Abstract
The use of abundant and cheap one carbon (C1) feedstocks to produce value-added chemicals is an important approach for achieving carbon neutrality and tackling environmental problems. The conversion of C1 feedstocks to high-value chemicals is dependent on efficient C1 assimilation pathways and microbial chassis adapted for efficient incorporation. Here, we opted to summarize the natural and synthetic C1 assimilation pathways and their key factors for metabolizing C1 feedstock. Accordingly, we discussed the metabolic engineering strategies for enabling the microbial utilization of C1 feedstocks for the bioproduction of value-added chemicals. In addition, we highlighted future perspectives of C1-based biomanufacturing for achieving a low-carbon footprint for the biosynthesis of chemicals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental and climate problems are becoming increasingly serious, with the continuous consumption of fossil fuels as one of the main causes. The mass of carbon in the atmosphere increased from 590 to 876 GtC (i.e., an increase of 48%) in 2020 [1]. To deal with this issue, recent advances have mainly focused on producing high-value chemicals from one-carbon (C1) feedstock, such as carbon dioxide (CO2), carbon monoxide (CO), methane (CH4), methanol (CH3OH), and formate (HCOOH), which is a win–win strategy to exploit carbon resources and alleviate global energy shortages and environmental problems. Many methods have been developed to efficiently utilize C1 feedstocks, including thermal catalytic conversion, electrocatalysis, photocatalysis, and photo-electrocatalysis. In electrochemical reactions, CO2 can proceed through a 2-electron, 4-electron, and other reduction pathways to generate various products, such as ethylene, ethanol, and propanol [2]. However, such processes are usually technically complicated and require energy-intensive process operations.
Many efforts have been dedicated to the use of C1-based biomanufacturing for the production of value-added products. The bio-utilization of CO and CH4 is mainly achieved by natural microorganisms. Methanotrophs, which can oxidize methane and utilize it as their sole carbon and energy source, are attractive hosts that can convert CH4 to bioplastics, biofuels, and other high-value chemicals [3]. Acetogens are ideal biocatalyst hosts owing to their energy-efficient Wood–Ljungdahl pathway [4]. Acetogens can be used to convert CO to C2 chemicals, such as ethanol and acetic acid. Methanol, CO2, and formate can be utilized for the bioproduction of chemicals by natural C1-utilizing bacteria and genetically modified industrial chassis. Numerous studies have been performed to understand the physiology of these C1-utilizing strains and engineer them to produce value-added chemicals from C1 feedstock. For example, the methylotroph, Methylobacterium extorquens AM1, is used for the biosynthesis of chemicals, such as mevalonate, butadiene, and 3-hydroxypropionic acid, from methanol. However, naturally occurring C1-utilizing microorganisms are limited by low pathway efficiency and lack of genetic tools for metabolic engineering [5]. An attractive alternative is to engineer model biotechnological microorganisms to utilize CO2, methanol, and formate. Such efforts are mainly dependent on the introduction of existing pathways or new routes into these strains. For example, the ribulose monophosphate cycle has been introduced into Escherichia coli to produce succinic acid, ethanol, and acetone. In particular, E. coli and Pichia pastoris have been achieved for the transformation from heterotrophic to autotrophic [6, 7].
In this review, the natural and synthetic pathways for C1 fixation and their unique characteristics were summarized. As a result, advances in metabolic engineering strategies for converting C1 feedstocks to high-value chemicals were discussed (Fig. 1). The biocatalysis of C1 feedstocks to chemicals is a key strategy for achieving carbon neutrality, and involves the exploitation of more suitable native or non-native C1 utilizers, ultimately promoting future development of biological C1 utilization platforms.
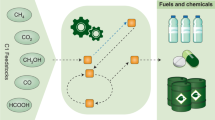
The C1-based biomanufacturing. C1 feedstocks from biomass, industrial exhaust, automobile exhaust, and fossil fuels can be converted into value-added chemicals such as bioplastics, foods, pharmaceuticals, biofuels by C1-fixing enzymes, pathways and cell factories. The C1-based biomanufacturing is an important approach to achieve carbon neutrality and tackle environmental problems
Natural pathways for C1 utilization
Natural C1-utilizing microorganisms can produce organic compounds from C1 feedstocks by consuming coenzymes, such as ATP and NAD(P)H. Herein, all native C1 assimilation pathways, including the Calvin–Benson–Bassham, cycle, the Wood–Ljungdahl pathway, the reductive glycine pathway, the reductive TCA cycle, the xylulose monophosphate cycle, the ribulose monophosphate cycle, the 3-hydroxypropionate bicycle, the 3-hydroxypropionate-4-hydroxybutyrate cycle, the dicarboxylate/4-hydroxybutyrate cycle, and the serine cycles, were systemically discussed.
The Calvin–Benson–Bassham cycle
The Calvin–Benson–Bassham (CBB) cycle, also known as the reductive pentose-phosphate cycle, is the most famous pathway used by plants, algae, proteobacteria, and cyanobacteria (Fig. 2j). In this cycle, only one molecule of CO2 is fixed by ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisco) in each round of the cycle. The key enzyme, RuBisco, catalyzes CO2 and ribulose-1,5-bisphosphate (RuBP) to generate two molecules of 3-phosphoglycerate (3PG). 3PG is then reduced to glycolaldehyde-3-phosphate (G3P) through two enzyme-catalyzed reactions by 3-phosphoglycerate kinase and G3P dehydrogenase. The 5-bisphosphate (Ru5P), which generates RuBP, is regenerated through the biological conversion of C3, C4, C5, C6, and C7 sugars. After three rounds of the CBB cycle, one molecule of 3PG is produced, and six NAD(P)H and nine ATP molecules are consumed, highlighting an energy-intensive process. This pathway has been introduced into E. coli to induce transformation from heterotrophy to autotrophy [8].
Natural pathways for C1 utilization. a The Wood-Ljungdahl pathway. b The reductive glycine pathway. c The serine cycle. d The ribulose monophosphate cycle. e The reductive TCA cycle. Green, rTCAI; Black, rTCAII. f The xylulose monophosphate cycle. g The 3-hydroxypropionate bicycle. h The 3-hydroxypropionate-4-hydroxybutyrate cycle. i The dicarboxylate/4-hydroxybutyrate cycle. j The Calvin-Benson-Bassham cycle. Each segment of the dotted lines shows a reaction; dot and dash lines show the anaplerosis of Ru5P and Xu5P though pentose phosphate pathway. Abbreviations of metabolites stand for: Ac-CoA, acetyl-CoA; Gly, glycine; Ser, Serine; pyru, pyruvate; hypyr, hydroxypyruvate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; mal-CoA, malyl-CoA; gly, glyoxylate; Ru5P, ribulose 5-phosphate; H6P, hexulose 6-phosphate; F6P, fructose-6-phosphate; suc-CoA, succinyl-CoA; 2-oglu, 2-oxoglutarate; isoc, isocitrate; cit, citrate; cit-CoA, citryl-CoA; Xu5P, xylulose 5-phosphate; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; Ma-CoA, malonyl-CoA; 3-HP, 3-hydroxypropionate; pro-CoA, propionyl-CoA; mem-CoA, (S)-methylmalonyl-CoA; mm-CoA, (2R,3S)-β-methylmalyl-CoA; cm-CoA, (3S)-citramalyl-CoA; acac-CoA, acetoacetyl-CoA; RuBP, ribulose 1, 5-bisphosphate; 3PG, 3-phosphoglycerate; BPG, 1,3-bisphosphoglycerate
The Wood–Ljungdahl pathway
The Wood–Ljungdahl pathway (WLP), also known as the reductive acetyl-CoA pathway in several types of microorganisms, such as homoacetogenic bacteria, is the most energy-efficient pathway for synthesizing acetyl-CoA from CO2 (Fig. 2a). In this pathway, CO2 is reduced to formate by formate dehydrogenase. Thereafter, formate is incorporated into tetrahydrofolate (THF) to produce 5,10-methylenetetrahydrofolate (5,10-CH2 = THF) by three enzymes: formate-tetrahydrofolate ligase, methenyltetrahydrofolate cyclohydrolase, and methylenetetrahydrofolate dehydrogenase. Methylenetetrahydrofolate reductase then catalyzes the conversion of 5,10-CH2 = THF to 5-methyltetrahydrofolate for condensing with CO reduced from another molecule of CO2. As a result, WLP outputs one molecule of acetyl-CoA after one round of this cycle, consuming four NAD(P)H and one ATP. Notably, the WLP is restricted to anaerobic conditions, and can be broadened to produce acetone and isopropanol from acetyl-CoA [9].
