Abstract
Lignocellulosic biomass (LCB) generated from various agro-waste can be effectively used to manufacture a broad range of value-added products cost-effectively. However, the high cost of cellulases is still a major challenge in producing biofuels and biochemicals from LCB on an industrial scale. The enzyme output and activity of cellulase in the fermentation broth are closely linked in terms of enzyme manufacturing costs. Therefore, research on efficient fermentation processes of hyperactive fungi, and cost-effective recovery systems have been directed toward lowering enzyme costs and increasing overall enzyme production. Penicillium funiculosum NCIM 1228 (P. funiculosum NCIM 1228) is a feasible cellulase-producing strain that possesses all four enzymes required to efficiently hydrolyse LCB. The primary objective of this study was to employ random mutagenesis to increase enzymes titer, yield, and productivity. The potential mutant D4 (derived by Ethyl methanesulfonate (EMS) mutation) exhibited 6.47, 3.05, 3.03, and 3.19-fold higher activities of FPase, CMCase, β-glucosidase, and xylanase, respectively, compared to the parent strain. Mutant D4 demonstrated a promising protein titer of 17.96 g/L at the 40 L fermenter scale, with productivities of 479, 4249, and 6987 U/L/day for FPase, CMCase, and Xylanase, respectively, on the tenth day. Interestingly, the crude form of enzymes from the mutant demonstrated promising saccharification, releasing 3.54% of glucose and achieving a 54.03% of cellulose conversion efficiency without formulation. In comparison, a commercially formulated enzyme exhibited 53.07% efficiency against pre-treated sugarcane bagasse, indicating its promising potential for future applications.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
According to the Indian Ministry of New and Renewable Energy, India produces approximately 500 and 600 million tonnes of agro-based LCB per year. A majority of the waste generated is utilized as feed and fuel for both home and industrial purposes [1]. However, over 100 million tonnes of agro-residual waste are burned each year, and it is alarming to note that the percentage of agricultural waste burned in India exceeds the total amount of agricultural waste produced in other nations [1, 2]. The concerns that such activities pose a threat to the environment by contributing to the alarming increase in greenhouse gas production cannot be ignored [3, 4]. Likewise, depositing agricultural-based lignocellulosic biomass as garbage is yet another ineffective approach for dealing with the crop’s leftovers [5]. Furthermore, the elimination of these residues from the fields requires new technological advancements and more expensive professional support [1]. As a result, alternative, cost-effective, and less hazardous methods of converting agro-residual waste into value-added goods are essential for an eco-friendly environment and sustainable development.
Microbial enzymes such as cellulase, laccase, and xylanase can break down LCB obtained from various agro-based residual wastes like sugarcane bagasse, rice straw, and corncob [3]. However, the three main enzymes involved in cellulose hydrolysis are exoglucanase (avicelase), endoglucanase (carboxymethyl cellulase, CMCase), and β-glucosidase. To completely degrade cellulose into soluble sugars, these enzymes must work together [6, 7]. Due to its widespread availability, various LCBs like wheat straw, rice straw, bagasse, and corn cob, etc. were successfully employed to make a wide range of value-added products. In this regard, cellulosic biorefinery has gained considerable attention for development of a wide range of platform chemicals, biofuels, and other biochemicals [8]. Products from biofuels are biorefinery can be used world-wide to reduce the use of gasoline and greenhouse gas emissions. Liu et al. estimated that the cost of one enzyme unit in the purchase mode ranged from 1.25 to 23.3 $/kg protein using the Aspen Plus model [9]. Therefore, optimizing the cost of the biorefinery process is imperative. Novel technologies should be integrated to produce additional high-quality platform chemicals and other biomaterials to overcome the constraints led by the biorefineries [10]. Peng et al. stated that cellulase is a protein generated by a filamentous fungus that aids in the conversion of LCB into soluble sugars for the production of chemicals and fuels. However, most filamentous fungi developed through spontaneous breeding have a limited ability to secrete cellulase, making them unsuitable for commercial production. Therefore, combining random mutagenesis with an adaptive laboratory evolution approach would certainly aid in an increased fungal enzyme output [11].
This study evaluates the potential of filamentous fungi P. funiculosum as an efficient cellulase and xylanase producer. To date, there is a dearth in the knowledge regarding the improvement of the cellulase production capacity by using P. funiculosum for commercial application. Strain improvement has traditionally been accomplished through mutation, selection, or genetic recombination [12, 13]. In this study, EMS, a chemical mutagen thought to primarily promote random nucleotide substitution in genetic material, was used to improve the cellulase and xylanase production by P. funiculosum NCIM 1228 [14]. This strain was the preferred choice due to its potential to produce the necessary enzymes which are required for the efficient hydrolysis of LCB. The necessary cocktail is already available in the secretome of the P. funiculosum NCIM 1228 [15]. This study explores the enhancement of efficient cellulase and xylanase enzyme production through strain improvement by chemical mutagenesis and its applications in the production of various high-value products from cellulose biorefineries. It also examines the commercial obstacles and economic viability of the process [16].
The discovery of high-titer cellulase-producing mutants significantly reduces the costs of cellulase synthesis and downstream processing in commercial-scale enzyme manufacturing. Interestingly, cellulase production was evaluated in the putative P. funiculosum mutant D4 in comparison to the parent strain. The mutant D4 exhibited enhancements of 647%, 305%, 303%, 319% of FPase, CMCase, β-glucosidase, and xylanase, respectively, as compared to the parent strain [15]. Furthermore, 18.81 g/L of extracellularly secreted protein was observed, which was 267.56% higher than the wild type. Isolating mutants with better growth and hydrolysis zones on cellulose-containing plates resulted in cellulase-hypersecreting mutants [17]. The mutant that grew the fastest on cellulose also had the most cellulase activity, implying that higher cellulase protein levels are associated with increased cellulase synthesis. In theory, our approach may be used to enhance the extracellular protein output of filamentous fungi. The cellulase enzyme of mutant D4 efficiently saccharified pre-treated bagasse, releasing 35.38 g/L of glucose at a dose of 6 FPU/g cellulose, while the commercial enzyme (Sacchariceb) from Advanced Enzymes, at the same dose, released 34.83 g/L of glucose. So, further, improvement in P. funiculosum mutant D4 can be employed in biorefinery processes for onsite enzyme production due to its high enzyme yield and great biomass hydrolysis capabilities of D4 crude enzyme. As a result, the overall cost for the second generation (2G) of ethanol production will be lower. Using filamentous fungi as hosts to produce enzymes and proteins is constrained by the low production titers that are typically reached [18]. Here, we describe a mutagenesis technique for isolating EMS mutants with increased enzyme and protein production titers.
Materials and methods
Penicillium funiculosum NCIM 1228
In this investigation, filamentous fungus P. funiculosum NCIM 1228 was used as the parent strain for generating mutations. After five days of incubation at 30 °C, sporulation was detected on potato dextrose agar (PDA). Spores containing glycerol stocks, were prepared and preserved at -80 °C for future experiments [15].
