Abstract
The subcortical maternal complex (SCMC) is a multiprotein complex in oocytes and preimplantation embryos that is encoded by maternal effect genes. The SCMC is essential for zygote-to-embryo transition, early embryogenesis, and critical zygotic cellular processes, including spindle positioning and symmetric division. Maternal deletion of Nlrp2, which encodes an SCMC protein, results in increased early embryonic loss and abnormal DNA methylation in embryos. We performed RNA sequencing on pools of meiosis II (MII) oocytes from wild-type and Nlrp2-null female mice that were isolated from cumulus-oocyte complexes (COCs) after ovarian stimulation. Using a mouse reference genome-based analysis, we found 231 differentially expressed genes (DEGs) in Nlrp2-null compared to WT oocytes (123 up- and 108 downregulated; adjusted p < 0.05). The upregulated genes include Kdm1b, a H3K4 histone demethylase required during oocyte development for the establishment of DNA methylation marks at CpG islands, including those at imprinted genes. The identified DEGs are enriched for processes involved in neurogenesis, gland morphogenesis, and protein metabolism and for post-translationally methylated proteins. When we compared our RNA sequencing data to an oocyte-specific reference transcriptome that contains many previously unannotated transcripts, we found 228 DEGs, including genes not identified with the first analysis. Interestingly, 68% and 56% of DEGs from the first and second analyses, respectively, overlap with oocyte-specific hyper- and hypomethylated domains. This study shows that there are substantial changes in the transcriptome of mouse MII oocytes from female mice with loss of function of Nlrp2, a maternal effect gene that encodes a member of the SCMC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the earliest divisions of preimplantation embryonic development, before the zygotic genome is activated, transcription is quiescent and depends on the action of stored RNAs and proteins expressed from maternal effect genes during oocyte development. The proteins that maternal effect genes encode abound in oocytes and early preimplantation embryos and play essential roles in early embryo development [1]. The subcortical maternal complex (SCMC) is a multiprotein complex localized below the cortex of oocytes that contains proteins encoded by maternal effect genes. The SCMC remains present at the periphery of the preimplantation embryo during the earliest cell divisions, but its expression gradually decreases after fertilization. In mice, it is no longer visible by the blastocyst stage [2, 3]. Nine SCMC proteins have been identified so far: OOEP (known as FLOPED), NLRP5, TLE6, KHDC3 (known as FILIA), ZBED3, NLRP2, PADI6, NLRP9, and NLRP4F [2, 4,5,6,7,8,9,10]. The SCMC is involved in multiple cellular processes, including control of spindle position, symmetric zygote division, organelle distribution, and cytoplasmic lattice formation [6, 11,12,13].
Studies in mice have shown that maternal loss of function of genes that encode SCMC proteins causes sterility or subfertility. Targeted disruption of maternal Ooep, Nlrp5, Padi6, or Tle6 results in complete embryonic arrest at the two-cell or early cleavage stages [2, 7, 11, 13, 14]. Although the maternal loss of Khdc3, Zbed3, or Nlrp2 also leads to delayed preimplantation development and reduced fertility, it can be compatible with later embryo development, and even with survival after birth in a small number of offspring [6, 15, 16]. SCMC genes and proteins are highly conserved in mammals. In humans, women with biallelic inactivating mutations in NLRP7 (which is not present in rodent genomes, but highly homologous with rodent Nlrp2), KHDC3L, and PADI6 develop biparental complete hydatidiform mole (BiCHM), which manifests in an abnormal pregnancy characterized by chorionic villi overproliferation and lack of embryonic/fetal tissue [17,18,19].
Molar trophoblast tissues lack methylation at maternally imprinted loci, including those with placenta-specific imprinted differentially methylated regions (DMRs) [20]. Imprinted loci contain genes that are expressed from only the maternally inherited or the paternally inherited allele. Their allele-specific expression is controlled by parent-of-origin dependent allele-specific DNA methylation at the DMRs that control their expression. When the methylation at imprinted DMRs is disrupted, lack of expression from the active allele or overexpression from the silenced allele of an imprinted gene can cause imprinting disorders; examples of imprinting disorders include Beckwith-Wiedemann syndrome or Angelman syndrome [21]. The recent finding that oocytes from a BiCHM patient with a homozygous inactivating mutation in maternal KHDC3L had a genome-wide loss of DNA methylation that persisted at several imprinted loci until post-implantation confirmed that the origin of the methylation defect is in the female germline [22]. Loss of several other SCMC genes (NLRP5, NLRP2, NLRP7, PADI6, OOEP, and KHDC3L) in humans also causes multi-locus imprinting disturbance, in which DNA methylation is lost at several multiple maternally imprinted loci, resulting in recurrent pregnancy loss and offspring with complex multi-locus imprinting disorders (MLID). These present with features of more than one imprinted disease in the affected individuals [23,24,25,26,27]. We previously reported that maternal loss of Nlrp2 in mice causes subfertility with variable reproductive outcomes ranging from early embryonic loss to stillborn pups to, rarely, liveborn offspring with myriad developmental defects. We also found altered methylation at the DMRs of selected imprinted genes [15].
The mechanism by which disruption of the SCMC, a cytoplasmic structure, affects DNA methylation, a nuclear process, remains poorly understood. The above-described observations in mice and humans suggest that the SCMC may have an epigenetic regulatory role. It may directly affect the epigenetic machinery or disrupt different oocyte functions which could in turn globally alter the transcriptome.