The reductive glycine pathway
The reductive glycine pathway (rGlyP) was designed as a novel pathway [10] prior to its discovery in Desulfovibrio desulfuricans [11] and Clostridium drakei [12]. The enzyme-catalyzed reactions from CO2 to 5,10-CH2 = THF in rGlyP are the same as those in the WLP (Fig. 2b). 5,10-CH2 = THF reacts with CO2 and NH3 to generate glycine followed by glycine to generate serine. Serine is then catalyzed by serine deaminase to produce pyruvate, which enters central metabolism. Overall, each round of rGlyP can fix three molecules of CO2 to form one molecule of pyruvate, consuming three NAD(P)H and two ATP molecules. Notably, this pathway has been used to engineer synthetic methylotrophic E. coli [13, 14].
The reductive TCA cycle
The reductive TCA cycle (rTCA) is the TCA cycle in the reverse direction found in some autotrophic eubacteria and archaea (Fig. 2e). The two natural rTCA pathways, rTCAI and rTCAII, are the same for most metabolites and enzymes. Two successive carboxylation reactions can fix two CO2 molecules with succinyl-CoA to generate C6 compounds. First, succinyl-CoA is transformed to 2-oxoglutarate, catalyzed by 2-oxoglutarate synthase in rTCAI and rTCAII. However, only one enzyme (isocitrate dehydrogenase) converts 2-oxoglutarate to isocitrate in rTCAI, and two enzymes (2-oxoglutarate carboxylase and oxalosuccinate reductase) are required for rTCAII. Isocitrate is then converted to citrate in rTCAI, but is converted to citryl-CoA by rTCAII. C6 compounds are split into acetyl-CoA and oxaloacetate. Each round of the rTCA cycle assimilates two molecules of CO2 and outputs one molecule of acetyl-CoA, consuming two ATP and four NAD(P)H. The rTCA pathway has been used for succinic acid production in Mannheimia succiniciproducens [15].
The xylulose monophosphate cycle and the ribulose monophosphate cycle
The ribulose monophosphate (RuMP) cycle and the xylulose monophosphate (XuMP) cycle are natural pathways for formaldehyde assimilation (Fig. 2d, f). In RuMP, hexulose phosphate synthase catalyzes the conversion of ribulose 5-phosphate (Ru5P) and formaldehyde to hexulose 6-phosphate (H6P), which is further transformed to fructose-6-phosphate (F6P) by phosphohexulose isomerase. F6P enters the central carbon metabolism to regenerate Ru5P. In XuMP, formaldehyde condenses with xylulose 5-phosphate (Xu5P) to generate dihydroxyacetone (DHA) and glyceraldehyde 3-phosphate (G3P). DHA and G3P can regenerate Xu5P via the pentose phosphate pathway (PPP). Notably, each round of XuMP or RuMP assimilates one formaldehyde molecule. XuMP and RuMP have been heterologously expressed to biosynthesize many chemicals, such as succinate [16].
Other C1-utilizing pathways
In addition to the above pathways, many other natural C1 assimilation pathways exist, including the 3-hydroxypropionate (3-HP) bicycle, the 3-hydroxypropionate-4-hydroxybutyrate (3-HP/4-HB) cycle, the dicarboxylate/4-hydroxybutyrate (DC/HB) cycle, and the serine cycle.
The 3-HP bicycle contains two cyclic CO2 fixation pathways (Fig. 2g). The reactions from acetyl-CoA to propionyl-CoA are common in both cycles. Acetyl-CoA fixes CO2 to generate malonyl-CoA by acetyl-CoA carboxylase, and malonyl-CoA is transformed to propionyl-CoA through five enzyme-catalyzed reactions. On one branch from the bifurcation point, propionyl-CoA fixes CO2 to generate methylmalonyl-CoA via propionyl-CoA carboxylase. Methylmalonyl-CoA is then transformed to malyl-CoA, which is split into acetyl-CoA for another round of cycle and glyoxylate for the reactions of other branches. On the other branch from the bifurcation point, propionyl-CoA combines with glyoxylate to form (2R,3S)-β-methylmalyl-CoA, which can be converted to (3S)-citramalyl-CoA by three enzyme-catalyzed reactions. Subsequently, (3S)-citramalyl-CoA is split into acetyl-CoA for another round of the cycle and pyruvate for the biosynthesis of other metabolites. One round of the 3-HP bicycle fixes three molecules of CO2, consuming five ATP and seven NAD(P)H.
In the 3-HP/4-HB cycle, the reactions from acetyl-CoA to methylmalonyl-CoA are the same as those in the 3-HP cycle (Fig. 2h). Methylmalonyl-CoA is transformed into 4-hydroxybutyrate (4-HB), which is further converted to acetoacetyl-CoA. Finally, acetoacetyl-CoA is split into two molecules of acetyl-CoA. One round of the 3-HP/4-HB cycle fixes two molecules of CO2, consuming six ATP and four NAD(P)H.
In the DC/HB cycle, acetyl-CoA fixes CO2 to generate pyruvate by pyruvate synthase, and pyruvate is transformed to phosphoenolpyruvate (PEP) by fixing another CO2, which is converted to oxaloacetate by phosphoenolpyruvate (PEP) carboxylase (Fig. 2i). Oxaloacetate is transformed to succinyl-CoA through four steps of rTCA, and succinyl-CoA is transformed into two molecules of acetyl-CoA through seven steps of 3-HP/4-HB. One round of the DC/HB cycle fixes two molecules of CO2, consuming five ATP and four NAD(P)H.
In the serine cycle, glycine reacts with 5,10-CH2 = THF to generate serine, which is then transformed to toxic hydroxypyruvate (Fig. 2c). Hydroxypyruvate is then converted to PEP to fix another CO2 to form oxaloacetate. Oxaloacetate is transformed into malate and is finally split into acetyl-CoA and glyoxylate to generate glycine. One round of the serine cycle fixes two molecules of CO2, consuming three ATP and three NAD(P)H.
Synthetic pathways for C1 utilization
Although natural C1 assimilation pathways exist in many microorganisms, these pathways might not be the most optimal as they contain many enzymes and require remarkable amounts of energy. Thus, many studies have focused on the design of novel pathways. These unnatural pathways, including the modified serine cycle, the homoserine cycle, the formyl-CoA elongation pathway, the artificial starch anabolic pathway, a synthetic acetyl-CoA pathway, the crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA cycle, the Gnd-Enter-Doudoroff cycle, the reductive glyoxylate and pyruvate synthetic cycle, malyl-CoA-glycerate pathway, and the POAP cycle, are described below.
