebacterium glutamicum is a Gram-positive bacterium widely used for the production of various amino acids, and its product is generally recognized as safe [6]. In a previous study, ido gene was introduced into C. glutamicum ssp. lactofermentum SN01, resulting in a de novo synthesis of 65.44 ± 2.27 mM 4-HIL from its own produced Ile [7]. Besides Ile, α-ketoglutaric acid (α-KG) and O2 are also required for the synthesis of 4-HIL in IDO reaction (Fig. 1). Recently, to increase the supply of co-substrates α-KG and O2 in an Ile responsive manner, the modified Ile biosensor Lrp-PbrnFEN was used to co-express the ido gene with odhI and vgb genes, increasing the 4-HIL titer to 135.3 mM [8]. odhI gene encodes the inhibitor of α-KG dehydrogenase complex and vgb gene encodes the Vitreoscilla hemoglobin VHb. But L-lysine (Lys) was still remained as the main by-product. To reduce the production of Lys, the Lys-OFF riboswitch was integrated before dapA gene, the key gene of Lys biosynthetic pathway, and Lys content decreased greatly in the resulting D-RS strain [9].
The C. glutamicum genome codes for 7 sigma subunits (factors) of RNA polymerase: primary sigma factor SigA (σA), primary-like SigB and 5 other alternative sigma factors (SigC, SigD, SigE, SigH and SigM). Each sigma factor is responsible for recognizing promoters of genes belonging to its regulon (sigmulon) involved in specific functions of the cell [10]. Among them, SigD (σD) is mainly responsible for recognizing promoters involved in membrane-associated genes such as mycolic acid (MA) synthesis genes (fadD2, pks), mycolyltransferase genes (cop1, cmt1, cmt2, cmt3) [11], L, D-transpeptidase gene (lppS), and others [12]. Recently, it has been shown that overexpression of sigD influences the cell envelopes of C. glutamicum strain, reduces the formation of foams during fermentation, and affects the expression of genes related to MA synthesis [13]. C. glutamicum and other Corynebacteria have special cell envelopes with a peptidoglycan layer, arabinogalactan [14], an MA layer, and a S layer sequentially arranged on the outside of its phospholipid bilayer (Fig. 2). The MA layer is composed of trehalose monocorynomycolate (TMCM) and trehalose dicorynomycolate (TDCM) formed by mycolic acid and trehalose. Comparison of the transcriptome data of the Ile-producing strain C. glutamicum SN01 [15] with that of C. glutamicum ATCC13032 showed that the sigD and many genes of SigD regulon, such as some mycolyltransferase genes were weakly expressed in SN01 during the exponential phase (Table S1). It was found that regulating the expression of some genes of SigD regulon altered the cell envelope properties of C. glutamicum ATCC13032 and affected cell growth [13]. Changes in MA content also have an effect on amino acid synthesis and efflux in C. glutamicum [13, 16]. The MA content of the cell wall permeability barrier was implicated in the amino acid excretion process [17].
C. glutamicum cell envelope and its mycolic acid synthesis pathway. The enzymes shown in red are regulated by SigD factors during gene transcription. AG: arabinogalactan, CA-CoA: carboxylated acyl-CoA, CMk: keto corynomycolic acid, FA: fatty acid, MA: mycomembrane, PB: phospholipid bilayer, PG: peptidoglycan, SL: S layer, TDCM: trehalose dicorynomycolate, TMCM: trehalose monocorynomycolate, TRE: trehalose
Considering that the expression of sigD and SigD regulon are usually downregulated at the stationary phase, and the sigD expression in SN01 was quite weak, several growth-regulated promoters (GRP) with different strengths such as PCP_2836 [18] were used here in D-RS strain to increase the expression strength of sigD gene during the exponential phase. The changes of membrane properties, cell growth, and 4-HIL production in these modified strains were then analyzed. Next, considering the importance of mycolyltransferase on covalent linkage of MA with other layers and formation of the entire cell envelope, we evaluated the effect of regulation of some crucial mycolyltransferase genes on 4-HIL production and cell envelope properties of C. glutamicum D-RS. Finally, the best strain PodhI-sigD was cultivated for 4-HIL production in the bioreactor.
Materials and methods
Strains, media and culture conditions
The strains used in this study are listed in Table 1. Escherichia coli JM109 was used as a host to construct the integrative plasmids and expression plasmid. E. coli was cultured in LB medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 37 °C, 200 rpm. C. glutamicum D-RS was engineered for synthesizing 4-HIL, while C. glutamicum ATCC13032 was used for amplifying target genes and promoters. C. glutamicum was cultured in LBB medium (18.5 g/L brain-heart infusion, 5 g/L tryptone, 2.5 g/L yeast extract, 5 g/L NaCl) using a rotary shaker at 30 ℃ and 200 rpm [19], and 30 mg/L kanamycin, 10 mg/L chloramphenicol, or 100 g/L sucrose were added to medium as needed.