Culture, inoculum preparation, and media
A spore suspension was prepared by aseptically harvesting spores of P. funiculosum NCIM 1228 from the PDA slant surface using sterile water and a glass rod. Subsequently, these spores were transferred into a sterile 50 mL falcon tube, and a serial dilution was performed to obtain a standard inoculum containing 1–10 × 106 spores per mL. The same stock was used to make glycerol stocks and preserve it at -80 °C for future use. The medium consisted of 1.4 g (NH4)2SO4, 2 g KH2PO4, 0.3 g Urea, 0.3 g CaCl2.2H2O, 0.3 g MgSO4·7H2O, 0.005 g FeSO4, 0.0016 g MnSO4, 0.0017 g ZnSO4.7H2O, 0.02 g CoCl2, 0.75 g Peptone, 0.5 g Yeast extract, 1 g Tween80, per liter and 5.0 g/L of the cellulose (Avicel) as a carbon source. Experiments were performed in 250 mL Erlenmeyer flasks with 100 mL of the above-mentioned media [15].
Production of enzymes in submerged fermentation (SmF)
Spores of P. funiculosum NCIM 1228 were inoculated into Czapek dox broth and kept at 30 °C for 48 h to generate the pre-seed inoculum. For seed inoculum generation, 10% of grown pre-seed was inoculated into modified Mandels and Reese medium containing: 2.0 g/L KH2PO4 0.3 g/L; Urea; 0.3 g/L CaCl2.2H2O; 0.3 g/L MgSO4.7H2O; 1.4 g/L (NH4)2SO4; 0.1 g/L Yeast extract; 0.25 g/L Peptone; 2.0 g/L Glucose; 1.0 g/L Tween 80; 0.0005 g/L FeSO4.7H2O; 0.0014 g/L ZnSO4.7H2O; 0.0016 g/L MnSO4; 0.002 g/L CoCl2; and 5.0 g/L Cellulose Acetate; pH 5.5 [19]. The seed inoculum was incubated at 30 °C under 150 rpm for 48 h on an incubating shaker (Innova 4080 Incubator Shaker, New Brunswick Scientific, USA). Further, 10% of the grown seed of P. funiculosum or mutants was inoculated into optimized formulated enzyme production media consisting of: 2.0 g/L KH2PO4; 5.0 g/L Urea; 1.0 g/L CaCl2.2H2O; 0.3 g/L MgSO4.7H2O; 0.5 g/L Yeast extract; 16.0 g/L Peptone; 1.0 g/L Tween 80 0.0005 g/L; FeSO4.7H2O; 0.0014 g/L ZnSO4.7H2O; 0.0016 g/L MnSO4; 0.002 g/L CoCl2; 10.0 g/L Corn steep liquor (CSL),;5.0 g/L Citric acid; 7.2 g/L Na2HPO4;10.0 g/L Wheat Bran and 30 g/L of microcrystalline cellulose (MCC) also called MCC101 [19, 20]. MCC101 which is obtained from Blanver Farmoquimica Ltd. (made in Brazil) was used as a substrate in the fermentation medium throughout this study. As a nitrogen source, Yeast extract, CSL, and Urea were employed. Wheat bran obtained from the local market was used as a carbon source. CSL was obtained from Rasayan Trading Co. Ambawadi, Ahmedabad, Gujarat, India, which is a complex, viscous, and a by-product of corn wet milling containing minerals, amino acids, vitamins, and other essential constituents of growth media. All these media constituents were distributed 100 mL in 500 mL Erlenmeyer flasks and then autoclaved at 121 °C for 15 min. After seed inoculations, all flasks were incubated at 30 °C at 150 rpm. The experiment was continued for twelve days and samples were collected every 24 h interval for enzyme assay of cellulases (FPase, CMCase, β-glucosidase, xylanases, and protein estimation) [15]. The pH was maintained at 5.5 ± 0.3 throughout the batch [21].
Chemical mutagenesis using EMS for possible overproduction of enzymes by P. funiculosum NCIM 1228
The first step of this approach is to determine the kill curve. Spores were visible on Malt Dextrose Agar after 3 to 4 days, whereas PDA required 10 to 12 days at 30 °C. Fungal spores were scraped from a Malt dextrose Agar slant after six days, suspended in 5 mL sterile distilled water, and mixed on a cyclomixer. To separate the mycelium, the suspension was filtered. The spore suspension of P. funiculosum NCIM 1228, prepared as described above was then serially diluted to achieve a final concentration of 2.22 × 106 spores/mL [22]. Took samples for spores to count microscopically. And used these spores for EMS Mutagenesis. The spore suspension was made with 106 spores per mL in 0.2 M phosphate buffer (pH 7.0). The spore suspension was transferred aseptically into a sterile test tube and subjected to EMS for 60 min at varied EMS concentrations of 2,4,6,8,10,15,20,25,30 mg/mL [23, 24]. No chemical mutagens were used in the control tube. The reaction was stopped with 10% sodium thiosulphate and the treated tubes were centrifuged (Eppendorf, 5810 R) the for 10 min at 5000 rpm SI Fig. 1. The cells were washed three times in sterile phosphate buffer before being resuspended in 5 mL of the same buffer. The samples were serially diluted in Phosphate buffer and plated on Potato Dextrose agar medium. The plates were then incubated for five days at 28 ± 2 °C. Several isolated mutant strains were randomly selected for further investigation. Mutant colonies were obtained on plates at four different concentrations. Rose Bengal carboxymethyl cellulose (RB-CMC) plates were used to screen all mutations [25]. Submerged fermentation was used to screen for FPase, CMCase, and Xylanase activity in mutants that showed positive findings. The initial growth conditions for submerged fermentation of strains B3 and D4 were optimized [26].
Sampling and data analysis
Experiments were carried out in triplicates. Therefore, 5 mL of fermented broth was collected every 24 h at regular intervals. The pH of the broth was measured. Subsequently, the samples were then centrifuged at 12,000 rpm for 10 min at 4 °C, following which the supernatant was used in triplicate for enzyme assays and protein estimation, with the means values reported. The 3,5-dinitrosalicylic acid (DNS) method was employed to evaluate the activities of CMCase, Avicel cellulase (Avicelase), filter-paper cellulase (FPase), and Xylanase [15]. Enzyme production was measured at 24-hour intervals for up to twelve days during cultivation. The broth was then centrifuged at 12,000 rpm for 10 min at 4 °C, and for enzyme testing, the supernatants were collected as crude enzymes.
Cellulase activity assay
Cellulases are a group of enzymes that work together to break down cellulose, a complex polysaccharide found in plant cell wall. The three main types of enzymes involved in cellulose degradation are FPase, CMCase, β-glucosidase and xylanases. The combined action of these three enzymes is essential for the complete hydrolysis of cellulose into glucose monomers. Therefore, to estimate hydrolysis of cellulose, FPase, CMCase, β-glucosidase, and xylanases enzyme assays are crucial.