The modified serine cycle and the homoserine cycle
The modified serine cycle, an adapted pathway for E. coli, avoids the toxic intermediate hydroxypyruvate. In the modified serine cycle, C1 compounds are converted to 5,10-CH2 = THF, which can condense with glycine to produce serine (Fig. 3a). Serine is then deaminized to generate pyruvate, which is further transformed into phosphoenolpyruvate. Phosphoenolpyruvate fixes another molecule of C1 compounds to generate oxaloacetate via phosphoenolpyruvate carboxylase. Oxaloacetate is transformed to malate, which splits into two C2 compounds: acetyl-CoA and glyoxylate. Glyoxylate combines with amidogen to regenerate glycine in the next round of this cycle. However, the modified serine cycle is ATP-inefficient compared with the homoserine cycle, which does not overlap with the central metabolism. In the homoserine cycle, glycine can be directly converted to serine via condensation with formaldehyde and threonine aldolase (LtaE) (Fig. 3b). Serine is deaminized to form pyruvate, which can further generate 4-hydroxy-2-oxobutanoate (HOB) by condensing formaldehyde with 4-hydroxy-2-oxobutanoate aldolase (HAL). The C4 compound, HOB, is then transformed to threonine through three enzyme-catalyzed reactions. Finally, LtaE catalyzes the cleavage of threonine into two C2 compounds: glycine for another cycle and acetaldehyde as a product.
Synthetic pathways for C1 utilization. a The modified serine cycle. b The homoserine cycle. c The formyl-CoA elongation pathway. Green, aldose elongation; Black, aldehyde elongation. d The artificial starch anabolic pathway. e A synthetic acetyl-CoA pahway. f The crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA cycle. g The Gnd-Enter-Doudoroff cycle. h The reductive glyoxylate and pyruvate synthetic cycle and the malyl-CoA-glycerate pathway. i The POAP cycle. Abbreviations of metabolites stand for: Gly, glycine; Ser, serine; pyru, pyruvate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; mal-CoA, malyl-CoA; gly, glyoxylate; Ac-CoA, actyl-CoA; HOB, 4-hydroxy-2-oxobutanoate; Thr, threonine; acede, acetaldehyde; form-CoA, formyl-CoA; meth, methanol; forde, formaldehyde; DHA, dihydroxyacetone; DHAP, DHA phosphate; GAP, D-glyceraldehyde 3-phosphate; FBP, D-fructose-1,6-Bisphosphate; G6P, glucose-6-phosphate; ADPG, ADP-glucose; form, formate; glyde, glycolaldehyde; Ac-pho, acetyl-phosphate; Acr-CoA, acryloyl-CoA; cro-CoA, crotonyl-CoA; etm-CoA, methymalonyl-CoA; mema-CoA, methylmalyl-CoA; pro-CoA, propionyl-CoA; Ru5P, ribulose 5-phosphate; 6PG, 6-phosphogluconate; 3PG, 3-phosphoglycerate; acac-CoA, acetoacetyl-CoA; cim-CoA, (S)-citramalyl-CoA; oxal, oxalate
The formyl-CoA elongation pathway
The formyl-CoA elongation (FORCE) pathway is an orthogonal system that converts methanol, formaldehyde, and formate to glycolate, ethanol, and other products (Fig. 3c). C1 substrates (formate, formaldehyde, and methanol) are activated to formyl-CoA for C1 elongation by 2-hydroxyacyl-CoA lyase (HACL). Formyl-CoA reacts with Cn aldehyde to form 2-hydroxy-Cn+1-acyl-CoA by HACL, which is further transformed to the corresponding 2-hydroxy aldehyde by acyl-CoA reductase. Thereafter, 2-hydroxy aldehyde reacts with formyl-CoA, entering the next round of carbon-chain elongation (i.e., aldose elongation). However, similar to fatty acid biosynthesis, 2-hydroxy aldehyde is reduced to 1,2-diol by diol oxidoreductase, and 1,2-diol is dehydrated to Cn+1 aldehyde. Finally, the Cn+1 aldehyde is extended using formyl-CoA (i.e., aldehyde elongation). The FORCE pathway not only produces various compounds in vivo and in vitro, but can also be used in a two-strain co-culture system to achieve synthetic methylotrophy.
The artificial starch anabolic pathway
The artificial starch anabolic pathway (ASAP) is a cell-free system that can synthesize starch from CO2 using a chemo-biohybrid approach (Fig. 3d) [17]. The ASAP is divided into four modules: C1, C3, C6, and Cn. In the C1 module, CO2 is hydrogenated with H2 to chemically form methanol, which is then oxidized to formaldehyde by alcohol oxidase (AOX). In the C3 module, formolase catalyzes the condensation of three molecules of formaldehyde to generate dihydroxyacetone (DHA), which is converted to D-glyceraldehyde 3-phosphate (GAP) through a two-step reaction catalyzed by dihydroxyacetone kinase (dak) and triosephosphate isomerase (tpi). In the C6 module, the C3 compounds, GAP and DHAP (phosphorylation of DHA by dak), are irreversibly converted to the desired C6 compound, D-fructose-1,6-Bisphosphate (F-1,6-BP). F-1,6-BP generates glucose-6-phosphate (G-6-P) through a series of gluconeogenic reactions. In the Cn module, C6 compound G-6-P is converted to ADP-glucose (ADPG), which can be condensed into starch by starch synthase (ss). The ASAP pathway produces amylose and amylopectin in vitro.
A synthetic acetyl-CoA pathway
A synthetic acetyl-CoA (SACA) pathway is the ATP-independent, shortest, oxygen-insensitive, and carbon-conserving pathway for acetyl-CoA biosynthesis (Fig. 3e) [18]. In this pathway, two molecules of formaldehyde are converted to one molecule of acetyl-CoA through three enzyme-catalyzed reactions. First, two molecules of formaldehyde are condensed into glycolaldehyde by glycolaldehyde synthase (GALS). Acetyl-phosphate synthase (ACPS) is used to transform glycolaldehyde to acetyl-phosphate, which generates acetyl-CoA by phosphate acetyltransferase (PTA). The SACA pathway can produce 5.5 mM acetyl-CoA with 1 g/L formaldehyde in vitro. However, the application of the SACA pathway to E. coli remains a challenge owing to the low enzyme activity of GALS and ACPS.
The crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA cycle
The crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA (CETCH) cycle, which combines the 3-HP/4-HB cycle and ethylmalonyl-CoA pathway, is more efficient than the CBB cycle (Fig. 3f) [19]. First, acryloyl-CoA fixes one molecule of CO2 to generate (S)-methylmalonyl-CoA via crotonyl-CoA carboxylase/reductase (CCR). Thereafter, (S)-methylmalonyl-CoA is converted to (R)-methylmalonyl-CoA, which is further transformed into crotonyl-CoA through four steps of the 3-HP/4-HB cycle. Crotonyl-CoA then fixes another molecule of CO2 by CCR to form methylmalonyl-CoA, which then generated methylmalyl-CoA in three steps of the ethylmalonyl-CoA pathway. Finally, methylmalyl-CoA is split into propionyl-CoA by β-methylmalyl-CoA lyase (MCL) to generate acrylyl-CoA and glyoxylate. The CETCH cycle has 17 enzymes that are only assembled in vitro to generate organic molecules at a rate of 5 nmol CO2/min/mg protein from the consumption of three NAD(P)H and two ATP by CO2 in each cycle.
The Gnd-Entner–Doudoroff cycle
The Gnd-Entner-Doudoroff (GED) cycle, an artificial carbon fixation pathway, was designed in silico and established using endogenous E. coli enzymes (Fig. 3g) [20]. First, ribulose 5-phosphate (Ru5P) is used to assimilate CO2 to generate 6-phosphogluconate (6PG) via 6-phosphogluconate dehydrogenase (Gnd). Thereafter, 6PG dehydratase (Edd) and 2-keto-3-deoxygluconate 6-phosphate aldolase (Eda) in the Entner–Doudoroff pathway break down 6PG into pyruvate and glyceraldehyde-3-phosphate (GAP). Through gluconeogenesis, pyruvate is converted into GAP, which is used to regenerate Ru5P via the pentose phosphate pathway. Of note, the GED shunt (a linear pathway variant) has only been applied in vivo.