Modification of promoters in the chromosome of SN01
The primers are showed in Table 2. According to our unreported transcriptome data, the five GRP promoters with the higher strength at the exponential phase and lower strength at the stationary phase, i.e. PodhI, Pcg1206, Pcg1633, Pcg2705, and Pcg3096 (Table S2), were selected to replace the promoter of sigD gene (PsigD, the upstream 157 bp of sigD gene). The pK18mobsacB and CRISPR-Cpf1-assisted genome editing system were used for promoter replacement [20]. The editing plasmid pk18-PsigD::PodhI was constructed by amplifying the upstream homologous arm of sigD, the PodhI promoter, and the downstream homologous arm of sigD using the primer pairs of sigD-U-F/sigD-U-R, PodhI-F/PodhI-R, and sigD-D-F/sigD-D-R, respectively. These fragments were overlapped and ligated into the pK18mobsacB vector. The other editing plasmids were constructed in a similar way. Each of the above constructed editing plasmids was then electrotransformed into C. glutamicum D-RS for replacing the PsigD promoter using the CRISPR-Cpf1-assisted SacB editing system. After single exchange, pCS-assisted double exchange and pCS curing, the corresponding sigD promoter replacement strains, i.e. D-RS PsigD::PodhI, D-RS PsigD::Pcg1206, D-RS PsigD::Pcg1633, D-RS PsigD::Pcg2705, and D-RS PsigD::Pcg3096 were constructed. Pcop1, Pcmt2, and Pcmt1 promoter replacement strains D-RS Pcop1::Pcg1633, D-RS Pcmt1::Pcg1633, D-RS Pcmt2::Pcg1633, D-RS Pcmt2::Ptuf and the D-RS Pcmt1-cmt1::Pcmt1-cmt1 (ATCC13032) were also constructed similarly (Table 1).
Construction of ido and vgb co-expression plasmid and strains
The plasmids are showed in Table 1. The PbrnFE7-ido fragment was amplified from pIL-7IU [9] by primer pairs PbrnFE7-U/ido-R. The PbrnFE7 and vgb fragments were amplified from pIL-7IU and pJYW-4-imi3-vgb [21] by primer pairs PbrnFE7-F/PbrnFE7-R and vgb-F/vgb-R, respectively. Then three fragments were overlapped, digested with BamHI, and ligated into BamHI-digested pJYW-4, generating the plasmid P7P7. The plasmid P7P7 was then transformed into strains D-RS, D-RS PsigD::Pcg1633, D-RS PsigD::Pcg1206, D-RS PsigD::Pcg2705, D-RS PsigD::PodhI, D-RS PsigD::Pcg3096, D-RS Pcop1::Pcg1633, D-RS Pcmt2::Pcg1633, D-RS Pcmt2::Ptuf, D-RS Pcmt1::Pcg1633 and D-RS Pcmt1-cmt1::Pcmt1-cmt1 (ATCC13032) to generate strain RS1, RSS1, RSS2, RSS3, RSS4, RSS5, RSC1, RSC2, RSC3, RSC4 and RSA1, respectively.
Analysis of cell hydrophobicity and permeability
Cell surface hydrophobicity was determined according to the following method [22]. Briefly, cell suspension was collected from the cultivated LBB medium at 8 h and 15 h, washed twice with PBS buffer (0.05 mM, pH 7.4), and resuspended in PBS buffer (0.05 mM, pH 7.4) to a final optical density at 562 nm (OD562) of 0.5 (recorded as OD562A). Then 2 mL xylene was added to 2 mL cell suspension, oscillated for 2 min and stood for 1 h. The OD562 of the water phase (recorded as OD562B) was determined after the xylene was removed. The cell hydrophobicity was calculated by formula (OD562A-OD562B)/OD562A.
The membrane permeability was determined by N-phenyl-1-naphthylamine (NPN) fluorescent probe method [23]. Briefly, cell suspension was collected from the cultivated LBB medium at 8 h and 15 h, washed twice with PBS buffer (0.05 mM, pH 7.4), and resuspended in PBS buffer (0.05 mM, pH 7.4) to a final OD562 of 0.5. Then 80 µL NPN (10 µmol/L) was added to 1.92 mL cell suspension in a 24-well plate. The fluorescence intensity was measured by microplate reader (Excitation wavelength 350 nm, emission wavelength 428 nm, slit width 5 nm), and the cell permeability was expressed by formula fluorescence intensity/OD562.