FPase activity assay
FPase Activity Units provide another way to measure cellulase activity [15]. In a solution comprising 0.5 mL enzyme supernatant and 1.0 mL citrate buffer (50 mM pH 4.8), a 1 × 6 cm strip of Whatman No.1 filter paper was added. The mixture was then incubated for 1 h at 50 °C. Subsequently, 3 mL of DNS solution was added, and heated for 5 min. After cooling, 20.0 mL of distilled water was added, and the absorbance was measured at 540 nm using glucose as a reference. Heat-inactivated (98 °C for 10 min) enzyme samples were used as controls in this enzyme assay [15]. “One unit of cellulase activity was defined as the amount of enzyme that released 1 mol of reducing sugar equal to glucose per minute under the conditions studied.” Units per milliliter (U/mL) were used to assess enzyme activity.
CMCase activity assay
The activity of CMCase was measured using the Ghose et al. method [27]. In Sodium citrate buffer (0.05 M, pH 5.5), CMCase activity was measured by combining 0.5 mL of a properly diluted crude enzyme with 0.5 mL of a 2% substrate (carboxymethyl cellulose, medium viscosity, Sigma). Subsequently, the enzyme-substrate mixture, was incubated for 30 min at 50 °C. The concentration of glucose, as the reducing sugar was measured using DNS to stop the process [22]. Three mL of DNS were added to the enzyme mixture and incubated for 5 min at 100 ± 2 °C. Then 20 mL of water was added. Subsequently, the test tube was submerged in cold water for 15 min. The amount of glucose present was determined by measuring the absorbance at 540 nm using a spectrophotometer (Shimadzu UV-2450 UV-VIS spectrophotometer). The breakdown of carboxymethyl cellulose to liberate glucose catalysed by CMCase activity in this experiment. The reaction was then stopped by introducing DNS, which reacted with glucose to produce a red-brown complex. “To measure CMCase activity, one unit (U) is defined as the quantity of enzyme that releases 1 mL of reducing sugar glucose per minute under the test conditions.” The activity of CMCase was determined using a glucose standard curve [15, 27].
β-glucosidase (cellobiase) assay
According to IUPAC guidelines, the cellobiase test was utilized to determine β-glucosidase [15, 27]“One unit of cellobiase activity was defined as the amount of enzyme necessary to liberate 2 mol of glucose per minute during cellobiose breakdown.” Cellobiase activity was determined by combining 1.0 mL 15 mM D (+) cellobiose produced in Sodium citrate buffer (0.05 M, pH 4.8) with 1.0 mL of a properly diluted crude enzyme in the same buffer. The enzyme mixture was then incubated at 50 °C for 30 min. Place the reaction mixture in a boiling water bath for 5 min to stop the process. Subsequently, add 10 mL of the aforementioned reaction mixture to a 1 mL Glucose oxidase - peroxidase (GOD POD) reagent and incubate for 15 min at 37 °C. Measure the OD at 500 nm to determine the amount of glucose produced [28].
Xylanases assay
Ghose’s approach was used to measure xylanase activity [15, 27]. In this assay, xylanase catalysed the hydrolysis of xylan to release xylose. “One unit (U) of xylanase activity is defined as the amount of enzyme that releases 1 mL of reducing sugar of xylose per minute under the test conditions.” The activity of xylanase was determined using the xylose standard curve. In this procedure, 0.5 mL of citrate buffer (0.05 M, pH 5.5), and 0.5 mL of supernatant (appropriately diluted) were mixed with 0.5 mL of 2% substrate (birch wood xylan, Hi-media) in citrate buffer (0.05 M, pH 5.5) to determine xylanase activity. Following this, the enzyme combination was incubated for 30 min at 50 °C. The concentration of xylose as the reducing sugar was measured using DNS to stop the reaction. Three mL of DNS was added to the enzyme mixture, which created a red-brown complex with xylose, and then, incubated for 5 min at 100 ± 2 °C. Following that, 20 mL of water was added. Subsequently, the test tube was submerged in cold water for 15 min. To estimate the amount of xylose present, the absorbance was measured at 540 nm using a spectrophotometer (UV-2450, UV-VIS spectrophotometer, Shimadzu) [15].
Protein estimation
Total protein was calculated by Folin-Ciocalteu method, with bovine serum albumin (BSA) used as a standard [29]. The Folin protein determination method was used to calculate the amount of protein present in supernatants of fermented samples based on the absorbance reading at a wavelength of 750 nm. The BSA standard curve was utilized to determine the soluble protein concentration of the cell-free broth [30].
Production of cellulase by EMS mutant D4 at 40 L scale
The extra-cellular cellulolytic and hemicellulolytic enzymes were produced during submerged fermentation of EMS mutant D4 on microcrystalline cellulose (MCC101) in a 40 L fermenter (New Brunswick™ BioFlo® 510 Sterilize-in-Place (SIP) Fermentor from Eppendorf) [29, 31]. A total of 5% (v/v) of 48 h old inoculum was used for the batch. The agitation speed was maintained at 150 rpm, temperature at 30 °C, and aeration was provided at a rate of 1 volume of air per volume of liquid per minute (vvm). The media used for seed cultivation and enzyme production were the same as those used in the shake flask. MCC101, MCC101, less expensive than Avicel from Sigma, was employed as the sole source of carbon for the induction of cellulase [12, 32]. After production, tangential flow filtration (TFF, Millipore, Billerica, MA) was used to concentrate the crude fungal enzyme in the supernatant containing proteins greater than 10 kDa. A Biomax membrane (polyethersulfone) with a 10 kDa molecular mass cut-off (MMCO) was employed for diafiltration [33]. The retentate was collected and compare the saccharification efficiency with commercial enzymes for the degradation of biomass (by Avicelase, carboxy methyl cellulase, β-glucosidase, and xylanase respectively).
Saccharification of pre-treated LCB by cellulase obtained from mutant D4
To compare the hydrolysis efficiency of the cellulase enzyme produced from the D4 mutant with the commercial cellulases (Sacchariseb and Cellic CTech3) from Advanced Enzymes and Novozyme, the same dose of FPU (Filter Paper Unit) for both enzymes was used for the hydrolysis of lignocellulosic biomass (sugarcane bagasse, corn cob, and rice straw). For bagasse, the D4 mutant enzyme was tested at doses of 3 FPU, 4 FPU, 5 FPU, and 6 FPU per gram of cellulose, while Advanced Enzymes used doses of 5 FPU and 6 FPU per gram of cellulose. For corn cob and rice straw, the FPU doses for the D4 mutant enzyme ranged from 2 FPU, 4FPU, 6 FPU, and 8 FPU per gram of cellulose, while Novozyme Enzyme used doses of 6 FPU and 8 FPU per gram of cellulose [15].