The reductive glyoxylate and pyruvate synthetic cycle and the malyl-CoA-glycerate pathway
The reductive glyoxylate and pyruvate synthetic cycle and the malyl-CoA-glycerate pathway (rGPS-MCG) cycle is a self-replenishing, oxygen-insensitive CO2 fixation system (Fig. 3h) [21]. This system consists of rGPS cycle and MCG pathway. rGS and rPS constituting the rGPS cycle. The rGS is a common segment of the rGPS and MCG pathways. In the rGS, pyruvate is used to generate phosphoenolpyruvate (PEP), which is condensed with CO2 to produce oxaloacetate (OAA) by phosphoenolpyruvate carboxylase. OAA is then used to generate malyl-CoA through two enzyme-catalyzed reactions. Finally, malyl-CoA lyase catalyzes the splitting of malyl-CoA to form glyoxylate and acetyl-CoA. In the MCG pathway, two molecules of glyoxylate are converted to tartronate semialdehyde, with the release of one molecule of CO2. Tartronate semialdehyde is then used to regenerate phosphoenolpyruvate to enter rGS through three enzyme-catalyzed reactions. In rPS, two molecules of acetyl-CoA are condensed to form acetoacetyl-CoA, which is then transformed into crotonyl-CoA. Through crotonyl-CoA carboxylase/reductase, crotonyl-CoA assimilates one molecule of CO2 to form one molecule of (S)-ethylmalonyl-CoA, which is further converted into (S)-citramalyl-CoA via multi-step reactions. Finally, (S)-citramalyl-CoA is split into pyruvate to enter the rGS and regenerate acetyl-CoA for rPS. Overall, each round of the rGPS-MCG cycle assimilates two molecules of CO2 to produce one molecule of acetyl-CoA. Compared to other in vitro pathways, the rGPS-MCG cycle has the highest CO2 fixation rate of up to 0.55 mM/h.
The POAP cycle
The POAP cycle contains only four enzymes [22]: pyruvate carboxylase, oxaloacetate acetylhydrolase, acetate-CoA ligase, and pyruvate synthase (Fig. 3i). In this minimized cycle, acetyl-CoA can fix one molecule of CO2 to generate pyruvate via pyruvate synthase (PFOR). Pyruvate is then converted to oxaloacetate via the fixing of another CO2 molecule with pyruvate carboxylase. Oxaloacetate is split by oxaloacetate acetylhydrolase to form oxalate and acetate. Oxalate is the final product and acetate is used to regenerate acetyl-CoA by acetate-CoA ligase for another cycle. A round of POAP assimilates two molecules of CO2, consuming 2 ATP and 1 NAD(P)H. By constructing POAP in vitro, the CO2 fixation rate reaches 8.0 nmol CO2/min/mg CO2-fixing enzymes under anaerobic conditions.
Engineering C1-utilizing pathways for chemical production
In-depth research on C1-based biomanufacturing, regardless of natural/synthetic C1 assimilation pathways or natural/non-natural C1-utilizing microorganisms, has both advantages and disadvantages. The main challenge involves the adaptation of microbial chassis and C1 utilization pathways to maximize carbon conversion efficiency. Here, an overview of the metabolic engineering of natural and non-natural C1-utilizing strains to efficiently utilize C1 feedstock to produce value-added chemicals is presented.
Engineering natural C1-utilizing pathways for chemical production
Many native microorganisms, such as Methylorubrum extorquens and Clostridium autoethanogenum, contain natural C1-utilizing pathways and can grow on C1 compounds as the sole carbon and energy source. However, the conversion efficiency of C1 compounds is relatively low and carbon flux is mainly used for cell growth, generally resulting in the accumulation of only a few metabolites. Accordingly, several attempts have been made to produce amino acids, malate, and other chemicals from C1 feedstocks using natural microorganisms. Furthermore, several strategies have been developed to screen non-traditional strains to identify efficient C1-utilizing host strains, developing new biotechnologies to rationally and efficiently engineer C1-utilizing strains, and engineering natural and non-natural C1-utilizing strains to balance metabolic flux for bioproduction.
The native C1-fixing strains still need to be fully exploited for further applications. Screening for potential strains is an ongoing task to meet the needs of industrial biomanufacturing for the production of different chemicals. To date, three effective strategies have been proposed: screening efficient natural C1-utilizing strains from the environment, applying random mutagenesis to screen efficient C1-utilizing strains, and rationally engineering C1-utilizing strains to enhance the biosynthetic efficiency of value-added chemicals (Fig. 4a). For example, a natural poly-β-hydroxybutyrate (PHB)-producing strain, Synechococcus elongatus UAM–C/S03, was isolated from an extreme environment [23]. This strain can capture CO2 at a rate of 674 mg/L/d, with the highest PHB productivity of up to 58.10 mg/L/d. However, the samples were collected from a small pond in Mexico, indicating that a well-behaved strain from nature can sometimes be obtained by chance. To overcome this issue, high-throughput screening and random mutagenesis can be used to screen efficient C1-utilizing strains. Previously, random mutagenesis and phenotypic selection were used to enhance light-to-biomass conversion efficiency in Chlorella sorokiniana [24]. The selected mutants had higher productivity and more efficient photon utilization, achieving 30% higher biomass yield. However, the screening process is time consuming and labor intensive. With the development of genetic tools, researchers can exploit the potential of strains using rational tools. The pathway for transforming acetyl-CoA to acetone and isopropanol was introduced into Clostridium autoethanogenum [9]. Following optimization of the pathway, strain, and process, acetone and isopropanol were produced by the gas fermentation process, with 1.78 kg CO2 e/kg and 1.17 kg CO2 e/kg reduction in greenhouse gas emissions, respectively. These examples indicate that strain performance can be improved through rational or irrational approaches. In the future, high-throughput screening technology and genetic tools must be developed to rapidly screen native C1-fixing strains with better phenotypes.
Engineering C1-utilizing pathways for chemicals production. a Engineering natural C1-utilizing pathways for chemicals production. b Engineering semi-synthetic C1-utilizing pathways for chemicals production. c Engineering synthetic C1-utilizing pathways for chemicals production. d Engineering energy pathways for chemicals production
The metabolic engineering of C1-utilizing strains to produce more chemicals from C1 compounds relies on the development of genetic tools. However, most native C1-fixing strains are non-model microorganisms that lack mature biological genetic manipulation tools. Three strategies were developed to solve this problem: in silico design and analysis to search for modification targets, building high-throughput workflows in vitro to accelerate design-build-test-learn cycles, and developing and optimizing genetic tools to improve the efficiency of genetic modification (Fig. 4a). For example, elementary mode analysis (EMA) and flux balance analysis (FBA) have been used to analyze metabolic pathways for isobutanol and hexadecanol production in autotrophic Cupriavidus necator H16 [25]. The in silico-designed mutants are predicted to yield 0.21–0.42 g/g isobutanol and 0.20–0.34 g/g hexadecanol from CO2. However, the targets screened by metabolic modeling have not been verified, and will serve as a valuable guidance for future metabolic engineering of C. necator H16. To improve the efficiency of screening target genes, in vitro prototyping and rapid optimization of biosynthetic enzymes (iPROBE) have been established to accelerate the design and optimization of biosynthetic pathways [26]. iPROBE was used to screen for different 3-hydroxybutyrate production (3-HB) pathways in cell-free systems. In fact, the highest-performing pathway was scaled up in vivo, and 3-HB production in Clostridium was found to increase by 20-fold to 14.63 g/ L. However, genetic tools for DNA construction and transformation are still required when validating the highest-performing pathways in vivo. An efficient, optimized CRISPRi system for M. extorquens AM1 was developed by balancing the expression level of dcas9 and sgRNA [27]. CRISPRi was used to repress the transcriptional level of the target gene, squalene-hopene cyclase (shc), resulting in a 1.9-fold increase in carotenoid production. Based on genetic tools and in silico modeling, the pros and cons of various combinations can be quickly identified to construct high-yield strains by mining all possible combinations. The development of genetic tools is a constant driving force for the sustainable development of biomanufacturing; thus, the development of genetic tools for natural C1-utilizing microorganisms remains a worth exploration to break the block of metabolic engineering in the future.