In human and mouse oocytes, transcription drives the establishment of DNA methylation [28, 29], and in mice, transcriptional aberrations disrupt oocyte-derived methylation [30]. We were interested in detecting the origin of decreased viability of embryos from Nlrp2-null females and understanding the possible connection to aberrant transcriptome in the oocytes. We hypothesized that transcriptional changes should occur in Nlrp2-null female oocytes and that these might underlie methylation defects in oocytes, embryos, or offspring. We began investigating this by performing RNA sequencing (RNA-Seq) on pooled oocytes isolated from cumulus-oocyte complexes (COCs) collected after superovulation of Nlrp2 females. Our results show that maternal Nlrp2 loss globally alters the oocyte transcriptome.
Materials and Methods
All experiments were approved by the Baylor College of Medicine (BCM) Institutional Animal Care and Use Committee (protocol AN-2035). Experiments were conducted according to institutional and governmental regulations concerning the ethical use of animals and the American Veterinary Medical Association (AVMA) and ARRIVE Guidelines were followed. All animal facilities are approved by the Association for Assessment and Accreditation for Laboratory Animal Care International (AAALAC).
Animals, Superovulation, and Sample Collection
Experiments were performed on the previously described Nlrp2-null (Nlrp2tm1a/tm1a) mouse model on a C57BL6/J background [15]. Superovulation was induced in 4-week-old wild-type (WT) (strain of origin: C57BL/6N) and Nlrp2-null mice by intraperitoneal injection of 5 units (U) of pregnant mare’s serum gonadotropin (PMSG; Fisher Scientific, USA) followed by injection of 5 U of human chorionic gonadotropin (hCG; EMD Millipore, USA) 48 h (h) later. The following day at midday, COCs were collected from the ampulla of the oviduct. The same timing was respected for all animals. Denuded oocytes were prepared by treating COCs with 3 mg/ml hyaluronidase (Sigma-Aldrich, USA) for 2 min at room temperature. Oocytes from 3 to 4 mice of the same genotype were pooled to obtain pools of approximately 150 oocytes in each sequenced RNA sample. RNA was isolated from four Nlrp2-null and three WT oocyte pools using the Qiagen miRNeasy kit (QIAGEN, US) and used for the preparation of RNA-Seq libraries.
Library Preparation and RNA Sequencing
Library preparation and sequencing were done at the genomic and RNA profiling core at BCM. Libraries were prepared with the NuGEN RNA-Seq library preparation protocol. Briefly, purified double-stranded (ds) cDNA was generated from approximately 20 ng of total RNA and amplified using both oligo d(T) and random primers. Five hundred nanograms of each sample’s ds-cDNA were sheared using the Covaris S2 focused ultrasonicator (Covaris, USA) to a target size of 400 bp. A double-stranded DNA library was then generated from 1 μg of sheared, double-stranded cDNA to prepare the fragments for hybridization to the flow cell. Using the concentration from the ViiA7™ qPCR machine, 14 pM of the equimolarly pooled library was loaded onto an Illumina HiSeq 2500 sequencing instrument (Illumina, USA). PhiX Control v3 adapter-ligated library was spiked-in at 2% to ensure balanced diversity and to monitor clustering and sequencing performance. A paired-end 100 cycle run is used to sequence the flow cell on a HiSeq Sequencing System in Rapid Mode with v2 chemistry. Bcls (Illumina sequencer’s base call files) were demultiplexed, and FASTQ files were generated. The average depth of sequencing per sample was 125 million read pairs.
RNA-Seq Data Analyses
For each sample, about 100 million pairs of 100-bp paired-end reads were generated. Following the standard approach, 10 bp were from the 5’ end of the raw reads to remove biases on certain nucleotides resulting from poor priming during library preparation. For the first analysis strategy (Fig. 1), which aimed to determine the abundance of transcripts expressed from annotated genes in the reference mouse genome, trimmed reads were aligned to the Mus musculus genome (GRcm38, p6.VM24) using TopHat v2.0.9 [31] with default parameters. The average mappability for the seven samples was 71.16% (70.40–71.90%). HTSeq [32, 33] was used to obtain read counts. Differential expression analysis between Nlrp2-null and WT superovulated oocytes was carried out based on the read counts using the DESeq package in the R environment [33]. The genes with adjusted p value < 0.05 were considered differentially expressed genes (DEGs).
Experimental design. Four-week-old WT and Nlrp2-null females were superovulated. The next day cumulus oocyte complexes (COCs) were collected from the ampulla of the oviduct. Total RNA sequencing was performed on RNA from pooled denuded oocytes. Left route: first strategy in which reads are mapped to the mouse reference genome. Right route: second strategy, in which reads are mapped to the mouse oocyte-specific transcriptome
For the second analysis strategy (Fig. 1), which aimed to determine transcript abundance compared to a recently published oocyte reference transcriptome, raw reads were mapped to the oocyte-specific transcriptome reported by Veselovska et al. [28] using STAR (version 2.5.0a) with default parameters [34]. The average mappability of this reference oocyte transcriptome for the seven samples was 79.5% (79.01–80.1%). Next, featureCounts (version 1.5.0-p1) [35] was used to quantify the reads for oocyte-specific transcripts based on the annotation gene transfer format (GTF) file provided by Veselovska et al. [28]. Based on the read counts, differential transcript analysis between Nlrp2-null and WT superovulated oocytes was performed using DESeq2 (version 1.22.2) in R [36]. We annotated the transcripts to their closest genes using HOMER [37], and a gene is considered for the following analyses if the transcript is within 10 kb upstream of the gene’s transcription start site. For both strategies, principal component analysis (PCA) was carried out using the normalized read counts for all genes or all transcripts in the R environment.