Briefly, for enzyme hydrolysis, pre-treated sugarcane bagasse (LCB) was taken in a 250 mL Erlenmeyer flask (50 g). A pH of 5.0 was maintained for commercial cellulases from Advanced Enzymes, and 4.8 pH was maintained for mutant D4 [22, 34]. Sampling was conducted at 0-h, 24-h, 48-h, and 72-h for high-performance liquid chromatography (HPLC) quantification of glucose, xylose, and arabinose. The HPLC set-up consisted of an Aminex HPX-87 H, 300 mm × 7.8 mm column (Bio-Rad, USA), and the mobile phase used for elution was 0.050 M H2SO4 at a flow rate of 0.6 mL min − 1 at 35 °C. Samples were injected using an autosampler injector with a fixed volume of 20 µl. A refractive index detector (Agilent HPLC 1260 series), was used for the detection of sugars and other metabolites. Glucose, xylose, and arabinosewere detected at a retention time of 9.06 min, 9.72 min, and 10.62 min respectively [35]. The FPase information only accounts for three primary enzymes, Cellobiohydrolase (CBH), Endoglucanase (EG), and β-glucosidase (BGL). Therefore, the protein concentration of the mutant D4 secretome was considered for enzyme loading versus FPase in a comprehensive LCB hydrolysis experiment. The saccharification performance of the cellulase from mutant D4 was measured using HPLC analysis. The amount of liberated free sugars from the saccharification sugarcane bagasse pre-treated was estimated with a ratio of 3:1, phosphoric acid: Sulphuric acid (PASA) [15].
Results and discussions
Production of cellulases by EMS treated mutant
EMS mutagenesis kill curve plot
Each of the experimental set comprised an initial spore count of 2.2 × 106/mL with various EMS concentrations, such as 2, 4, 6, 8, 10, 15, 20, 25, and 30 mg/mL, labelled as B1 to B9 respectively. The experimental sets were exposed to varying concentrations of EMS for 60 min. EMS induces point mutations in DNA by alkylating guanine residues, particularly at the O6 position, leading to G:C to A:T transition mutations. Understanding the mode of action of EMS mutation is crucial for its effective use in mutagenesis studies aimed at generating genetic diversity and studying gene function. The control set (C)was not exposed to EMS. Table 1 [36]. For each set of EMS-exposed concentrations, serial dilutions were made up to 10− 5, followed by their spreading them onto the PDA plates. The experiments were performed in triplicates, and the resulting plates were incubated at 28 ± 2 ºC for 4 ± 1 days. After incubation, the colonies obtained on the plates in triplicates were counted. The average of colony count from each experimental set was used to plot the kill curve Table 1. Colonies that could grow on the plates post-EMS treatment were termed as mutants. These isolated mutants were plated on Production media containing Rose Bengal dye and carboxy methyl cellulose (PM-RB-CMC) medium for primary screening [17, 25, 37].
EMS primary screening
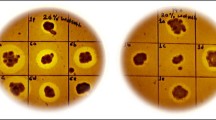
A total of 334 mutants were screened on PM-RB-CMC plates to identify the most potent cellulase producing mutant in comparison to its parent strain. These mutants were selected based on the maximum zone of cellulose hydrolysis on the PM-RB-CMC agar medium plates. The isolates that demonstrated the ratio of zone size to colony size, greater than that of the parent strain, were termed as higher cellulase producers. Out of 334 mutants,47 strains showed promising zones of clearance defined by a Zone Ratio (X-Y)/Y greater than 0.75 cm. This selection criterion has been categorized into three distinct levels marked by +++ (best) with a zone of hydrolysis greater than 1 cm, ++ (good) with a zone of hydrolysis in the range of 0.75 to 1.0 cm, and + (avg.) with a zone of hydrolysis between 0.5 and 0.75 cm. Since the parent strain showed the zone ratio of approximately 0.5 cm, this value has been set as the critical benchmark for comparison with the mutants (SI Tables 1 and 2).
The maximum number of the hits exhibiting the zone ratio in the category > 1.0 cm and 0.75-1.0 cm was obtained with doses of 25 mg/mL and 15 mg/mL of EMS, respectively. The consideration here is that the number of hits selected after exposure for primary screening was 53 and 198, respectively; thus, the relative occurrence was considered. Maximum relative occurrence of the best and good hits, based on zone ratio, was obtained in the case of a 30 mg/mL dose of EMS (SI Table 3).
Production of FPase
EMS mutants from primary screening were selected for extracellular enzyme production based on the plate-clearing zone assay used in primary screening. The stimulation of extracellular cellulase enzyme synthesis was carried out in submerged fungus cultures [38]. Agar blocks were inoculated into Erlenmeyer flasks (250 mL) containing 50 mL of the production medium used to test cellulose digestion by specific fungi from one-week-old mutant colonies cultivated on PDA at 28 ± 2 °C (SI Fig. 2). The mutants were incubated in an orbital shaker incubator for twelve days at 28 ± 2 °C. Following twelve days of cultivation, the fermented broth was centrifuged at 10,000 rpm for 10 to 15 min at 4 °C to remove solids. The clear supernatant was then analysed for the extracellular or free enzymes activities of the samples by Ghose’s method [27]. All of the experiments were conducted in triplicate. In a 500 mL Erlenmeyer flask, 100 mL of medium was placed, and average values were considered for the analysis. The results obtained till 11 days of incubation, top 17 mutants results have been tabulated (SI Table 4). The highest FPase activity observed was 6.66 U/mL by the D4 mutant followed by 5.55 U/mL produced by mutant B3 (Fig. 1 and SI Fig. 3). The highest sp. activity observed was 0.35 by mutant D4 which is 2.33-fold higher than the wild type (Table 3).
a) Production of cellulase by D4 mutant in a 40-L scale fermenter. b) SDS PAGE analysis of mutant D4 extracellular protein at 40 L scale fermenter and before and after TFF samples. Lane-1: 3rd -day supernatant, Lane-2: 4th -day supernatant, Lane-3: 5th -day supernatant, Lane-4: 6th -day supernatant, Lane-5: 7th -day supernatant, Lane-6: 8th -day supernatant, Lane-7: 9th -day supernatant, Lane-8: 10th -day supernatant of cell free enzyme broth. Lane-9 and 10: 10th day before TFF sample, Lane-11 and 12: 1:1 diluted concentrated broth after TFF sample and Lane M:Molecular markers from the top are72kDa, 55 kDa, 43 kDa, 34 kDa and 26 kDa
Production of CMCase
To begin, endoglucanases, notably carboxymethyl cellulase (CMCase), act on cellulose in between the chains to release tiny cellulose fragments with both reducing and non-reducing ends. Mutant B3 and D4 produced CMCase activities of 37 and 37.37 U/mL, respectively, which is 3.02 and 3.05-fold greater than the wild type (Fig. 1). The specific activities of B3 and D4 were 2.20 and 1.99, respectively (Table 4). Irfan et al. demonstrated that, the highest enzyme titer by strain Bacillus subtilis K-18 is 3.50 ± 0.11 IU/mL, obtained at 2% substrate concentration, 2% inoculum size, 1% yeast extract, pH 5.0, incubation temperature of 50 °C for 24 h during the fermentation period. The enzymatic hydrolysis of acid/alkali-treated pine needles exhibited 54.389% saccharification in acid-treated pine needles, demonstrating the effectiveness of the enzyme [39]. Tandon et al. reported a 12.81% hdrolysis rate of NaOH + H2O2 treated pine needles using indigenously produced cellulase and xylanase from P. notatum-102, which is lower than our results [40]. In this study, we observed a CMCase cell-free activity of 37.37 U/mL and a hydrolysis rate of approximately 49% for PASA-treated bagasse.