Many native C1-fixing strains have been used to produce chemicals, such as acetone, butanol, and α-humulene, from C1 feedstocks. However, some strains have no natural C1-fixing pathways for the production of the target chemicals, and some strains with natural C1-fixing pathways have weak metabolic flux toward the target chemicals. Three strategies were developed to solve this problem: (1) introduction of biosynthetic pathways to enable native C1-fixing strains to produce chemicals (Fig. 4a). For example, when the biosynthetic pathway of lovastatin was constructed in the methylotrophic yeast, P. pastoris, lovastatin production increased to 419.0 mg/L [28]. (2) Enhancement of the precursor or cofactor supply for bioproduction. The biosynthetic pathway of chondroitin sulfate A (CSA) was introduced into P. pastoris to biosynthesize CSA from methanol [29]. The supply of 3’-phosphoadenosine-5’-phosphosulfate (PAPS) was further strengthened by overexpressing two genes encoding adenosine-5’-triphosphate sulfurylase (ATPS) and adenosine-5’-phosphosulfate kinase (APSK) to increase CSA production from 182.0 mg/L to 2.1 g/L. (3) Deletion of the by-product branches to accumulate the target products. Two unwanted byproducts, 2,3-butanediol and 3-hydroxybutyrate, were eliminated by knocking out 0553, 1586, 1524, and 2932 in Clostridium autoethanogenum, resulting in a 27-fold increase in acetone production of up to 74.58 mM [9]. These examples indicate that the reinforcement of metabolic flux toward target chemicals is an useful strategy for producing chemicals from C1 compounds. Further process optimization to enhance cell growth, including optimization of culture conditions and fermentation processes, especially gas fermentation, to enhance cell growth is worth studying to achieve a high production of chemicals.
Engineering semi-synthetic C1-utilizing pathways for chemical production
Although many microorganisms can intrinsically achieve net C1 utilization, such microorganisms cannot be easily engineered to redirect carbon flux for bioproduction owing to the lack of genetic engineering tools. In contrast, model microorganisms for industrial production, such as E. coli and Saccharomyces cerevisiae, possess many advantages, including mature genetic tools, fast growth, and a clear genetic background. However, these microorganisms cannot normally use the C1 feedstock as the sole carbon source. Thus, natural C1 fixation pathways must be introduced into the industrial chassis to build new C1 fixation platforms. To achieve this hypothesis, much of the effort in the engineering of these semi-synthetic pathways has focused on three industrial strains: E. coli, S. cerevisiae, and Corynebacterium glutamicum.
E. coli is an important microbial chassis owing to its diverse genetic manipulation tools and clear genetic background. Three natural C1-fixing pathways are mainly used in synthetic methylotrophic E. coli: (1) the RuMP pathway. By introducing this pathway into E. coli, methanol can be used as an auxiliary substrate for the production of chemicals. For example, the yield of succinic acid increases from 0.91 g/g to 0.98 g/g under anaerobic fermentation using methanol and glucose as co-substrates [16]. In addition, the final titers of ethanol and 1-butanol are up to 4.6 g/L and 2.0 g/L, respectively, with methanol and xylose co-assimilation at a molar ratio of approximately 1:1 [30]. In these studies, the RuMP pathway enabled the use of methanol as an auxiliary substrate; however, acquiring synthetic methylotrophic E. coli is a long process. Thus, successful metabolic rerouting [31] is important for converting E. coli to a fully synthetic methylotrophic mode. To achieve this aim, adaptive laboratory evolution (ALE) is used, and the evolved E. coli can grow solely on methanol at a growth rate comparable to that of natural methylotrophs [7]. (2) the CBB cycle: fully autotrophic E. coli has been achieved using the CBB cycle, with CO2 and formate as the sole carbon and energy sources, respectively [8]. (3) The rGlyP pathway: by introducing this pathway into E. coli, the OD600 reaches 7.38 after 450 h of growth on CO2 and formic acid alone through a series of optimizations [32, 33]. The engineered E. coli can grow on formate and CO2, with a doubling time of less than 8 h, and its biomass yield is improved to 2.3 gCDW/mol formate [14, 34]. These examples suggest that as an industrial chassis, E. coli is widely used for C1 utilization. Although E. coli can grow on C1 as the sole carbon source, its production capacity is not suitable for industrial production. Thus, the underlying mechanism of autotrophic E. coli must be understood and more suitable industrial chassis must be further explored as a bioproduction platform to produce value-added chemicals from C1 compounds (Fig. 4b).
S. cerevisiae has many advantages, such as simple culture conditions, diverse genetic manipulation tools, and a clear genetic background. The genome of a widely used eukaryotic model microorganism has been completely sequenced. S. cerevisiae can tolerate high concentrations of formaldehyde and methanol [35] and has the native capacity for methanol utilization [36]. Thus, S. cerevisiae has great potential for C1 utilization. Three C1-fixing pathways are heterologously expressed in S. cerevisiae: (1) XuMP and RuMP pathways. After integration of the methanol oxidation pathway into the chromosome of S. cerevisiae, the cell growth (OD600) of the recombinant strain increased by 3.13% with 1.04 g/L methanol as the sole carbon source [37]. (2) The rGlyP pathway. When the heterogeneous rGlyP pathway is introduced into S. cerevisiae, a high activity is displayed [38], suggesting that the recombinant S. cerevisiae is a potential strain for C1 fixation. These examples indicate that S. cerevisiae is a potential model strain for C1 fixation owing to its natural advantages, such as high tolerance to C1 feedstocks. Further, ALE of S. cerevisiae containing C1 assimilation pathways can be conducted to improve the growth phenotype and refactor the metabolic network to achieve highly efficient production of chemicals from C1 feedstocks (Fig. 4b).
Corynebacterium glutamicum has attracted attention owing to its high glutamic acid production. The RuMP pathway is heterogeneously expressed in C. glutamicum; however, synthetic methylotrophic C. glutamicum is yet to be obtained. For example, by introducing the RuMP pathway into C. glutamicum with the expression of lysine decarboxylase (Fig. 4b), the production of cadaverine was up to 1.5 g/L in shake flasks using glucose, ribose, and methanol as co-substrates [39]. Further, by expressing the RuMP pathway in C. glutamicum, the production of glutamate reached 90 mg/L using methanol and xylose as carbon sources [40, 41]. ALE was then adopted to improve its tolerance to methanol, resulting in a higher glutamate production up to 230 mg/L [47]. These examples indicate that C. glutamicum assimilating C1 compounds is still challenging; thus, increasing the tolerance of some toxic C1 compounds is an effective strategy, and is beneficial for chemical production from C1 fixation.
Engineering synthetic C1-utilizing pathways for chemical production
Some natural assimilation pathways are used for the transformation of industrial chassis into new platforms for producing chemicals from C1. Recently, E. coli and P. pastoris were converted from heterotrophic to autotrophic microorganisms [6]. However, these natural pathways still have many limitations, such as high energy consumption, oxygen sensitivity, and low enzyme activity. Thus, much work has been performed on the synthetic pathways for chemical production. These synthetic pathways can be divided into three classes: cyclic C1 fixation pathways, linear C1 fixation pathways, and synthetic pathways containing C1 fixation reactions for chemical biosynthesis.