Gene Ontology and Pathway Enrichment
Function and pathway enrichment analysis was performed using three online tools to analyze the biological process, cell component, and molecular function of the DEGs; Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/home.jsp), and the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt Toolkit) (http://www.webgestalt.org/) [38, 39]. The STRING (https://string-db.org/) online database was applied to obtain a description of know protein–protein interaction (PPI). Cytoscape software platform (https://cytoscape.org/) was used to visualize the network.
Validations by RT-qPCR
RNA samples used for validation were isolated from independent pools of superovulated oocytes, and 500 ng of RNA was reverse transcribed into cDNA using the qScript cDNA SuperMix (Quanta Biosciences #95,048). Quantitative RT-PCR of reverse-transcribed cDNAs was done with PerfeCTa® SYBR® Green FastMix ROX (Quanta Biosciences #95,073) using the primers listed in Supplementary Table 2. Gene expression analysis was performed using the ΔΔCt method and normalized to the expression of Rpl19 mRNA.
Overlap of Identified Transcripts with Oocyte Hyper- and Hypomethylated Domains
Hypermethylated domains (HyperDs) and hypomethylated domains (HypoDs) identified by Veselovska, et al. [28] were used as references. For the first strategy, a gene was considered to be into a HyperD or HypoD domain if at least 50% of the gene body (from transcription start to transcription end) overlaps with the region. Similarly, for the second strategy, a transcript is considered in the domain if at least 50% of the transcript overlaps with the region. Intersect function from bedtools (v2.25.0) was used to identify the overlaps.
Results
Differentially Expressed Genes in Nlrp2-null Compared to WT Oocytes
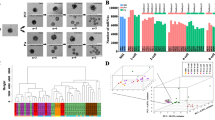
We isolated meiosis II (MII) oocytes from the COCs of superovulated wild-type (WT) and Nlrp2-null female mice. To profile the oocyte transcriptomes for both genotypes and examine changes in transcript abundance caused by maternal loss of Nlrp2, we performed RNA-Seq on three pools of ~ 150 oocytes from WT females and four pools of ~ 150 oocytes from Nlrp2-null females (Fig. 1). We first analyzed the RNA-Seq data by aligning obtained sequences to the mouse reference genome. After filtering out genes that were covered by fewer than 10 average read counts, we retained 18,951 genes for further analysis. We performed principal component analysis (PCA) to characterize the internal group separation, detect any outliers, and determine the degree of variation between WT and Nlrp2-null samples and between replicates for each. This showed that transcriptome data from the four Nlrp2-null pools clustered with each other and separated from the WT pool data (Fig. 2a). We then used DESeq to find differentially expressed genes (DEGs) between the WT and Nlrp2-null oocyte pools, which yielded 231 DEGs (adjusted p value < 0.05) (Supplementary Table 1). Of these, 123 were upregulated, and 108 were downregulated in Nlrp2-null oocyte pools (Fig. 2b).
Principal component analysis and MA plot. a Principal component analysis of normalized transcript counts from WT oocyte pools (red) and Nlrp2-null oocyte pools (blue) cluster separately. b MA plot representing the Log2 fold change (Y-axis) versus normalized mean expression (X-axis) between WT and Nlrp2-null. Unchanged genes are in grey; differentially expressed genes are in red, adjusted p value < 0.05
Altered Biological Processes and Pathways in Nlrp2-null Oocytes
We next analyzed the biological functions of the 231 genes that are differentially expressed between Nlrp2-null and WT oocyte pools. Based on Database for Annotation, Visualization, and Integrated Discovery (DAVID) analysis [40,41,42] (Table 1), both neurogenesis and differentiation were enriched among genes with altered expression in Nlrp2-null oocytes. Within the neurogenesis category, the main significant enriched terms were axon guidance and axon midline choice point recognition. Within the differentiation category, the main enriched terms were multicellular organism development, nervous system development, and axon guidance. There were two significantly enriched molecular function categories: (1) developmental protein, which included significant multicellular organism development and nervous system development subcategories, and (2) chaperone, which included significant enrichment of four subcategories (ATPase activity, protein processing in the endoplasmic reticulum, positive regulation of telomerase activity, and COP9 signalosome). Enriched cellular components included cytoplasm and tight junction. Enriched tight junctions included cell adhesion molecules, bicellular tight junction, cell junction, and cell–cell adhesion. We performed functional analysis using the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) [43] (Table 2) for enriched biological processes (with p value < 0.05 and FDR < 0.05) and found the following enriched processes: protein metabolism, gland morphogenesis, post-translational protein modification, cell morphogenesis that occurs during neuron differentiation, axonogenesis, plasma membrane–bounded cell projection organization, cellular protein localization, cellular macromolecule localization, cell projection organization, and neuron projection morphogenesis pathways. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [44] (Table 3) revealed the following top 10 enriched pathways: gland morphogenesis, biological regulation, cellular process regulation, axonogenesis, neuron projection morphogenesis, biological process regulation, cell morphogenesis that occurs during neuron differentiation, regulation of cellular response to growth factor stimulus, multicellular organism development, and neuron differentiation. Enrichments that were identified by all three tools (DAVID, WebGestalt, and STRING analysis) (Tables 1, 2, and 3) include processes and pathways involved in neurogenesis, nervous system development, axonogenesis, axon guidance, cell morphogenesis that occurs during neuron differentiation, and neuron projection morphogenesis. Interestingly, scientists observed neurogenesis enrichment in another study that evaluated DNA methylation in meiosis II (MII) stage oocytes exposed to different ovarian stimulation protocols used in assisted reproductive technologies [45].