Production of β-glucosidase
The major enzyme component of cellulase is β-glucosidase, which completes the final phase of cellulose hydrolysis by converting cellobiose to glucose. In a recent study, Chen et al. reported the cloning of the BGL gene from Bacillus licheniformis into Escherichia coli and the production of the β-glucosidase enzyme with an activity of 45.44 U/mL [41]. We observed the highest β-glucosidase activity produced by mutant D4 was 32.1 U/mL (Fig. 1). In a study by Mallerman et al., Flammulina velutipes CFK 3111, a white rot fungus, was found to produced 1.6 U/mL of β-glucosidase with a glucose output of 10 g/L [42]. In this study, we observed that P. funiculosum NCIM1228, EMS was employed to produce cellulase overproducing mutant. Sixteen of the mutant derivatives improved FPase production from cellulose in batch cultures, out of a total of 334. When compared to the parental strain, D4, a stable EMS mutant, produced the highest protein production of 18.81 mg/mL, a 2.67-fold increase (Table 3). de Oliveira Rodrigues et al. reported that Aspergillus fumigatus produced 27.5 U/mL and 5.68 U/mL β-glucosidase when grown on Microcrystalline cellulose (Avicel) and Kraft pulp, respectively [43]. In terms of batch fermentation kinetic parameters, the mutation increased FPase specific activity by 2.33 times and improved CMCase and β -glucosidase specific activities by 1.14 and 1.13 times, respectively, in the D4 mutant (Tables 4 and 5).
Production of xylanases by EMS treated mutant
The EMS-treated mutant D4 and B3 produced the highest maximum xylanase activity 83.2 U/mL and 114.49 U/mL, respectively, which are 2.85 and 3.13-fold higher compared to the Parent P. funiculosum NCIM 1228 (Table 6) Interestingly, other mutants exposed to different concentrations of EMS (15 mg/mL to 30 mg/mL) for one hour achieved a relatively higher maximum xylanase activity, ranging from 1.07 to 2.74-fold. The profile of xylanase activity obtained by the non-treated control and EMS-treated mutants in SmF is shown in Fig. 1. In this investigation, mutants A23, B35, and D139 exhibited the lowest maximal xylanase activity, having been exposed to 30, 25, and 15 mg/mL of EMS for 60 min, respectively. An activity increment of 313% was observed in mutant C55 when compared to the parent strain (Table 6). The mutant C55 exhibited a substantially higher maximal xylanase activity of 121.53 U/mL, derived from a spore concentration of 1 ± 10 × 106 spores/mL and a total cell-free protein concentration of 14.23 mg/mL at medium pH of 5.5. EMS treatment, as described by Qin et al. produces base-pair insertions or deletions [44]. Indeed, EMS causes lasting lesions that leads to DNA base sequence changes. EMS treatment resulted in enhanced xylanase activity from the mutation D4 and B3 of the parent strain P. funiculosum NCIM 1228 in this investigation. When compared to the wild strain, the EMS mutant D4 produced 1.96 times the amount of xylanase. However, in certain circumstances, overproduction resulting from EMS-treated mutants was shown to be positive. The increase in xylanase activity in the mutant strain is attributed to a change in the gene sequences encoding for the xylanase enzyme, which enhances gene expression after exposure to the EMS mutagen. Radha et al. used a mutant of A. niger that underwent EMS treatment and found that it exhibited a maximal protease activity of 7.12 U/mL [45]. Yousaf et al. reported a similar result, indicating that the maximal yield of protease activity produced by the EMS mutant of A. oryzae was 662.61 ± 0.36 U/gds, representing a 4.03-fold improvement as compared to the wild type [46].
We investigated that FPase, CMCase, β-glucosidase, and xylanase activities were about three to six times greater in the EMS mutants than that in the parent strain (SI Table 4) [15]. The highest enzyme activity obtained in the EMS mutants was 37.37 U/mL CMCase in D4, 6.66 U/mL FPase in D4, 121 U/mL xylanases in the C55 mutant, and 32.1 U/mL -glucosidase in D4. The extracellular protein in the broth was found to be 19.86 mg/mL and 18.81 mg/mL in mutant D60 and D4 respectively.
Production of cellulases by promising EMS mutants of P. funiculosum at 40 L scale fermenter
The Fig. 2 (a) represents the average of three batches, demonstrating that the synthesis of enzymes and proteins is linked to growth. Improved enzyme production patterns were observed significantly. Enzyme activities and extracellular protein amounts in the culture broth of the fungi at the end of cultivation are shown in Fig. 2 (a). Carvalho et al. and their colleague reported that the growth process was conducted in batches in the fermenter. After an optimizing bioreactor, operational settings of 220 rpm agitation and 0.6 vvm aeration resulted in an enzyme pool with the activities of 0.508 U/mL for FPase, 9.204 U/mL for endoglucanase, and 2.395 U/mL for β -glucosidase. The sequential optimization technique was successful, resulting in cellulase output that was 3.6 to 9.5 times higher than under non-optimized conditions [47]. In this study, extracellular protein production in fermenter cultures was observed as promising. Enzyme activities were improved from 2-fold to 5-fold as compared to parent fungi. Moreover, the D4 mutant boosted the activity of cellulolytic enzymes in fed-batch culture; this is the first time that it has been documented that the fermenter system used in this study considerably improved cellulase production in addition to protein production. The FPase activity of 4.79 U/mL was achieved at a 40 L scale of submerged fermentation in this study. In contrast, Ogunyewo et al. reported a maximum FPase production of 3.41 U/mL in a 20 L scale fermenter with the assistance of recombinant P. funiculusum PfMig188 [12] Prasanna et al. obtained the best activities on an optimised medium with Penicillium sp., achieving yields of 25 U/mL for CMCase and 9.52 U/mL for β-glucosidase. These values represented increases of 9.2, 5.9, and 43.8-fold over unoptimized medium titers [19]. In this study, we achieved 42.49 U/mL and 21.17 U/mL of CMCase and β-glucosidase, respectively, at a 40 L fermenter scale Fig. 2 (a). Sridevi et al. reported Penicillium sp. that produced xylanase via submerged fermentation with xylose as the major substrate, and the enzyme yield was measured on a regular basis [48]. On the fourth day of incubation, xylanase production reached 123.1 U/mL. In this work, Mutant D4 achieved a maximum xylanase production of 101.87 U/mL on the sixth day at the fermenter scale, along with a promising cellulase titer. On the tenth day at the 40 L fermenter scale, it was noted that the protein productivity was 1.79 g/L/day and the FPase productivity was 479 U/L/day (Table 7). According to Gang Hu et al., the molecular mass of isozymes of cellulases from Trichoderma reesei (Celluclast) and Aspergillus niger ranged from around 18 kDa to about 120 kDa. The sensitivity of staining methods determines the amount of proteins that may be observed. The gels stained with the silver stain kit revealed eight protein bands in his study [49]. An almost similar pattern was observed in this study with mutant D4 of P. funiculosum Fig. 2 (b). Goyal et al. reported the results are similar to those described by Gang Hu et al. who reported molecular mass in the range of 45 to 100 kDa for Trichoderma reesei cellulases. Purified CBHI from multiple sources has a molecular mass ranging from 42 to 72 kDa, CBHII has a molecular mass ranging from 50 to 58 kDa, endoglucanase-I is a significant cellulase component has 54 kDa, endoglucanase-II has 48 kDa, and endoglucanase-III has a molecular mass ranging from 20 to 23.5 kDa [50]. McGregor et al. predicted a molecular mass major xylanase band at ~ 57 kDa and a xylanase band of 111 kDa may represent a β-xylosidase [51]. We observed that the secretome of mutant D4 has roughly nine bands in the range of 28 kDa to 100 kDa, with the most intense band between 45 kDa and 58 kDa. TFF was used to concentrate the crude extracellular enzyme by 1.40 folds. The FPase concentration was 4.79 U/mL before TFF and increased to 6.71 U/mL after TFF. Fang et al. reported 2-fold concentrated by using TFF [33].