Some cyclic C1 fixation pathways have been developed, including CETCH, ROAP, and FORCE. However, most of the synthetic cyclic pathways have been verified in vitro; thus, further studies are needed to achieve bioproduction in vivo. Here, we mainly discussed three synthetic cyclic pathways: (1) the CETCH cycle (Fig. 4c), which can be used to produce terpenes from CO2 by coupling with the β-hydroxy aspartate cycle and biosynthetic pathways for terpenes in vitro [42]. The productivity of terpenes from CO2 reaches approximately 0.2 mg/L/h. (2) The modified serine cycle [43], which can be used in E. coli to increase ethanol production via the co-utilization of methanol with xylose. In addition, this pathway can be used to support pyruvate synthesis by assimilating two formate molecules with one bicarbonate molecule. (3) Homoserine cycles [44]. The in vivo feasibility of the homoserine cycle was tested by coupling the activity of different pathway segments with the growth of several gene-deleted E. coli strains. The homoserine cycle can support a high yield of a wide array of products derived from acetyl-CoA, such as ethanol, acetone, butyrate, and 1-butanol. These examples demonstrate that the output of products and accumulation of biomass can be increased by optimizing these synthetic cyclic pathways in vitro to broaden the product spectrum and improve the concentration and activity of key enzymes. In the future, the applications of these cyclic C1 fixation pathways must be further improved to construct C1 utilization platform strains for industrial biomanufacturing, such as the construction of high-efficiency autotrophic E. coli.
Linear C1 fixation pathways are relatively short compared with cyclic pathways and generally have little interference with central metabolism. Several in vivo synthetic linear pathways have been developed. (1) Three linear C1 fixation pathways, FLS [45, 46], HWLS [47], and SMGF [48], were designed based on formolase. When the FLS pathway is assembled in E. coli, it can be used to synthesize biomass from formate. HWLS and SMGF were also designed for CO2 sequestration, which is an auxiliary approach to reach the theoretical yield of L-malate and butyrate, achieving carbon-efficient bioproduction. (2) The SACA pathway. When the SACA pathway was introduced into E. coli, cell growth (OD600) increased to 0.2 in 26 h with methanol as the sole carbon source [18]. These examples suggest that the C1 assimilation ability of linear synthetic pathways in vivo is inefficient, possibly owing to the influence of limiting enzymes in the C1 fixation pathways. Thus, identifying the limiting steps in the C1 fixation pathways is key to enhancing carbon flux to support cell growth, with C1 compounds as the sole carbon source.
To develop C1-based industrial platforms for industrial biomanufacturing, the utilization efficiency of C1 compounds must be improved. Thus, the design of biosynthetic pathways involving C1 assimilation reactions is an important method for C1 utilization. As these pathways involve different C1-fixing enzymes, three strategies were developed (Fig. 4c): (1) The transplantation of natural C1-utilizing enzymes into biosynthetic pathways. For example, the biosynthetic pathway of succinate is redesigned via coupling with a part of the 3-HP cycle, which contains two CO2 fixation reactions catalyzed by acetyl-CoA carboxylase and propionyl-CoA carboxylase (PCC) [49]. By introducing the PCC mutant into the redesigned pathway, succinate production reaches 2.66 g/L using glucose and CO2 as dual carbon sources under fed-batch conditions. (2) Mining of natural enzymes that can catalyze C1 assimilation reactions in nature. For example, the novel biosynthetic pathway of 1,3-propanediol (PDO) was redesigned by introducing deoxyribose-5-phosphate aldolase (DERA) from Escherichia coli, which can condense HCHO and acetaldehyde to form 3-hydroxypropionaldehyde. The final production of PDO reaches 1.32 g/L with ethanol and formaldehyde in resting E. coli cells [50]. (3) Design of artificial C1-utilizing enzymes via a computational approach. Glycolyl-CoA carboxylase (GCC) is a good example as it catalyzes glycolyl-CoA and CO2 to form (S)-tartronyl-CoA [51]. Based on the GCC, the newly designed TaCo pathway can convert glycolate into glycerate in vitro at a rate of 27 nmol/min/mg protein. Based on these examples, C1-fixing enzymes are usually the limiting steps for realizing the carbon-negative production of value-added chemicals. Thus, mining natural enzymes or redesigning artificial enzymes may significantly contribute to C1 fixation in the future.
Engineering energy pathways for chemical production
Metabolic engineering changes the intracellular balance of redox and energy, which may inhibit cell growth and chemical production. Thus, increasing the energy supply to rebuild this balance is important, especially for producing target chemicals using C1 feedstock. Many strategies have been developed to balance energy systems to achieve optimal NADH/NAD+ and ATP/ADP ratios, such as generating energy from chemical energy through the exogenous addition of carbon sources or the endogenous generation of metabolites, capturing energy from electric energy through microbial electrosynthesis, and capturing energy from sunlight by light-harvesting systems.
Using C1 compounds as the sole carbon source for heterotrophic microorganisms requires a large energy supply. However, intracellular imbalance in metabolic flux leads to an insufficient supply of energy. Thus, directing metabolic flux to pathways, such as PPP for generating energy and reducing power, can increase the NADH/NAD+ and ATP/ADP ratios. Accordingly, three effective strategies were proposed (Fig. 4d): (1) the use of C1 feedstock and other carbohydrates as co-substrates to generate energy. For example, the CBB cycle was introduced into E. coli for sugar biosynthesis from CO2 by supplying pyruvate to generate ATP and reducing equivalents [52]. After 150 chemostat generations, the evolved E. coli could grow in a minimal medium. Furthermore, a fully autotrophic E. coli strain was obtained, which can generate all biomass from CO2. Simultaneously, formate is supplied to generate reducing power and energy by introducing formate dehydrogenase (FDH) [8]. (2) The channeling of native metabolic flux to generate energy. The RuMP cycle was incorporated into M. extorquens AM1, which has an intrinsic serine cycle. Central carbon metabolism was rewired to improve NADPH/NADP+ to enhance C1 fixation. The biosynthetic pathway of 3-HP was introduced into M. extorquens, leading to a 3.1-fold increase in the titer of 3-HP up to 91.2 mg/L compared with the control strain in shake flasks. (3) The optimization of the incubation temperature to improve the electron transport chain. Some researchers can solve this energy deficiency by controlling the incubation temperature [13]. At low temperatures, the expression level of cytochrome bo3 ubiquinol oxidase (Cyo) increases and the expression level of cytochrome bd-I ubiquinol oxidase decreases, enabling more efficient conversion from reducing power to ATP. Thus, E. coli can grow from an initial OD600 of 0.31 to 2.84 on CO2 and formate alone, which is higher than that of the control strain. Based on these examples, intermediate metabolites can be efficiently converted to energy generation or target products only when these two parts are balanced. Thus, future work must focus on fine-tuning metabolic flux to achieve continuous and efficient production of chemicals through the dynamic regulation of reducing power and energy levels.
The use of clean energy has rapidly increased worldwide in recent decades [53]. Recycling CO2 into bioproducts through microbial electrosynthesis (MES) has been studied for a long time. The combination of clean electricity with C1 fixation can achieve the green bioproduction of high-value chemicals. Accordingly, three effective strategies were proposed (Fig. 4d). (1) Electron acquisition in electroautotrophs to produce chemicals. For example, the heterologous biosynthesis pathway of α-humulene is expressed in the electroautotrophic C. necator for α-humulene production [54]. In this process, CO2 is fixed by the CBB cycle with reduction equivalents from the electrochemical process. Engineered C. necator was cultivated with a bioelectrochemical system, which produces 10.8 mg/L α-humulene under a gas atmosphere of CO2/H2/O2 in septum flasks. (2) Development of inorganic cathodic catalysts to increase the mass transfer rate. A biocompatible perfluorocarbon nanoemulsion was exploited and introduced into Sporomusa ovata as an H2 carrier [55]. The average titer of acetate showed a 190% increase up to 6.4 g/ L in four days due to the kinetics of H2 transfer and subsequent oxidation, which increased by more than 3.0-fold. (3) Electrons are taken up by symbiotic microorganisms to avoid large overpotentials. The Fe(0)-corroding Desulfopila corrodens strain IS4 was used as a biocathode to form intermediate molecular hydrogen, and Methanococcus maripaludis and Acetobacterium woodii were used for the hydrogenotrophic synthesis of methane and acetate, respectively [56]. Co-cultures of D. corrodens IS4 and M. maripaludis achieved an electromethanogenesis rate of 0.6–0.9 μmol/cm2/h at -500 mV. Acetate is formed at a rate of 0.57–0.74 μmol/cm2/h at − 500 mV through co-cultures, D. corrodens IS4 and A. woodii. These examples indicate that the combination of electrochemistry with biology is effective for bioproduction; however, the overall energy efficiency still needs to be improved. In the future, more efforts are needed to illustrate the detailed mechanisms, such as the electron uptake mechanism, and develop further applications.