Chromatography of lipids
Total lipids were extracted according to methods in the literature [24]. Briefly, wet cells were harvested from the fermentation medium, then sequentially resuspended in a mixture of 10 mL CHCl3/CH3OH (1:2, v/v), 10 mL CHCl3/CH3OH (1:1, v/v), and 10 mL CHCl3/CH3OH (2:1, v/v) for 16 h and the supernatants of each suspension were collected and mixed. The CHCl3 and CH3OH was removed from the mixed supernatants by rotary evaporation to obtain the crude lipid extract. Then 10 mL of CHCl3/H2O mixture (1:1, v/v) was added to the crude lipid extract, fully mixed and stratified. After collecting the organic phase, the solvent was removed by rotary evaporation to obtain the total lipid extract. Finally, 50 µL CHCl3/CH3OH (2:1, v/v) was added to dissolve the lipid extract and 2 µL solution was spotted onto thin-layer chromatography (TLC) silica gel plates and chromatographed for 20 min with the spreading agent CHCl3/CH3OH/H2O/ammonia (65:25:1:3, v/v/v/v).
4-HIL fermentation in shake flasks and bioreactor
For producing 4-HIL via fermentation, the recombinant C. glutamicum were streaked onto plates containing the LBB medium and incubated at 30 °C for 48 h. And then the grown bacteria were inoculated in a 500 mL baffled flask containing 40 mL of seed medium and cultivated at 30 °C and 200 rpm for 17 h. The seed cultures were inoculated into a 500 mL baffled flask containing 30 mL of fermentation medium until the final OD562 was 1.8 and then shook at 200 rpm and 30 °C for 144 h by a cyclotron shaker [25]. During the fermentation, 1 mL culture samples were taken regularly to determine the OD562, residual glucose and the Ile and 4-HIL concentration.
For producing 4-HIL via fermentation in a bioreactor, the cultivated culture of C. glutamicum strain RSS4 in shake flask were inoculated at 10% volumetric inoculum into a 2 L fermenter (T&J-Minbox 2 L*4; T&J Bioengineering Co., Ltd., Shanghai, China) containing 800 mL medium. The medium contains 100 g/L glucose, 20 g/L (NH4)2SO4, 15 g/L corn syrup, 1 g/L KH2PO4, 1.5 g/L MgSO4·7H2O, 1.1 g/L FeSO4·7H2O, 0.3*10− 4 g/L protocatechuic acid, 3 g/L yeast extract, 1*10− 3 g/L vitamin B1, and 1.5*10− 3 g/L betaine. Fermentation was initially carried out at pH 7.0–7.2, 800 rpm, 30 °C and 2 vvm of aeration rate. After 72 h, the pH, rotational speed and aeration rate were controlled at pH 6.5–7.0, 800 rpm and 1.5 vvm, respectively. Relative dissolved oxygen was controlled at 80–100% for the first 24 h, after which it was coupled to the stirring speed and controlled at 30%. Samples were taken every 12 h to determine the OD562 value and residual glucose. Glucose was fed to the final concentration of 40 g/L when the residual glucose concentration in the medium dropped below 20 g/L.
Real-time PCR analysis
Total RNA was extracted from C. glutamicum cells growing in LBB medium at 8 h. After removing residual DNA and reversely transcribing RNA into cDNA, real-time (RT)-PCR was performed as described previously [15]. The relative abundance of 16 S rRNA was used as a standard control. The relative gene expression analysis was quantified by the cycle threshold (Ct) and 2−∆∆CT value.
Result and discussion
Regulation of sigD expression by growth-regulated promoters
A previously constructed strain, C. glutamicum D-RS, was selected as the chassis strain for modifying sigD expression. In strain D-RS, a Lys-OFF riboswitch was chromosomally inserted at the upstream of dapA gene of strain SN01 to reduce the accumulation of by-product Lys [9]. According to transcriptomic data, the sigD gene expression of D-RS was about 80% weaker than that of C. glutamicum ATCC13032 (Table S1). Because the growth of ATCC13032 is better than that of SN01, and the expression of sigD and SigD regulon in C. glutamicum ATCC13032 is usually downregulated at the stationary phase, so in order to enhance sigD gene expression at the exponential growth phase of strain C. glutamicum D-RS, the GRP with the higher strength at the exponential phase was exploited. Previously, PCP_2836 was applied as an exponential up-regulated promoter to promote L-valine production [18], PodhI was used to regulate glutamate decarboxylase expression for γ-aminobutyric acid production and showed high transcription level at exponential phase and significantly low transcription level at stationary phase [26]. Besides PodhI and the identical promoter of PCP_2836 in C. glutamicum ATCC13032, i.e. Pcg3096, several GRP that were up-regulated at the exponential phase and down-regulated at the stationary phase, i.e. Pcg1206, Pcg1633, Pcg2705, were identified from the transcriptome data (Table S2). These 5 GRPs were used to replace the original promoter of the sigD, generating 5 GRP-modified strains such as D-RS PsigD::PodhI. Overexpression of vgb genes has been demonstrated to be effective to increase O2 supply and 4-HIL production [8, 25]. Then the P7P7 plasmid carrying ido and vgb genes regulated by an Ile-responsive promoter PbrnFE7 was electrotransformed into chassis strain D-RS and these GRP-modified strains to obtain the control strain of RS1 and the sigD-regulated strains of RSS1–RSS5. These strains were cultured in shake flasks, and the fermentation results were shown in Fig. 3a, b.