LCB hydrolysis by cellulases from EMS mutants of P. funiculosum vs Advanced enzymes
The composition of untreated and PASA-treated bagasse, rice straw as well as corn cob is detailed in Table 8. Pre-treated LCB has been suggested as a viable alternative for the conversion of LCB into fermentable sugars, biofuels, and other platform chemicals such as 5-hydroxymethylfurfural (5-HMF), xylitol, sugar alcohols, lactic acid, 2,3BDO, levulinic acid, phenols, succinic acid, and furfurals through enzymatic degradation by fungal enzymes [52,53,54]. Peng et al. reported in their study that the crude enzyme from MEA-12 demonstrated good enzymatic hydrolysis efficiency against three different biomasses (cornstalk, bamboo, and reed), when supplemented with cellulase from T.reesei Rut-C30 [11]. In this study, we found that without supplementation of an external enzyme, the crude secretome of the D4 mutant of P. funiculosum effectively saccharified the pre-treated bagasse, rice straw and corn cob into soluble sugars. Figure 3a,b and c is the average of the three values. The results showed that after saccharification with the mutant enzyme, 3.54%, 5.13% and 4.54% of soluble sugars were liberated from pre-treated bagasse, corn cob and rice straw (SI Tables 5, 6 and 8).
Bussamra et al. utilized an optimised enzyme cocktail for efficient hydrolysis, comprising T.reesei fraction (80%), endoglucanase (10%), and β-glucosidase (10%), theoretically converted 72% of the cellulose in hydrothermally pre-treated bagasse. In comparison, commercial Celluclast 1.5 L converts 49.11% [55]. By applying the synergism principle and statistical analysis, a rational enzyme mixture was capable of enhancing biomass saccharification. The enzyme from P. funiculosum NCIM 1228 exhibited a hydrolysis efficiency of 51.30% on sugarcane bagasse when administered at a dosage of 7 FPase units per gram of cellulose [15]. However, when we employed doses of 6 FPU, 8 FPU, and 8 FPU per gram of cellulose in this trial on bagasse, corn cob and rice straw respectively, we achieved hydrolysis efficiencies of 54.03%, 71.4% and 76.87% (Fig. 3a, b, and c). This indicates that no formulation was required as the necessary cocktail is already available in the secretome of the P. funiculosum D4 mutant. In this way, D4 mutant is a potential choice for efficient LCB hydrolysis and other biocatalysis-dependent processes.
Conclusions
In this study, the extracellular cellulase activity of various mutants from filamentous fungus P. funiculosum NCIM 1228 was investigated to enhance extracellular cellulase production. EMS-based mutagenesis was employed to improve the strain for better performance, especially for increasing enzyme production. To the best of our knowledge, no definitive reports or studies are highlighting 6.47, 3.05, 3.03, and 3.19-fold higher activities of FPase, CMCase, β-glucosidase, and xylanase, respectively, compared to the parent strain, indicating its promising future. We believe that this study is crucial, and the substantial results can serve as a baseline to carried out further studies to meet the demand of practical application. Submerged fermentation of the promising strain D4 was conducted in a 40 L fermenter and the saccharification efficiency of the cellulase enzyme secreted by mutant D4 was compared with commercial cellulase enzymes using 6 FPU per gram of cellulose dosage. The mutant D4 enzyme demonstrated hydrolysis efficiencies of 54.03%, 71.44%, and 76.87% on bagasse, corn cob, and rice straw, respectively, whereas the commercial enzyme showed hydrolysis efficiencies of 53.07%, 78.06%, and 78.66% on the same substrates. These results clearly show that the mutant strains produced efficient cellulases that are comparatively promising. Additionally, compared to the parent strain, the EMS mutant D4 also exhibited high extracellular FPase, CMCase, xylanase, and protein in the broth, which could be promising in reducing the overall cellulase production costs at a large scale. Further process improvement could result in a more suitable alternative for industrial scale. Furthermore, the P. funiculosum D4 mutant and its crude enzyme extract rich in cellulase efficiently degraded pre-treated LCB without any modifications in the secretome. A promising hyper-producing mutant and its cellulase, efficient in hydrolysing common biomass bagasse, corn cob, and rice straw provide a new option for bioresource processing in the future.
References
Bhuvaneshwari S, Hettiarachchi H, Meegoda JN. Crop Residue Burning in India: Policy challenges and potential solutions. Int J Environ Res Public Health 2019. 2019;16:832. https://doi.org/10.3390/IJERPH16050832.
Pandey A, Soccol C, Nigam P. Biotechnology for agro-industrial residues Utilisation. Dordrecht: Springer Netherlands; 2009.
Sarsaiya S, Jain A, Kumar Awasthi S, et al. Microbial dynamics for lignocellulosic waste bioconversion and its importance with modern circular economy, challenges and future perspectives. Bioresour Technol. 2019;291. https://doi.org/10.1016/j.biortech.2019.121905.
Paul PK, Al Azad S, Rahman MH, et al. Catabolic profiling of selective enzymes in the saccharification of non-food lignocellulose parts of biomass into functional edible sugars and bioenergy: an in silico bioprospecting. J Adv Vet Anim Res. 2022;9:19–32. https://doi.org/10.5455/javar.2022.i565.
Jiang C, Cheng Y, Zang H, et al. Biodegradation of lignin and the associated degradation pathway by psychrotrophic Arthrobacter sp. C2 from the cold region of China. Cellulose. 2020;27:1423–40. https://doi.org/10.1007/S10570-019-02858-3.