Engineering bacteria to directly use light energy is attractive as sunlight is abundant and sustainable. However, the conversion rate of solar energy into biomass is typically less than 3% in engineered bacteria. To maximize the utilization of light energy in phototrophic systems, three effective strategies have been proposed: engineering photoautotrophs to improve natural light-harvesting systems, transplanting natural light-harvesting systems for heterotrophs, and designing new light-harvesting carriers to further enhance photon absorption (Fig. 4d). For example, malate production can be improved by balancing the capacity to generate energy in the photoautotrophic bacteria, Synechococcus elongatus, the ATP-consuming RuBisCO shunt in the CBB cycle, and the ATP-generating carboxylation reaction in central metabolism [57]. The biosynthesis pathway of malate is integrated into the chromosome of S. elongatus, leading to malate production of up to 260 μM, with a 110% increase in the CO2-fixing rate. However, the production capacity of chemicals in S. elongatus is not comparable to that in E. coli. Thus, light-capturing rhodopsin is expressed in E. coli, which allows this heterotrophic microorganism to synthesize ATP for the efficient production of target chemicals [58]. Of note, rhodopsin does not provide reducing power and cannot support full autotrophy alone. As a result, an inorganic-biological hybrid system is developed by combining photosynthetic Rhodopseudomonas palustris with CdS nanoparticles [59]. Using this system, NADPH is increased under visible light irradiation, leading to a higher yield from CO2. Consequently, the production of poly-β-hydroxybutyrate (PHB), solid biomass, and carotenoids increased by 147%, 148%, and 122%, respectively. These examples show that the use of light energy is effective for bioproduction; however, many challenges must be addressed before these applications can become a reality. Future work should focus on parsing electron transduction mechanisms in depth and building general systems for the biological utilization of sunlight.
Concluding remarks
Food shortages, energy crises, and unsustainable production are the urgent risks. The sustainable production of high-value chemicals from C1 feedstocks with renewable energy can enable the emergence of a sustainable bio-economy, which will significantly contribute to alleviating these risks. Here, we reviewed the natural and synthetic pathways of C1 utilization, and the applications of these pathways in natural C1-fixing microorganisms and industrial chassis for bioproduction. Although the use of C1 feedstocks as the sole carbon source or auxiliary substrates can lead to the production of many chemicals, most cannot meet the needs of industrial production owing to the low catalytic efficiency of some key C1-fixing enzymes, inefficient pathways for C1 assimilation, and weak metabolic capacity of C1-utilizing strains. Therefore, further research should be conducted in the following directions to address these challenges.
Designing and mining new C1-fixing enzymes with high catalytic efficiency must be carried out. Currently, the C1 assimilation pathways rely on a very limited number of C1-fixing enzymes, which blocks the development of new C1 assimilation pathways. Thus, recent studies have focused on the computational design and development of new functional enzymes for further applications in C1 fixation. In addition, directed evolution methods can be adopted to generate potential enzymes to increase the C1 assimilation efficiency. A classic example is the computational design of a formolase. A positive mutant formolaseS420L/G449A/G452V was obtained by high-throughput screening, and its activity was found to be 2.3-fold higher than that of formolase. Furthermore, the efficiency of the FLS pathway was increased by 2.4-fold in this mutant [48].
The design and development of novel metabolic pathways with high pathway efficiency are required to convert C1 feedstock to chemical production. In recent years, several novel C1 assimilation pathways have been proposed. However, most natural and synthetic pathways have not been fully applied in industrial production. These reported pathways must be applied to build process extensions for the production of one or more types of chemicals. For example, the product spectrum of CETCH has been broadened by coupling this C1-fixing pathway with the biosynthetic pathway of terpenes and polyketides [42]. However, high-efficiency synthetic pathways should be developed using powerful computer tools. For example, some novel C1-fixing pathways have been systematically designed, such as the reductive citramalyl-CoA cycle (rCCC) and the glycolaldehyde assimilation (GAA) pathway, based on reactions in databases, such as MetaCyc [60] and ATLAS [61], and algorithms, such as the COBRApy toolbox [62], the enzyme cost minimization (ECM) algorithm [63] and the parameter balancing algorithm [64].
C1-utilizing microorganisms must be engineered to increase the cell tolerance and conversion efficiency of C1 feedstocks. Few industrial platform strains are available for the production of value-added chemicals from C1. On one hand, the industrial C1-utilizing microorganisms can be further engineered for the biosynthesis of more products from some key intermediate metabolites. For example, the production of acetone and isopropanol from CO2 is derived from acetyl-CoA in Clostridium autoethanogenum through three- and four-enzyme-catalyzed reactions, respectively [9]. However, the tolerance of industrial chassis to C1 feedstocks, which limits the efficiency of C1 utilization, must be improved. For example, ALE can be used to increase the tolerance of C. glutamicum to methanol up to 15 g/L, resulting in a higher production of glutamate.
References
Friedlingstein P, Jones MW, O’Sullivan M, et al. Global carbon budget 2021. Earth Syst Sci Data. 2022;14:1917–2005.
Jouny M, Luc W, Jiao F. General techno-economic analysis of CO2 electrolysis systems. Ind Eng Chem Res. 2018;57:2165–77.
Gesicka A, Oleskowicz-Popiel P, Lezyk M. Recent trends in methane to bioproduct conversion by methanotrophs. Biotechnol Adv. 2021;53: 107861.
Pavan M, Reinmets K, Garg S, et al. Advances in systems metabolic engineering of autotrophic carbon oxide-fixing biocatalysts towards a circular economy. Metab Eng. 2022;71:117–41.
Cotton CA, Claassens NJ, Benito-Vaquerizo S, et al. Renewable methanol and formate as microbial feedstocks. Curr Opin Biotechnol. 2020;62:168–80.
Gassler T, Sauer M, Gasser B, et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat Biotechnol. 2020;38:210–6.
Chen FYH, Jung H-W, Tsuei C-Y, et al. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell. 2020;182:933.
Gleizer S, Ben-Nissan R, Bar-On YM, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell. 2019;179(1255–1263): e1212.
Liew FE, Nogle R, Abdalla T, et al. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat Biotechnol. 2022;40:335–44.
Bar-Even A, Noor E, Milo R. A survey of carbon fixation pathways through a quantitative lens. J Exp Bot. 2012;63:2325–42.
Sanchez-Andrea I, Guedes IA, Hornung B, et al. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat Commun. 2020;11.
Song Y, Lee JS, Shin J, et al. Functional cooperation of the glycine synthase-reductase and Wood-Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proc Natl Acad Sci U S A. 2020;117:7516–23.
Bang J, Hwang CH, Ahn JH, et al. Escherichia coli is engineered to grow on CO2 and formic acid. Nat Microbiol. 2020;5:1459.
Kim S, Lindner SN, Aslan S, et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. 2020;16:538.
Ahn JH, Seo H, Park W, et al. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat Commun. 2020;11:1970.
Zhang W, Zhang T, Song M, et al. Metabolic engineering of Escherichia coli for high yield production of succinic acid driven by methanol. ACS Synth Biol. 2018;7:2803–11.
Cai. T, Sun. H, Qiao. J, et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science. 2021.
Lu XY, Liu YW, Yang YQ, et al. Constructing a synthetic pathway for acetylcoenzyme A from one-carbon through enzyme design. Nat Commun. 2019;10.