Both WebGestalt and STRING analysis revealed that gland morphogenesis had the highest enrichment ratio with a false discovery rate (FDR) of 0.01. Among the genes annotated to gland morphogenesis were Notch2, Ctnnd1, and Tet2, which have higher expression in Nlrp2-null oocyte pools. These genes are of interest because of their known function and involvement in mouse oocyte growth and embryonic development [46,47,48,49,50,51,52]. Quantitative RT-PCR with primers listed in Supplementary Table 2 confirmed their upregulation in independently collected oocyte pools from Nlrp2-null (Fig. 3a–c). TET2 is essential for survival during the early postnatal period [53]. Tet2 deficiency decreases oocyte development and quality and accelerates age-associated infertility [47]. Tet1-3 deficient embryos arrest at the two-cell stage; the most severe phenotype is linked to Tet2 deficiency [54]. NOTCH2 regulates ovarian follicle formation and coordinates follicular growth [54], and Notch2 deficiency results in embryonic death by E11.5, which is related to placental defects [48]. CTNND1 plays a crucial role in the developing mouse brain such that its ablation results in aberrant embryonic morphology and defective neurogenesis and neural tube formation in mid-gestation embryos [49, 55].
Network Analysis Shows Enrichment for Proteins that Contribute to Post-Translational Methylation
We then used the STRING online database [44] to identify proteins that interact with or are functionally associated with proteins that upregulated DEGs encode. When we used the highest confidence setting in STRING, one “Annotated Keyword” from the UniProt database, methylation (KW-0488, gene count = 16, FDR = 0.026), a subcategory of post-translational modification (PTM; KW-9991), was enriched. We then used Cytoscape [56] to extract and visualize the gene network based on this STRING protein interaction enrichment (Fig. 4a). We further expanded this network to include adjacent genes, which yielded 75 upregulated genes (Fig. 4b). This expanded network also contains Notch2, Ctnnd1, and Tet2, which we found annotated to the gland morphogenesis GO term. The expanded network further contains Kdm1b, which had higher expression in Nlrp2-null oocytes and was confirmed by qRT-PCR (Fig. 3d). Kdm1b is also the highest differentially expressed gene in the enriched “metabolism of proteins” pathway. The KDM1B enrichment is of interest because it is a H3K4 histone demethylase that is required during oocyte development for the establishment of DNA methylation marks at CpG islands, including at imprinted genes [57]. Oocytes from Kdm1b-null females have increased H3K4 methylation and fail to set up proper DNA methylation marks at some imprinted genes. Kdm1b-null mice pregnancies show severe placental defects and embryos with myriad developmental abnormalities [57].
Gene network analysis. a STRING protein–protein interaction and pathway enrichment analysis of upregulated differentially expressed genes result in one “Annotated Keyword” from the UniProt database, methylation (KW-0488, gene count = 16, FDR = 0.026). b Direct interaction between the genes implicated in oocyte growth and embryonic development (selected for qRT-PCR) and methylation (KW-0488)
Data Analysis According to the Reference Oocyte-Specific Transcriptome Identified Many New Transcripts
We next aligned our sequence reads to a previously reported de novo assembled reference mouse oocyte-specific transcriptome. This mouse oocyte-specific transcriptome was obtained using strand-specific deep RNA-seq with de novo transcriptome assembly at different follicular growth stages. It contains 82,939 transcripts corresponding to 39,099 expressed genes, more than half of which were not known prior to that study [28]. We mapped the reads to the oocyte-specific transcriptome and, by including a minimum average read count of 10 in our samples, identified and further studied 18,430 transcripts. PCA analysis using these transcripts revealed larger variations among samples and showed that one Nlrp2-null sample did not cluster with the other Nlrp2-null samples (Fig. 5a–b). We therefore removed this sample, which resulted in three WT and three Nlrp2-null samples available for differential expression analysis. There were 228 transcripts with differential expression at FDR < 0.05 between oocyte pools from WT and Nlrp2-null females (Supplementary Table 3). Of these, 90 had lower expression, and 138 had higher expression in Nlrp2-null mice oocytes (Fig. 5c). Since only 47.7% of the oocyte transcriptome corresponded to the mouse reference annotation [28], we did not expect all 228 differentially expressed transcripts to correspond to known genes in the reference genome. To further understand these transcripts’ characteristics as to known canonical functions, we next inspected the genomic alignment of the 228 differentially expressed transcripts for known reference genes that have a transcription start site (TSS) located within 10 kb of the aligned oocyte-specific transcripts. This identified 159 known reference genes (Supplementary Table 4). WebGestalt GO enrichment analysis of these 159 genes showed that most of them are involved in the TGF-beta signaling pathway and negative regulation of the hydrogen peroxide metabolic process.