Fermentation data and cell envelope properties of sigD-regulated strains. (a) cell growth and glucose consumption, Square: RS1, circle: RSS2, upward triangle: RSS1, downward triangle: RSS3, diamond: RSS4, left triangle: RSS5, (b) 4-HIL and Ile concentration, (c) cell hydrophobicity at 8 h and 15 h, (d) cell permeability at 8 h and 15 h
The growth and glucose consumption of sigD-regulated strains were not affected, except for RSS5 (Fig. 3a), so RSS5 was discarded for subsequent studies. RSS1–RSS4 grew quickly to OD562 of 60–70 before 48 h and slowly thereafter. Glucose was consumed quickly and exhausted at 72 h. The RSS4 strain grew better than the RS1 in the first 48 h and achieved a 4-HIL titer of 142.45 ± 3.69 mM, which was 20.73% higher than that of RS1 (117.99 ± 0.34 mM) (Fig. 3b). The growth of other three modified strains were not significantly affected and their 4-HIL titers (102.51–120.72 mM) were similar to RS1. However, the 4-HIL titer of RSS4 was only 7.15 mM higher than that of a previous strain ST17 (135.30 mM) [8], in which the ido, odhI and vgb genes were dynamically overexpressed by the Ile biosensor Lrp-PbrnFE7. Therefore, altering the cell envelope properties by modifying sigD expression may not contribute much to cell growth and 4-HIL production in shake flask fermentation. Subsequently, in order to further understand the metabolic performance of these strains in the bioreactor, some cell properties such as the cell hydrophobicity and permeability of the sigD-regulated strains growing in LBB medium were investigated during the exponential (8 h) and stationary phases (15 h), and the results are shown in Fig. 3c, d. Firstly, the hydrophobicity of C. glutamicum D-RS was much stronger than that of ATCC13032. This indicates that with the increase of cell density of D-RS strain, cells will tend to aggregate and adhere to the wall of flasks or bioreactors. Upon comparison between D-RS and sigD-modified strains, the hydrophobicity of both RSS2 and RSS4 decreased by about 17% at the exponential phase, while that of the rest of the strains was similar with the control strain RS1. The permeability of RSS4 increased by 12% at the exponential phase, while that of the remaining strains especially RSS2 was lower than that of RS1. In addition, all the permeability of RS1 and RSS1–RSS4 at 15 h were higher than their permeability at 8 h, also with the permeability of RSS4 the highest. As at exponential phase, Pcg1206, Pcg1633 and Pcg2705 were all moderate promoters with higher strength than PsigD, it can be assumed that the induction of sigD expression with moderate promoter (RSS1–RSS3) will lead to an increase in cell envelope synthesis and a decrease in cell permeability. Meanwhile, their hydrophobicity did not reduce and finally their 4-HIL production did not increase. The much higher cell hydrophobicity would cause cell adhesion and attachment and was not conducive to fermentation [27], while the increased cell permeability would facilitate the excretion of amino acids [7]. Therefore, RSS4 with decreased hydrophobicity and increased permeability was considered as a suitable strain for further detecting the 4-HIL titer in a fermenter.
Total lipids of RSS4 strain were extracted and compared to that of the control strain RS1 and a strain unable to synthesize MA, i.e., ATCC13869 Δpks [23]. Two kinds of lipid component in C. glutamicum will be shown by chromatography: one is the phospholipids (below the dashed line in Fig. 4a), and the other is the substances with MA (above the dashed line in Fig. 4a). As shown in Fig. 4a, ATCC13869 Δpks did not possess MA, while RS1 and RSS4 contained MA. The MA content of RSS4 increased and its phospholipid content was similar as compared to RS1. Moreover, the sigD transcription level of RSS1–RSS5 was basically in accordance with the strength of their modified promoters, as verified by RT-PCR (Fig. 4b). The sigD transcription of RSS4 with strong PodhI-regulated sigD was the highest, about 12 times that of D-RS, while that of RSS5 with weak Pcg3096-regulated sigD was the lowest, a little lower than that of D-RS. RSS3 and RSS1 with moderate Pcg2705 and Pcg1633-regulated sigD showed 5.2 and 2.4-fold level of D-RS. Therefore, the up-regulation of sigD expression during the exponential phase by PodhI can effectively improve the content of MA in the cell envelopes, which might be helpful for increasing the titer of 4-HIL. With sigD gene overexpression at exponential phase, the hydrophobicity of RSS4 cell envelope decreased. The increase of MA content promoted the growth performance of strain and the production of amino acids (Fig. 3). In this study, enhancing the expression of genes related to MA synthesis was beneficial to cell growth, as indicated in previous report [28]. The increased permeability also contributed to the production of 4-HIL.