Zabidi NAM, Foo HL, Loh TC, et al. Enhancement of versatile extracellular cellulolytic and hemicellulolytic enzyme productions by Lactobacillus plantarum RI 11 isolated from Malaysian food using renewable natural polymers. Molecules. 2020;25. https://doi.org/10.3390/molecules25112607.
Randhawa A, Ogunyewo OA, Eqbal D, et al. Disruption of zinc finger DNA binding domain in catabolite repressor Mig1 increases growth rate, hyphal branching, and cellulase expression in hypercellulolytic fungus Penicillium Funiculosum NCIM1228. Biotechnol Biofuels. 2018;11:15. https://doi.org/10.1186/s13068-018-1011-5.
Chavan S, Shete A, Mirza Y, Dharne MS. Investigation of cold-active and mesophilic cellulases: opportunities awaited. Biomass Convers Biorefin. 2021;1–24. https://doi.org/10.1007/s13399-021-02047-y.
Liu G, Zhang J, Bao J. Cost evaluation of cellulase enzyme for industrial-scale cellulosic ethanol production based on rigorous Aspen Plus modeling. Bioprocess Biosyst Eng. 2016;39:133–40. https://doi.org/10.1007/S00449-015-1497-1.
Song Y-H, Lee K-T, Baek J-Y, et al. Isolation and characterization of a novel glycosyl hydrolase family 74 (GH74) cellulase from the black goat rumen metagenomic library. Folia Microbiol (Praha). 2017;62:175–81. https://doi.org/10.1007/s12223-016-0486-3.
Peng ZQ, Li C, Lin Y et al. (2021) Cellulase production and efficient saccharification of biomass by a new mutant Trichoderma Afroharzianum MEA-12. Biotechnol Biofuels 14:. https://doi.org/10.1186/S13068-021-02072-Z.
Ogunyewo OA, Randhawa A, Joshi M, et al. Engineered Penicillium funiculosum produces potent lignocellulolytic enzymes for saccharification of various pretreated biomasses. Process Biochem. 2020;92:49–60. https://doi.org/10.1016/j.procbio.2020.02.029.
Taylor LE, Knott BC, Baker JO, et al. Engineering enhanced cellobiohydrolase activity. Nat Commun. 2018;9. https://doi.org/10.1038/s41467-018-03501-8.
Chen T, Huang L, Wang M, et al. Ethyl Methyl Sulfonate-Induced Mutagenesis and its effects on Peanut Agronomic, Yield and Quality traits. Agronomy. 2020;10:655–68. https://doi.org/10.3390/AGRONOMY10050655.
Chavan SB, Shete AM, Dharne MS. (2023) Bioprocess Optimization of Penicillium funiculosum NCIM 1228 for Improved Production and Hydrolytic Efficiency of Cellulases on Sugarcane Bagasse. Sugar Tech 2023 1–19. https://doi.org/10.1007/S12355-023-01324-6.
Liu F, Wang Z, Manglekar RR, Geng A. Enhanced cellulase production through random mutagenesis of Talaromyces Pinophilus OPC4-1 and fermentation optimization. Process Biochem. 2020;90:12–22. https://doi.org/10.1016/j.procbio.2019.11.025.
Ram L, Kaur K, Sharma S. Screening isolation and characterization of Cellulase Producing micro-organisms from Soil. Int J Pharm Sci Invent. 2014;3:12–8.
Ye Y, Li X, Cao Y, et al. A Β-xylosidase hyper-production Penicillium Oxalicum mutant enhanced ethanol production from alkali-pretreated corn stover. Bioresour Technol. 2017;245. https://doi.org/10.1016/j.biortech.2017.08.155.
Prasanna HN, Ramanjaneyulu G, Reddy BR. (2016) Optimization of cellulase production by Penicillium Sp. 3 Biotech 6:. https://doi.org/10.1007/S13205-016-0483-X.
Bhat S, Bhat KM. Food Macromolecular Science Department, Institute of Food Research Reading Laboratory, Earley Gate, Whiteknights Road, Reading, RG6 6BZ, United Kingdom. Biotechnol Adv. 1997;15:583–620.
Zhang X, Li Y, Zhao X, Bai F. Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour Technol. 2017;223. https://doi.org/10.1016/j.biortech.2016.10.083.
Burlacu A, Israel-roming F, Cornea CP. Fungal strains improvement for Xylanase over Production through Physical. Agrolife Sci J. 2017;6:40–7.
Javed MM, Ikram-Ul-Haq, Mariyam I. Multistep mutagenesis for the over-expression of cellulase in humicola insolens. Pak J Bot. 2011;43:669–77.
Yang B, Zhou S, Ou L, et al. A novel single-base mutation in CaBRI1 confers dwarf phenotype and brassinosteroid accumulation in pepper. Mol Genet Genomics. 2019. https://doi.org/10.1007/s00438-019-01626-z.
Mahmoud M, El-zyat S, El-sayed M. (2018) Cellulolytic activity of cellulose-decomposing fungi isolated from Aswan hot desert soil, Egypt.
Venkatachalam M, Shum-Chéong-sing A, Dufossé L, Fouillaud M. (2020) Statistical Optimization of the Physico-Chemical Parameters for Pigment Production in Submerged Fermentation of Talaromyces albobiverticillius 30548. Microorganisms 2020, Vol 8, Page 711 8:711. https://doi.org/10.3390/MICROORGANISMS8050711.
Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987. https://doi.org/10.1351/pac198759020257.
Contreras F, Pramanik S, Rozhkova AM, et al. Engineering Robust cellulases for tailored lignocellulosic degradation cocktails. Int J Mol Sci. 2020;21:1–25. https://doi.org/10.3390/ijms21051589.
Das A, Paul T, Halder SK, et al. Study on regulation of growth and biosynthesis of cellulolytic enzymes from newly isolated aspergillus fumigatus ABK9. Pol J Microbiol. 2013;62:31–43. https://doi.org/10.33073/pjm-2013-004.
Bashirova A, Pramanik S, Volkov P, et al. Disulfide Bond Engineering of an endoglucanase from Penicillium Verruculosum to improve its thermostability. Int J Mol Sci. 2019;20:1602. https://doi.org/10.3390/ijms20071602.
Khokhar ZU, Syed Qul, Wu A, Athar J MA. On-site cellulase production by Trichoderma reesei 3EMS35 mutant and same vessel saccharification and fermentation of acid treated wheat straw for ethanol production. EXCLI J. 2014;13:82–97. https://doi.org/10.17877/DE290R-15480.
Patel AK, Singhania RR, Sim SJ, Pandey A. Thermostable cellulases: current status and perspectives. Bioresour Technol. 2019;279:385–92. https://doi.org/10.1016/j.biortech.2019.01.049.