Principal component analysis. a Principal component analysis of normalized transcript counts from WT oocyte pools (red) and Nlrp2-null oocyte pools (blue). b PCA after removal of sample 2820. c MA plot representing the Log2 fold change (Y-axis) versus normalized mean expression (X-axis) between WT and Nlrp2-null samples compared to reference oocyte transcriptome [28]. Unchanged genes are in grey; differentially expressed genes are in red, adjusted p value < 0.05
DEGs Overlap with Hypermethylated and Hypomethylated Domains
The average oocyte methylation level is \(\sim\) 40% with a bimodal distribution of CpG methylation, and hypermethylated CpGs are primarily located within the transcribed regions of some genes [58, 59]. In addition to defining the mouse oocyte transcriptome, Veselovska, et al. also demonstrated that transcription drives DNA methylation establishment in female germ cells. They further discovered that methylated CpGs are not randomly distributed in oocytes but instead tend to cluster together. They defined 21,044 hypermethylated domains (HyperDs) and 25,165 hypomethylated domains (HypoDs) within the oocyte DNA methylome [28]. A HyperD is a region that, due to active transcription, acquires methylation on the gene body, while a HypoD is depleted of methylation because it lacks transcription [28].
We first examined the overlap between HyperDs and HypoDs of genes with altered expression from RNA-Seq data alignment to the reference mouse genome (Fig. 6a, Supplementary Table 5). Of the 231 DEGs, 156 (68%) overlapped with HyperDs or HypoDs. Given the known correlation between gene transcription and DNA methylation at HyperDs, we next examined the relationship between transcript levels in Nlrp2-null versus WT oocytes and mapping to HyperDs. We found that 83 of 108 (77%) underexpressed genes and 55 of 123 (45%) overexpressed genes in Nlrp2-null oocytes overlap with HyperDs. It is significantly more likely for underexpressed genes to overlap with HyperDs than for overexpressed genes (p = 0.0043, Fisher’s Exact test). In contrast, only 4 of the 108 (4%) underexpressed genes and 14 of the 123 (11%) overexpressed genes overlap with HypoDs.
Overlap of differentially expressed genes with HyperDs and HypoDs. a Overlap of differentially expressed genes from RNA-Seq data that were aligned to the mouse reference genome. b Overlap of differentially expressed genes from RNA-Seq data were aligned to the reference oocyte transcriptome. The top Venn diagrams for each panel represents the overlap of overexpressed differentially expressed genes with HyperDs and HypoDs and the bottom panel represents the overlap of underexpressed differentially expressed genes with HyperDs and HypoDs; hypergeometric test; p < 0.05
Next, we analyzed HyperDs and HypoD overlap with DEGs identified from alignment to the oocyte reference transcriptome. Of the 228 DEGS, 127 (56%) overlapped with either HyperDs (89 transcripts) or HypoDs (38 transcripts) (Fig. 6b, Supplementary Table 6). We found that 51 of 90 (57%) underexpressed genes mapped to HyperDs compared to only 38 of 138 (28%) overexpressed genes. Again, it is much more likely for underexpressed genes to overlap with HyperDs than overexpressed genes (p = 0.0018, Fisher’s exact test). When we examined overlap with HypoDs, we found that 10 of 90 (11%) underexpressed genes and 28 of 138 (20%) overexpressed genes in Nlrp2-null oocytes overlapped with HypoDs. We do not have genome-wide methylation profiling data on these oocytes, but considering that transcription drives methylation in oocytes, it will be interesting to see if methylation levels at the HyperDs and HypoDs correlate with over- and underexpression of their overlapping transcripts. This could provide mechanistic insight into the consequences of maternal Nlrp2 loss on oocytes and, consequently, offspring DNA methylation patterns at specific loci.
Discussion
In this study, we found that maternal loss of Nlrp2, a gene that encodes an oocyte SCMC protein, causes altered levels of hundreds of transcripts in superovulated MII-stage oocytes. Considering that the zygote and early preimplantation embryo depend on the actions of stored transcripts and proteins expressed during oocyte development (until the zygotic genome is activated) for all their functions [60], these results could elucidate mechanisms by which disrupting the SCMC (through loss of a component protein) can affect the earliest stages of embryonic development. The SCMC is conserved among mammals and is essential for mammalian preimplantation development. In addition to described roles in several important biological processes during oocyte-to-embryo transition, the SCMC affects epigenetic reprogramming and regulation of mRNAs [13]. Yet, despite extensive research, its function remains incompletely understood.
Women who carry biallelic pathogenic variants in one of several genes encoding SCMC proteins are healthy but have an increased risk of embryonic lethality and reproductive failure. This failure is often due to early developmental arrest. Early embryonic arrest is one of the major causes of recurrent assisted reproduction failure. It was first reported in 2015 that most TLE6-mutant oocytes failed to fertilize and that embryos resulting from those that did fertilize arrested at the zygote, two or four cells stage [61]. In addition to causing female infertility, maternal inactivating variants in these genes can also cause imprinting disorders in offspring. Mutations in some genes that encode SCMC proteins have been associated with multi-locus imprinting disturbance (MLID) and biparental complete hydatidiform mole (BiCHM) [17, 18, 23, 27]. This suggests that SCMC proteins play a role in establishing and possibly maintaining differential DNA methylation at maternally imprinted loci.