Regulation of cmt2 and cop1 expression
Cmt2 and Cop1, the two mycolyltransferases that are functionally close to each other, induces the conversion between TDCM and TMCM. Cop1 has been reported to transfer corynomycolate from TMCM to arabinogalactan layer of C. glutamicum [29]. The genes expressing these two enzymes are regulated by SigD factor during transcription (Fig. 2). Considering that the regulation of sigD expression may influent the transcription of all genes in SigD regulon, to exploit the effect of only a single crucial gene, the MA transferase genes cop1 and cmt2 were selected to be regulated. The transcriptome data showed that the expression intensity of cmt2 and cop1 in SN01 was about 90% lower than that in ATCC13032 (Table S1). Therefore, the growth-dependent promoter Pcg1633 and the strong constitutive promoter Ptuf were used in this study to regulate the cmt2 and cop1 expression. However, as the promoter replacement process carried out, it was found that the cop1 gene could not be regulated by the strong promoter Ptuf, and thus only three strains, RSC1, RSC2, and RSC3, were finally constructed (Table 1).
The results of 144 h fermentation showed that RSC1 with Pcg1633-regulated cop1 was unable to grow properly and it was unable to utilize glucose for growth in exponential phase (Fig. 5a). The TLC image in Fig. 4c showed that the MA content of RSC1 was very low. This may be due to the non-normal expression of cop1, which affects the linkage between MA and AG layers. According to a previous report, a deficiency in AG layer biosynthesis may affect the MA layer stability and is unfavorable to the growth of C. glutamicum strain [14]. In addition, as shown by Fig. 5b, there was nearly no 4-HIL titer in RSC1. The growth of other two Pcg1633- or Ptuf-modified cmt2 strains (RSC2 and RSC3) were not significantly affected and their 4-HIL titers (88.25–96.44 mM) were a little lower than that of RS1 (Fig. 5a, b). The cell hydrophobicity and cell permeability of RSC2 changed a little compared with RS1 in exponential phase (Fig. 5c, d). However, the cell permeability of RSC3 reduced by 14% in exponential phase, which might lead to the 33.70% decrease in its 4-HIL titer compared to RS1. In addition, the MA content of RSC2 and RSC3 was similar with that of RS1 (Fig. 4c), indicating that the regulation of cmt2 has little effect on MA content. Mycoloyltransferases has many important functions for the physiological process of Corynebacteriales [30], the integrity of the cell envelope of C. glutamicum is essential for its survival. In summary, the regulation of cmt2 gene expression did not significantly affect the 4-HIL production, but the regulation of cop1 expression seriously retarded cell growth and 4-HIL production.
Fermentation data and cell envelope properties of cop1- and cmt2-regulated strains. (a) cell growth and glucose consumption, Square: RS1, circle: RSC1, upward triangle: RSC2, downward triangle: RSC3, (b) 4-HIL and Ile concentration, (c) cell hydrophobicity at 8 h and 15 h, (d) cell permeability at 8 h and 15 h
Regulation of cmt1 expression
Cmt1 also belongs to a type of MA transferase, but its function is distinct from that of Cmt2 and Cop1. Cmt1 is responsible for ligating the corynomycolate of TDCM and TMCM to porin [29, 31] (Fig. 2), and the promoter of the cmt1 gene is likewise subjected to regulation by the SigD factor. Transcriptome data showed that the expression intensity of cmt1 gene in SN01 was 84% lower than that in ATCC13032 (Table S1). Comparison of genomic sequences revealed that the sequence of the cmt1 gene and its promoter in SN01 differed significantly from the sequence of the corresponding gene in ATCC13032, with little continuous identity (Table S3). A difference in sequence may make the function of the enzyme after expression very different. In addition, it can also be seen from Fig. 3c that the hydrophobicity of ATCC13032 is particularly low compared with D-RS strain. Therefore, during the experiment, the cmt1 gene of D-RS was first regulated by growth-dependent promoter Pcg1633, generating the strain RSC4. On the other hand, the Pcmt1-cmt1 gene sequence in D-RS strain was replaced by the corresponding sequence of ATCC13032, generating the strain RSA1.
As shown in Fig. 6a, b, by the results of 144 h shake flask fermentation it can be seen that the growth of the modified strains was not affected, the final OD562 value is between 80 and 90. The 4-HIL titer of modified strains (103.88–106.80 mM) did not increase and the titer was even not as high as that of RS1 (117.99 ± 0.34 mM). In terms of cell properties, the cell hydrophobicity of RSA1 was much higher than that of ATCC13032 and was 19% lower than that of RS1 in exponential phase, as shown in Fig. 6c, suggesting that direct substitution of cmt1 sequences did not change cell hydrophobicity in exponential phase. The cell permeability of RSC4 and RSA1 increased slightly compared with RS1 (Fig. 6d), and did not promote 4-HIL production (Fig. 6b). TLC image showed a slight reduction in MA content in RSA1 and RSC4 (Fig. 4c). Thereby, regulation of cmt1 expression does not significantly influence cell hydrophobicity and permeability of strain, and may not promote the production of 4-HIL.