Schwander T, Schada von Borzyskowski L, Burgener S, et al. A synthetic pathway for the fixation of carbon dioxide in vitro. Science. 2016;354:900–4.
Satanowski A, Dronsella B, Noor E, et al. Awakening a latent carbon fixation cycle in Escherichia coli. Nat Commun. 2020;11.
Luo S, Lin PP, Nieh L-Y, et al. A cell-free self-replenishing CO2-fixing system. Nat Catal. 2022;5:154–62.
Xiao L, Liu G, Gong F, et al. A minimized synthetic carbon fixation cycle. ACS Catal. 2021;12:799–808.
Gonzalez-Resendiz L, Sanchez-Garcia L, Hernandez-Martinez I, et al. Photoautotrophic poly(3-hydroxybutyrate) production by a wild-type Synechococcus elongatus isolated from an extreme environment. Bioresour Technol. 2021;337: 125508.
Cazzaniga S, Dall’Osto L, Szaub J, et al. Domestication of the green alga Chlorella sorokiniana: reduction of antenna size improves light-use efficiency in a photobioreactor. Biotechnol Biofuels. 2014;7:157.
Unrean P, Tee KL, Wong TS. Metabolic pathway analysis for in silico design of efficient autotrophic production of advanced biofuels. Bioresour Bioprocess. 2019;6.
Karim AS, Dudley QM, Juminaga A, et al. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat Chem Biol. 2020;16:912–9.
Mo XH, Zhang H, Wang TM, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Appl Microbiol Biotechnol. 2020;104:4515–32.
Liu Y, Bai C, Xu Q, et al. Improved methanol-derived lovastatin production through enhancement of the biosynthetic pathway and intracellular lovastatin efflux in methylotrophic yeast. Bioresour Bioprocess. 2018;5.
Jin X, Zhang W, Wang Y, et al. Biosynthesis of non-animal chondroitin sulfate from methanol using genetically engineered Pichia pastoris. Green Chem. 2021;23:4365–74.
Chen C-T, Chen FYH, Bogorad IW, et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production. Metab Eng. 2018;49:257–66.
Meyer F, Keller P, Hartl J, et al. Methanol-essential growth of Escherichia coli. Nat Commun. 2018;9:1508.
Bang J, Lee SY. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc Natl Acad Sci U S A. 2018;115:E9271–9.
Bang J, Ahn JH, Lee JA, et al. Synthetic formatotrophs for one-carbon biorefinery. Adv Sci. 2021;8.
Yishai O, Bouzon M, Doering V, et al. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli. ACS Synth Biol. 2018;7:2023–8.
Yasokawa D, Murata S, Iwahashi Y, et al. Toxicity of methanol and formaldehyde towards Saccharomyces cerevisiae as assessed by DNA microarray analysis. Appl Biochem Biotechnol. 2010;160:1685–98.
Espinosa MI, Gonzalez-Garcia RA, Valgepea K, et al. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat Commun. 2020;11:5564.
Dai Z, Gu H, Zhang S, et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour Technol. 2017;245:1407–12.
Gonzalez de la Cruz J, Machens F, Messerschmidt K, et al. Core catalysis of the reductive glycine pathway demonstrated in yeast. ACS Synth Biol. 2019;8:911–7.
Lessmeier L, Pfeifenschneider J, Carnicer M, et al. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate. Appl Microbiol Biotechnol. 2015;99:10163–76.
Wang Y, Fan L, Tuyishime P, et al. Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun Biol. 2020;3:217.
Tuyishime P, Wang Y, Fan L, et al. Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab Eng. 2018;49:220–31.
Sundaram S, Diehl C, Cortina NS, et al. A modular in vitro platform for the production of terpenes and polyketides from CO2. Angew Chem Int Ed Engl. 2021;60:16420–5.
Yu H, Liao JC. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat Commun. 2018;9.
Hai He RH, Dodenhöft M, Marlière P, et al. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli. Metab Eng. 2020;60:1–13.
Wang X, Wang Y, Liu J, et al. Biological conversion of methanol by evolved Escherichia coli carrying a linear methanol assimilation pathway. Bioresour Bioprocess. 2017;4.
Siegel JB, Smith AL, Poust S, et al. Computational protein design enables a novel one-carbon assimilation pathway. Proc Natl Acad Sci U S A. 2015;112:3704–9.
Hu G, Li Z, Ma D, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals. Nat Catal. 2021;4:395–406.
Hu G, Guo L, Gao C, et al. Synergistic metabolism of glucose and formate increases the yield of short-chain organic acids in Escherichia coli. ACS Synth Biol. 2022;11:135–43.
Liu X, Feng X, Ding Y, et al. Characterization and directed evolution of propionyl-CoA carboxylase and its application in succinate biosynthetic pathway with two CO2 fixation reactions. Metab Eng. 2020;62:42–50.
Meng H, Wang C, Yuan Q, et al. An aldolase-based new pathway for bioconversion of formaldehyde and ethanol into 1,3-propanediol in Escherichia coli. ACS Synth Biol. 2021;10:799–809.
Scheffen M, Marchal DG, Beneyton T, et al. A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation. Nat Catal. 2021;4.
Antonovsky N, Gleizer S, Noor E, et al. Sugar synthesis from CO2 in Escherichia coli. Cell. 2016;166:115–25.
Zhang L, Chen Z, Yang C, et al. Global supply risk assessment of the metals used in clean energy technologies. J Clean Prod. 2022;331.
Krieg T, Sydow A, Faust S, et al. CO2 to terpenes: autotrophic and electroautotrophic alpha-humulene production with Cupriavidus necator. Angew Chem Int Ed Engl. 2018;57:1879–82.
Rodrigues RM, Guan X, Iñiguez JA, et al. Perfluorocarbon nanoemulsion promotes the delivery of reducing equivalents for electricity-driven microbial CO2 reduction. Nat Catal. 2019;2:407–14.
Deutzmann JS, Spormann AM. Enhanced microbial electrosynthesis by using defined co-cultures. ISME J. 2017;11:704–14.
Hu G, Zhou J, Chen X, et al. Engineering synergetic CO2-fixing pathways for malate production. Metab Eng. 2018;47:496–504.
Toya Y, Hirono-Hara Y, Hirayama H, et al. Optogenetic reprogramming of carbon metabolism using light-powering microbial proton pump systems. Metab Eng. 2022;72:227–36.
Wang B, Jiang Z, Yu JC, et al. Enhanced CO2 reduction and valuable C2+ chemical production by a CdS-photosynthetic hybrid system. Nanoscale. 2019;11:9296–301.
Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471-480.
Hadadi N, Hafner J, Shajkofci A, et al. ATLAS of biochemistry: a repository of all possible biochemical reactions for synthetic biology and metabolic engineering studies. ACS Synth Biol. 2016;5:1155–66.
Ebrahim A, Lerman JA, Palsson BO, et al. COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst Biol. 2013;7:74.
Noor E, Flamholz A, Bar-Even A, et al. The protein cost of metabolic fluxes: prediction from enzymatic rate laws and cost minimization. PLoS Comput Biol. 2016;12: e1005167.
Lubitz T, Schulz M, Klipp E, et al. Parameter balancing in kinetic models of cell metabolism. J Phys Chem B. 2010;114:16298–303.
Acknowledgements
This study was supported by the Provincial Outstanding Youth Foundation of Jiangsu Province (BK20211529), the National Science Fund for Excellent Young Scholars (22122806) and the Fundamental Research Funds for the Central Universities (JUSRP22031).
Author information
Authors and Affiliations
Contributions
JZ and XLC collected the information and wrote the manuscript, XLC, LG, CG, WS, JW and LML revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
All authors agree with their participation in this paper.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Guo, L., Gao, C. et al. Metabolic engineering strategies for microbial utilization of C1 feedstocks. Syst Microbiol and Biomanuf 3, 122–136 (2023). https://doi.org/10.1007/s43393-022-00135-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00135-2