Maternal knockouts of different genes that encode SCMC proteins in mice most frequently result in early embryo arrest but no observed alterations in oogenesis [13]. Female mice lacking Nlrp2 experience preimplantation or later embryonic loss and produce fewer offspring that have wide-ranging developmental phenotypes and abnormal DNA methylation at imprinted loci. Nlrp2-null females also produced more unhealthy and degenerating follicles compared to WT females. [15]. Kuchmiy and colleagues found that oocytes from 22 to 25 weeks old Nlrp2-mutant mice that underwent parthenogenetic activation progressed asynchronously and with significantly reduced rates of early embryonic development compared to WT [62]. To our knowledge, at the time of submission, no studies assessing the oocyte transcriptome in any mouse model knockout for genes encoding the SCMC exist. In a recent study, DNA methylation and gene expression were assessed in 2-cell embryos and MII oocytes from Nlrp14-knockout (KO) and WT female mice. Nlrp14 is a maternal effect gene with similar reproductive roles as all reproduction-related NLRP genes present in both mouse and human genome. Authors reported infertility in Nlrp14-KO female mice. These mice had normal oogenesis and oocyte ovulation, but embryos from Nlrp14-KO females arrested at the two-cell stage. DNA methylation and gene expression were impaired in embryos from Nlrp14-KO compared to WT. However, DNA methylation levels were comparable between MII oocytes from Nlrp14-KO and WT. Furthermore, 163 genes were up-regulated, and 172 were downregulated in MII oocytes from Nlrp14-KO compared to WT. These data highlight the importance of further investigation of the epigenetic regulatory role of all NLRP genes [63].
One study on oocytes from a human patient with loss of KHDC3L, an SCMC protein associated with hydatidiform moles and MLID, revealed genome-wide DNA methylation deficit in the KHDC3L-deficient oocytes. This supported the conclusion that KHDC3L is indispensable for establishing de novo DNA methylation in the growing human oocyte [22].
We performed functional analysis to gain deeper insight into the biological functions of the DEGs and construct that transcript networks that are dysregulated in the absence of Nlrp2. Functional annotation using DAVID analysis revealed that neurogenesis (GO:0,007,399) and differentiation (GO:0,030,154) are the two most enriched biological processes. Interestingly, NLRP2 is expressed in the central nervous system and its inactivation affects the transcription of genes implicated in nervous system development [64, 65]. Chaperone (KW-9992) is the most significantly enriched molecular function among DEGs. This function includes protein processing in the endoplasmic reticulum (mmu04141), a process that has been implicated in the oocyte-to-embryo transition upon the change of endoplasmic reticulum architecture [66]. The SCMC proteins NLRP5, OOEP, and PADI6 are required for the formation of oocyte cytoplasmic lattices (CPL), with which they colocalize. CPL regulates organelle distribution including endoplasmic reticulum, oocyte maturation, and microtubule dynamics, and stores components of the maternal protein synthesis machinery, including ribosomes, to contribute to the early embryo [4, 12, 14, 67]. Since NLRP2 is an SCMC protein that interacts with OOEP and NLRP5 [15], its absence may disrupt CPL function.
In our study, KDM1B was upregulated in Nlrp2-null oocytes. KDM1B is the major factor that removes methylated H3K4 in mouse oocytes and is essential to most imprinted genes and CpG islands (CGIs) that acquire methylation in oocytes [68]. In female germ cells, transcription drives most of the de novo DNA methylation establishment [29, 30]. Therefore, changes in gene expression that occur during oocyte maturation could alter the DNA methylation pattern. Another important event that occurs in a growing oocyte during de novo DNA methylation establishment is the exclusion of H3K4me2/me3 by KDM1B from regions that are destined to become methylated. The methylated H3K4 histone mark must be removed to recruit DNA methyltransferases 3A/3L [68]. The failure to set up DNA methylation marks at multiple maternally imprinted loci in KDM1B-deficient female oocytes confirms KDM1B’s role in this process [57].
Next, to better understand the significance of KDM1B upregulation in Nlrp2-null cells and investigate its possible connection to altered DNA methylation, we examined the expression changes of genes that map to annotated oocyte HyperDs and HypoDs. Because active transcription drives DNA methylation in oocytes, we hypothesize that altered transcription of genes that localize to annotated HyperDs and HypoDs can change the methylation status of those domains and alter the distribution of HyperDs and HypoDs in Nlrp2-null female oocytes. Specifically, less transcription of genes that map to HyperDs could cause those HyperDs to acquire less or no methylation, turning them into HypoDs. Conversely, more transcription of genes that map to HypoDs could cause those HypoDs to acquire more methylation, turning them into HyperDs (Fig. 7). We observed that among the four possible overlaps (1) underexpressed DEGs with HyperDs, (2) overexpressed DEGs with HyperDs, (3) underexpressed DEGs with HypoDs, and (4) overexpressed DEGs with HypoDs the most significant was underexpressed DEGs and HyperDs in both analyses (83 for the reference genome and 51 for the oocyte reference transcriptome). We, therefore, propose that less expression of transcripts that map to annotated HyperDs in Nlrp2-null oocytes could result in loss of methylation in those HyperDs. Although we do not have DNA methylation or histone profiling data for these oocytes, recent methylation profiling data from human oocytes deficient in KHDC3L revealed a genome-wide DNA methylation deficit [22]. It is therefore plausible that deleting NLRP2 and other SCMC members causes the same deficiency, but this requires further investigation.