4-HIL production in fed-batch fermentation
Considering that the 4-HIL production was relatively low in flasks, fed-batch fermentation of RSS4 shall be carried out to evaluate its potential in scale production. Therefore fed-batch fermentation of RSS4 was carried out in a 2 L fermenter.
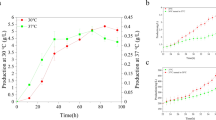
As shown in Fig. 7, at 72 h, the OD562 value was close to the value at the same period of shaking flask fermentation and the residual glucose concentration was close to 20 g/L. Then glucose was supplemented to the fermenter till about 40 g/L to continue the fermentation. During the 84–96 h, the strain consumed glucose fast, and the OD562 reached a maximum of 137, which was 1.41 times of the highest OD562 of the shake flask. On the one hand, the strain was not easy to stick to the wall of fermenter, likely due to the reduction of cell hydrophobicity [32], and on the other hand, the enhanced cell permeability also helps the strain to use nutrients and excrete products [33]. Therefore, the RSS4 strain developed in this study has certain practical significance for the production of 4-HIL. The modified strain RSS4 achieved a 4-HIL titer of 372.56 mM at the bioreactor level, which is the highest 4-HIL titer reported so far, at the bioreactor fermentation level. Due to the quite slow cell growth before 36 h, 4-HIL was not produced at this stage, while during 36–60 h and 84–96 h, 4-HIL accumulated quickly, in accordance with the quick cell growth during these periods. Finally, 372.56 mM 4-HILwas accumulated. In addition, only little by-products (15.31 mM Lys and 8.97 mM L-alanine) were accumulated at the end of fermentation. Compared with the 4-HIL titer of C. glutamicum strain HIL18 (34.2 g/L, i.e. 232.65 mM) modified by eight-step metabolic engineering in bioreactor previously reported [34], the 4-HIL titer of C. glutamicum RSS4 with only PodhI-sigD modification in bioreactor here increased by 60.13%. Through the mere regulation of sigD gene expression, the cell envelope properties of the strain were changed, which played a good role in supporting and promoting the production of 4-HIL. The modification of other genes involving in cell envelope synthesis such as accD2 or fadD2, cmrA and aftB and their potentials for enhancing 4-HIL production and stability of C. glutamicum strains shall be investigated in the future. To further improve 4-HIL production in RSS4, dynamic engineering of the Ile synthesis and 4-HIL conversion pathways shall be researched. Moreover, integration of multiple-copy ido genes in C. glutamicum chromosome will be exploited in the future to produce 4-HIL economically.
Conclusion
This study focused on the rearrangements in regulating sigD expression by GRP and thereby promoted the 4-HIL production from the Ile-producing strain D-RS. Finally, an improved strain RSS4 with PodhI-regulated sigD was obtained. Not only its cell permeability increased by 12%, but also its cell hydrophobicity reduced by 17%. This change of cell envelope properties was favorable to its fermentation in the shake flask and bioreactor, and finally the 4-HIL titer reached the highest titer of 372.56 mM so far under the bioreactor condition, and there were only little by-products. Thereby, it is feasible to improve metabolite production by changing the properties of cell envelope.
Data availability
All data associated with this work is embedded within the manuscript.
Code Availability
Not applicable.
References
Fuller S, Stephens JM. Diosgenin, 4-hydroxyisoleucine, and fiber from fenugreek: mechanisms of actions and potential effects on metabolic syndrome. Adv Nutr. 2015;6:189–97. https://doi.org/10.3945/an.114.007807.
Rawat AK, Korthikunta V, Gautam S, Pal S, Tadigoppula N, Tamrakar AK, Srivastava AK. 4-Hydroxyisoleucine improves insulin resistance by promoting mitochondrial biogenesis and act through AMPK and Akt dependent pathway. Fitoterapia. 2014;99:307–17. https://doi.org/10.1016/j.fitote.2014.10.006.
Gao F, Jian L, Zafar MI, Du W, Cai Q, Shafqat RA, Lu F. 4-Hydroxyisoleucine improves insulin resistance in HepG2 cells by decreasing TNF-α and regulating the expression of insulin signal transduction proteins. Mol Med Rep. 2015;12(5):6555–60. https://doi.org/10.3892/mmr.2015.4298.
Smirnov SV, Kodera T, Samsonova NN, Kotlyarova VA, Rushkevich NY, Kivero AD, Sokolov PM, Hibi M, Ogawa J, Shimizu S. Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine. Appl Microbiol Biotechnol. 2010;88(3):719–26. https://doi.org/10.1007/s00253-010-2772-3.
Hibi M, Kawashima T, Kodera T, Smirnov SV, Sokolov PM, Sugiyama M, Shimizu S, Yokozeki K, Ogawa J. Characterization of Bacillus thuringiensis l-isoleucine dioxygenase for production of useful amino acids. Appl Environ Microbiol. 2011;77(19):6926–30. https://doi.org/10.1128/AEM.05035-11.
Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng. 2018;50:122–41. https://doi.org/10.1016/j.ymben.2018.07.008.
Shi F, Niu T, Fang H. 4-Hydroxyisoleucine production of recombinant Corynebacterium glutamicum ssp. lactofermentum under optimal corn steep liquor limitation. Appl Microbiol Biotechnol. 2015;99(9):3851–63. https://doi.org/10.1007/s00253-015-6481-9.
Tan S, Shi F, Liu H, Yu X, Wei S, Fan Z, Li Y. Dynamic control of 4-hydroxyisoleucine biosynthesis by modified l-isoleucine biosensor in recombinant Corynebacterium glutamicum ACS Synth Biol. 2020;9(9):2378–89. https://doi.org/10.1021/acssynbio.0c00127.
Lai W, Shi F, Tan S, Liu H, Li Y, Xiang Y. Dynamic control of 4-hydroxyisoleucine biosynthesis by multi-biosensor in Corynebacterium glutamicum Appl Microbiol Biotechnol. 2022;106(16):5105–21. https://doi.org/10.1007/s00253-022-12034-6.
Patek M, Nesvera J. Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol. 2011;154(3):101–13. https://doi.org/10.1016/j.jbiotec.2011.01.017.
Brand S, Niehaus K, Puhler A, Kalinowski J. Identification and functional analysis of six mycolyltransferase genes of Corynebacterium glutamicum ATCC 13032: the genes cop1, cmt1, and cmt2 can replace each other in the synthesis of trehalose dicorynomycolate, a component of the mycolic acid layer of the cell envelope. Arch Microbiol. 2003;180(1):33–44. https://doi.org/10.1007/s00203-003-0556-1.
Dostálova H, Busche T, Holátko J. Overlap of promoter recognition specificity of stress response sigma factors SigD and SigH in Corynebacterium glutamicum ATCC 13032. Front Microbiol. 2019;9:3287. https://doi.org/10.3389/fmicb.2018.03287.
Taniguchi H, Busche T, Patschkowski T, Niehaus K, Patek M, Kalinowski J, Wendisch VF. Physiological roles of sigma factor SigD in Corynebacterium glutamicum BMC Microbiol. 2017;17:158. https://doi.org/10.1186/s12866-017-1067-6.
Bou R, Meniche X, De Sousa-d’Auria C, Chami M, Salmeron C, Tropis M, Labarre C, Daffe M, Houssin C, Bayan N. A deficiency in arabinogalactan biosynthesis affects Corynebacterium glutamicum mycolate outer membrane stability. J Bacteriol. 2010;192(11):2691–700. https://doi.org/10.1128/JB.00009-10.
Chen R, Xiang Y, Shi F. Comparative transcriptome analysis of global effect of ddh and lysE deletion on 4-hydroxyisoleucine production in Corynebacterium glutamicum. Syst Microbiol Biomanuf. 2022;2:542–54. https://doi.org/10.1007/s43393-022-00085-9.
Li H, Xu D, Zhang D, Tan X, Huang D, Ma W, Zhao G, Li Y, Liu Z, Wang Y, Hu X, Wang X. Improve l-isoleucine production in Corynebacterium glutamicum WM001 by destructing the biosynthesis of trehalose dicorynomycolate. Microbiol Res. 2023;272:127390. https://doi.org/10.1016/j.micres.2023.127390.
Gebhardt H, Meniche X, Tropis M, Kramer R, Daffe M, Morbach S. The key role of the mycolic acid content in the functionality of the cell wall permeability barrier in Corynebacterineae. Microbiology. 2007;153(5):1424–34. https://doi.org/10.1099/mic.0.2006/003541-0.
Ma Y, Cui Y, Du L, Liu X, Xie X, Chen N. Identification and application of a growth-regulated promoter for improving l-valine production in Corynebacterium glutamicum Microb Cell Fact. 2018;17:185. https://doi.org/10.1186/s12934-018-1031-7.
Shi F, Zhang M, Li Y, Fang H. Sufficient NADPH supply and pknG deletion improve 4-hydroxyisoleucine production in recombinant Corynebacterium glutamicum. Enzyme Microb Technol. 2018;115:1–8. https://doi.org/10.1016/j.enzmictec.2018.04.003.
Chen R, Shi F, Xiang Y, Lai W, Ji G. Establishment of CRISPR-Cpf1-assisted gene editing tool and engineering of 4-hydroxyisoleucine biosynthesis in Corynebacterium glutamicum. World J Microbiol Biotechnol. 2023;39(10):266. https://doi.org/10.1007/s11274-023-03705-1.
Shi F, Zhang S, Li Y, Lu Z. Enhancement of substrate supply and ido expression to improve 4-hydroxyisoleucine production in recombinant Corynebacterium glutamicum ssp. lactofermentum Appl Microbiol Biotechnol. 2019;103(10):4113–4124. https://doi.org/10.1007/s00253-019-09791-2.