Fang H, Kandhola G, Rajan K, et al. Effects of oligosaccharides isolated from pinewood hot water pre-hydrolyzates on recombinant cellulases. Front Bioeng Biotechnol. 2018;6:55. https://doi.org/10.3389/FBIOE.2018.00055/BIBTEX.
Aanchal AN, Kanika, et al. Response surface methodology for optimization of microbial cellulase production. Rom Biotechnol Lett. 2016;21:11832–41.
Patel M, Patel HM, Dave S. Determination of bioethanol production potential from lignocellulosic biomass using novel Cel-5m isolated from cow rumen metagenome. Int J Biol Macromol. 2019. https://doi.org/10.1016/j.ijbiomac.2019.10.240.
Ribeiro O, Magalhães F, Aguiar TQ, et al. Random and direct mutagenesis to enhance protein secretion in Ashbya Gossypii. Bioengineered. 2013;4. https://doi.org/10.4161/bioe.24653.
Paula CCPD, Montoya QV, Meirelles LA, et al. High cellulolytic activities in filamentous fungi isolated from an extreme oligotrophic subterranean environment (Catão cave) in Brazil. Acad Bras Cienc. 2019;91. https://doi.org/10.1590/0001-3765201920180583.
Kumar AK, Parikh BS, Singh SP, Shah D. Use of combined UV and chemical mutagenesis treatment of aspergillus terreus D34 for hyper-production of cellulose-degrading enzymes and enzymatic hydrolysis of mild-alkali pretreated rice straw. Bioresour Bioprocess. 2015. https://doi.org/10.1186/s40643-015-0062-8.
Irfan M, Mushtaq Q, Tabssum F, et al. Carboxymethyl cellulase production optimization from newly isolated thermophilic Bacillus subtilis K-18 for saccharification using response surface methodology. AMB Express. 2017;7:1–9. https://doi.org/10.1186/S13568-017-0331-3/FIGURES/7.
Tandon D, Sharma N, Kaushal R. Saccharification of pine needles by a potentialcelluolytic and hemicellulolyic strain of Penicilium notatum 102 isolated from forest soil - Google search. Int J Biol Pharm Allied Sci. 2012;1:1344–55.
Chen Z, Meng T, Li Z, et al. Characterization of a beta-glucosidase from Bacillus licheniformis and its effect on bioflocculant degradation. AMB Express. 2017;7:197. https://doi.org/10.1186/S13568-017-0501-3.
Mallerman J, Papinutti L, Levin L. Characterization of β-glucosidase produced by the white rot fungus Flammulina velutipes. J Microbiol Biotechnol. 2015;25:57–65. https://doi.org/10.4014/JMB.1401.01045.
de Oliveira Rodrigues P, dos Santos BV, Costa L, et al. Xylanase and β-glucosidase production by aspergillus fumigatus using commercial and lignocellulosic substrates submitted to chemical pre-treatments. Ind Crops Prod. 2017;95:453–9. https://doi.org/10.1016/J.INDCROP.2016.10.055.
Qin X, Li R, Luo X, et al. Deletion of ligD significantly improves gene targeting frequency in the lignocellulolytic filamentous fungus Penicillium Oxalicum. Fungal Biol. 2017;121:615–23. https://doi.org/10.1016/j.funbio.2017.04.005.
Radha S, Himakiran Babu R, Sridevi A, et al. Development of mutant fungal strains of aspergillus Niger for enhanced production of acid protease in submerged and solid state fermentation. Eur J Exp Biol. 2012;2:1517–28.
Yousaf M, Irfan M, Khokhar Z et al. (2010) Enhanced production of protease by mutagenized strain of Aspergillus Oryzae in solid substrate fermentation of Rice Bran. 22:119–23.
De Carvalho A, Carvalho ML, De Barros Gomes DF et al. E, (2014) Optimisation of cellulase production by Penicillium funiculosum in a stirred tank bioreactor using multivariate response surface analysis. Enzyme Res 2014:. https://doi.org/10.1155/2014/703291.
Sridevi A, Narasimha G, Suvarnalatha Devi P. Production of Xylanase by Penicillium Sp. and its biobleaching efficiency in Paper and Pulp Industry a. Int J Pharm Sci Res. 2017;7:266. https://doi.org/10.13040/IJPSR.0975-8232.10(3).1307-11.
Hu G, Heitmann JA, Rojas OJ, et al. Monitoring cellulase protein adsorption and recovery using SDS-PAGE. Ind Eng Chem Res. 2010;49:8333–8. https://doi.org/10.1021/IE100731B.
Goyal A, Ghosh B, Eveleigh D. Characteristics of fungal cellulases. Bioresour Technol. 1991;36:37–50. https://doi.org/10.1016/0960-8524(91)90098-5.
McGregor NGS, de Boer C, Santos M et al. (2022) Activity-based protein profiling reveals dynamic substrate-specific cellulase secretion by saprotrophic basidiomycetes.
Xie X, Madadi M, Al Azad S, et al. Unraveling the mechanisms underlying lignin and xylan dissolution in recyclable biphasic catalytic systems. Fuel. 2024;363:130890. https://doi.org/10.1016/J.FUEL.2024.130890.
Fang X, Yano S, Inoue H, Sawayama S. Strain improvement of Acremonium cellulolyticus for cellulase production by mutation. J Biosci Bioeng. 2009. https://doi.org/10.1016/j.jbiosc.2008.11.022.
Szengyel Z, Zacchi G, Varga A, Réczey K. Cellulase production of Trichoderma reesei rut C 30 using steam- pretreated spruce. Hydrolytic potential of cellulases on different substrates. Appl Biochem Biotechnol - Part Enzyme Eng Biotechnol. 2000;84–86:679–91. https://doi.org/10.1385/ABAB:84-86:1-9:679.
Bussamra BC, Freitas S, da Costa AC. Improvement on sugar cane bagasse hydrolysis using enzymatic mixture designed cocktail. Bioresour Technol. 2015;187:173–81. https://doi.org/10.1016/J.BIORTECH.2015.03.117.
Acknowledgements
SC is thankful to the Directors of CSIR-NCL, Pune., Academy of Scientific and Innovative Research (AcSIR), Ghaziabad- 201002, India, and Praj Matrix - R & D Center (a division of Praj Industries Limited), Pune 412115 for infrastructure and support. The manuscript has been checked for plagiarism using iThenticate software.
Funding
This research did not have any financial support.
Author information
Authors and Affiliations
Contributions
SC: performed conceptualization, methodology, data curation, formal analysis, writing original draft preparation, visualization, and editing. MD and AS performed editing, reviewing, visualization, and supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or vertebrates.
Consent for publication
Not applicable.
Competing interests
The author declares that he/she has no competing interest.
Consent to participate
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chavan, S., Shete, A. & Dharne, M.S. Strain improvement for cellulolytic enzymes for effective saccharification of lignocellulosic biomass by mutant of Penicillium funiculosum NCIM 1228. Syst Microbiol and Biomanuf 4, 716–730 (2024). https://doi.org/10.1007/s43393-024-00247-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-024-00247-x