Re-distribution of HyperDs and HypoDs in absence of Nlrp2. a Distribution of HyperDs and HypoDs within the WT oocytes genome. Black and white lollipops represent the presence and absence of DNA methylation, respectively. Gray blocks represent regions not identified as HyperDs/HypoDs. b, c Transition and distribution of HyperDs and HypoDs within the Nlrp2-null oocytes genome. Red X shows the underexpressed differentially expressed genes within HyperDs and triple black arrow shows overexpressed differentially expressed genes within HypoDs. Altered transcription may cause change in the methylation status and consequently re-distribution of HyperDs/HypoDs (c)
We generated transcriptome data from Nlrp2-null and WT oocytes at MII stage. Transcription is globally silenced in the final stages of oocyte growth and does not significantly resume until the 2-cell embryo stage in mice. Therefore, oocytes store large numbers of transcripts and proteins to support the processes of fertilization and early development before zygotic/embryonic genome activation. Thus, at the MII stage, when WT oocytes are transcriptionally silent, RNA-seq data reflect the abundance of stored transcripts. We do not currently know what exactly drives the changes in the Nlrp2-null MII oocyte transcriptome, but one or more of the following could be involved. There may be higher or lower transcription activity at earlier stages of oocyte maturation, the stored transcripts may be more stable or more prone to degradation or the programmed silencing of transcription before the MII oocyte stage may be disrupted such that genes continue to be transcribed longer or are silenced earlier. Which one of these is the culprit and whether different mechanisms are involved depending on the genomic features of interest is currently unknown. We note that NLRP2 is a protein of the SCMC and that its loss disrupts the SCMC, and may consequently disrupt structures in the cytoplasm, such as the cytoplasmic lattices that are involved in binding subsets of mRNAs.
In our study, MII oocytes were collected from superovulated mice, and there is increasing evidence that superovulation alters DNA methylation and gene expression [45, 69]. Ovarian stimulation also impairs oocyte/embryo quality, decreases the implantation rate, and increases embryo aneuploidy and fetal malformation rates [70,71,72]. Although a recent study showed very little difference in the DNA methylation profile between oocytes from superovulated adult females and naturally ovulated oocytes [45], we do not know if oocytes from Nlrp2 females could be more sensitive to the effects of ovarian stimulation. To address this, similar studies on oocytes collected without prior superovulation and closer to the timing of methylation establishment will be needed.
In conclusion, this study shows that the loss of maternal Nlrp2 has widespread consequences on the transcriptome of mature oocytes. This may in part underlie adverse reproductive outcomes and abnormal embryo development observed in Nlrp2-null females.
Data Availability
Data is publicly available from the Gene Expression Omnibus (GEO) accession number: GSE213059.
Code Availability
Not applicable.
References
Wolf JB, Wade MJ. What are maternal effects (and what are they not)? Philos Trans R Soc Lond B Biol Sci. 2009. https://doi.org/10.1098/rstb.2008.0238.
Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008. https://doi.org/10.1016/j.devcel.2008.07.010.
Lu X, Gao Z, Qin D, Li L. A Maternal Functional Module in the Mammalian Oocyte-To-Embryo Transition. Trends Mol Med. 2017. https://doi.org/10.1016/j.molmed.2017.09.004.
Qin D, Gao Z, Xiao Y, Zhang X, Ma H, Yu X, Nie X, Fan N, Wang X, Ouyang Y, Sun QY, Yi Z, et al. The subcortical maternal complex protein Nlrp4f is involved in cytoplasmic lattice formation and organelle distribution. Development. 2019. https://doi.org/10.1242/dev.183616.
Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, Wang Y, Liu J, Zhang B, Quan F, Guo Z, Zhang Y. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0030344.
Gao Z, Zhang X, Yu X, Qin D, Xiao Y, Yu Y, Xiang Y, Nie X, Lu X, Liu W, Yi Z, Li L. Zbed3 participates in the subcortical maternal complex and regulates the distribution of organelles. J Mol Cell Biol. 2018. https://doi.org/10.1093/jmcb/mjx035.
Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000. https://doi.org/10.1038/81547.
Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development. 2008. https://doi.org/10.1242/dev.011445.
Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, Robben TJ, Coonrod S, Gossen JA. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007. https://doi.org/10.1016/j.mce.2007.05.005.
Kanzaki S, Tamura S, Ito T, Wakabayashi M, Saito K, Kato S, Ohta Y, Sekita Y, Kimura T. Involvement of Nlrp9a/b/c in mouse preimplantation development. Reproduction. 2020. https://doi.org/10.1530/REP-19-0516.
Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, Ou-Yang Y, Wang ZB, Zheng P, Zhu MS, Wang H, Sun QY, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014. https://doi.org/10.1038/ncomms5887.
Kim B, Zhang X, Kan R, Cohen R, Mukai C, Travis AJ, Coonrod SA. The role of MATER in endoplasmic reticulum distribution and calcium homeostasis in mouse oocytes. Dev Biol. 2014. https://doi.org/10.1016/j.ydbio.2013.12.025.
Bebbere D, Albertini DF, Coticchio G, Borini A, Ledda S. The subcortical maternal complex: emerging roles and novel perspectives. Mol Hum Reprod. 2021. https://doi.org/10.1093/molehr/gaab043.