Ichikawa T, Hirata C, Takei M, Tagami N, Murasawa H, Ikeda R. Cell surface hydrophobicity and colony morphology of Trichosporon asahii clinical isolates. Yeast. 2017;34(3):129–37. https://doi.org/10.1002/yea.3220.
Gao Y, Hu X, Wang J, Li H, Wang X. Impact of mycolic acid deficiency on cells of Corynebacterium glutamicum ATCC13869. Biotechnol Appl Biochem. 2018;65(3):435–45. https://doi.org/10.1002/yea.3220.
Li H, Xu D, Tan X, Huang D, Huang Y, Zhao G, Hu X, Wang X. The role of trehalose biosynthesis on mycolate composition and l-glutamate production in Corynebacterium glutamicum Microbiol Res. 2023;267:127260. https://doi.org/10.1016/j.micres.2022.127260.
Shi F, Fan Z, Zhang S, Wang Y, Tan S, Li Y. Optimization of ribosomal binding site sequences for gene expression and 4-hydroxyisoleucine biosynthesis in recombinant Corynebacterium glutamicum Enzyme Microb Technol. 2020;140:109622. https://doi.org/10.1016/j.enzmictec.2020.109622.
Shi F, Luan M, Li Y. Ribosomal binding site sequences and promoters for expressing glutamate decarboxylase and producing gamma-aminobutyrate in Corynebacterium glutamicum AMB Express. 2018;8:61. https://doi.org/10.1186/s13568-018-0595-2.
Liu Y, Yang SF, Li Y, Xu H, Qin L, Tay J. The influence of cell and substratum surface hydrophobicities on microbial attachment. J Biotechnol. 2004;110(3):251–256. https://doi.org/10.1016/j.jbiotec.2004.02.012.
Toyoda K, Inui M. Extracytoplasmic function sigma factor σD confers resistance to environmental stress by enhancing mycolate synthesis and modifying peptidoglycan structures in Corynebacterium glutamicum Mol Microbiol. 2018;107(3):312–29. https://doi.org/10.1111/mmi.13883.
Puech V, Bayan N, Salim K, Leblon G, Daffé M. Characterization of the in vivo acceptors of the mycoloyl residues transferred by the corynebacterial PS1 and the related mycobacterial antigens 85. Mol Microbiol. 2000;35(5):1026–41. https://doi.org/10.1046/j.1365-2958.2000.01738.x.
Dautin N, De Sousa-d’Auria C, Constantinesco-Becker F, Labarre C, Oberto J, Li de la Sierra-Gallay L, Dietrich C, Issa H, Houssin C, Bayan N. Mycoloyltransferases: A large and major family of enzymes shaping the cell envelope of Corynebacteriales Biochim Biophys Acta Gen Subj. 2017;1861(1):3581–92. https://doi.org/10.1016/j.bbagen.2016.06.020.
Rath P, Demange P, Saurel P, Tropis M, Daffe M, Dotsch V, Ghazi A, Bernhard F, Milon A. Functional expression of the PorAH channel from Corynebacterium glutamicum in cell-free expression systems: implications for the role of the naturally occurring mycolic acid modification. J Biol Chem. 2011;286(37):32525–32. https://doi.org/10.1074/jbc.M111.276956.
Liu Y, Yang X, Yin Y, Lin J, Chen C, Pan J, Si M, Shen X. Mycothiol protects Corynebacterium glutamicum against acid stress via maintaining intracellular pH homeostasis, scavenging ROS, and S-mycothiolating MetE. J Gen Appl Microbiol. 2016;26(3):144–45. https://doi.org/10.2323/jgam.2016.02.001.
Aliyath S, Nampoothiri KM. The implication of sortase E in the morphology and physiology of Corynebacterium glutamicum FEMS Microbiol Lett. 2022;369(1):fnac080. https://doi.org/10.1093/femsle/fnac080.
Zhang C, Li Y, Ma Y, Liu Y, He J, Li Y, Zhu F, Meng J, Zhan J, Li Z, Zhao L, Ma Q, Fan X, Xu Q, Xie X, Chen N. High production of 4-hydroxyisoleucine in Corynebacterium glutamicum by multistep metabolic engineering. Metab Eng. 2018;49:287–98. https://doi.org/10.1016/j.ymben.2018.09.008.
Funding
This work was supported by the program of State Key Laboratory of Food Science and Technology (SKLF-ZZA-201904).
Author information
Authors and Affiliations
Contributions
FS conceived and designed research. CH, RC and YX conducted experiments. CH and FS analyzed data. FS, CH, RC and YX wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, C., Shi, F., Chen, R. et al. Rearrangement in the regulation of sigD gene expression promotes 4-hydroxyisoleucine production in Corynebacterium glutamicum. Syst Microbiol and Biomanuf (2024). https://doi.org/10.1007/s43393-024-00277-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43393-024-00277-5