Tashiro F, Kanai-Azuma M, Miyazaki S, Kato M, Tanaka T, Toyoda S, Yamato E, Kawakami H, Miyazaki T, Miyazaki J. Maternal-effect gene Ces5/Ooep/Moep19/Floped is essential for oocyte cytoplasmic lattice formation and embryonic development at the maternal-zygotic stage transition. Genes Cells. 2010. https://doi.org/10.1111/j.1365-2443.2010.01420.x.
Mahadevan S, Sathappan V, Utama B, Lorenzo I, Kaskar K, Van den Veyver IB. Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Sci Rep. 2017. https://doi.org/10.1038/srep44667.
Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci USA. 2009. https://doi.org/10.1073/pnas.0900519106.
Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, Carr I, Rittore C, Touitou I, Philibert L, Fisher RA, Fallahian M, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011. https://doi.org/10.1016/j.ajhg.2011.08.002.
Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006. https://doi.org/10.1038/ng1740.
Qian J, Nguyen NMP, Rezaei M, Huang B, Tao Y, Zhang X, Cheng Q, Yang H, Asangla A, Majewski J, Slim R. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018. https://doi.org/10.1038/s41431-018-0141-3.
Sanchez-Delgado M, Martin-Trujillo A, Tayama C, Vidal E, Esteller M, Iglesias-Platas I, Deo N, Barney O, Maclean K, Hata K, Nakabayashi K, Fisher R, et al. Absence of maternal methylation in biparental hydatidiform moles from women with NLRP7 maternal-effect mutations reveals widespread placenta-specific imprinting. PLoS Genet. 2015. https://doi.org/10.1371/journal.pgen.1005644.
Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014. https://doi.org/10.1101/cshperspect.a018382.
Demond H, Anvar Z, Jahromi BN, Sparago A, Verma A, Davari M, Calzari L, Russo S, Jahromi MA, Monk D, Andrews S, Riccio A, et al. A KHDC3L mutation resulting in recurrent hydatidiform mole causes genome-wide DNA methylation loss in oocytes and persistent imprinting defects post-fertilisation. Genome Med. 2019. https://doi.org/10.1186/s13073-019-0694-y.
Docherty LE, Rezwan FI, Poole RL, Turner CL, Kivuva E, Maher ER, Smithson SF, Hamilton-Shield JP, Patalan M, Gizewska M, Peregud-Pogorzelski J, Beygo J, et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun. 2015. https://doi.org/10.1038/ncomms9086.
Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, Woods CG, Reik W, Maher ER. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet. 2009. https://doi.org/10.1371/journal.pgen.1000423.
Eggermann T, Kadgien G, Begemann M, Elbracht M. Biallelic PADI6 variants cause multilocus imprinting disturbances and miscarriages in the same family. Eur J Hum Genet. 2021. https://doi.org/10.1038/s41431-020-00762-0.
Eggermann T. Maternal effect mutations: a novel cause for human reproductive failure. Geburtshilfe Frauenheilkd. 2021. https://doi.org/10.1055/a-1396-4390.
Begemann M, Rezwan FI, Beygo J, Docherty LE, Kolarova J, Schroeder C, Buiting K, Chokkalingam K, Degenhardt F, Wakeling EL, Kleinle S, Gonzalez Fassrainer D, et al. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J Med Genet. 2018. https://doi.org/10.1136/jmedgenet-2017-105190.
Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Mehouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015. https://doi.org/10.1186/s13059-015-0769-z.
Yan R, Gu C, You D, Huang Z, Qian J, Yang Q, Cheng X, Zhang L, Wang H, Wang P, Guo F. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell. 2021. https://doi.org/10.1016/j.stem.2021.04.012.
Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009. https://doi.org/10.1101/gad.495809.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013. https://doi.org/10.1186/gb-2013-14-4-r36.
Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015. https://doi.org/10.1093/bioinformatics/btu638.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010. https://doi.org/10.1186/gb-2010-11-10-r106.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013. https://doi.org/10.1093/bioinformatics/bts635.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014. https://doi.org/10.1093/bioinformatics/btt656.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014. https://doi.org/10.1186/s13059-014-0550-8.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010. https://doi.org/10.1016/j.molcel.2010.05.004.
Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013. https://doi.org/10.1093/nar/gkt439.
Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005. https://doi.org/10.1093/nar/gki475.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009. https://doi.org/10.1038/nprot.2008.211.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009. https://doi.org/10.1093/nar/gkn923.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022. https://doi.org/10.1093/nar/gkac194
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gkz401
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021. https://doi.org/10.1093/nar/gkaa1074
Saenz-de-Juano MD, Ivanova E, Billooye K, Herta AC, Smitz J, Kelsey G, Anckaert E. Genome-wide assessment of DNA methylation in mouse oocytes reveals effects associated with in vitro growth, superovulation, and sexual maturity. Clin Epigenetics. 2019. https://doi.org/10.1186/s13148-019-0794-y.
Vanorny DA, Prasasya RD, Chalpe AJ, Kilen SM, Mayo KE. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol Endocrinol. 2014. https://doi.org/10.1210/me.2013-1288.
Wang X, Rosikiewicz W, Sedkov Y, Martinez T, Hansen BS, Schreiner P, Christensen J, Xu B, Pruett-Miller SM, Helin K, Herz HM. PROSER1 mediates TET2 O-GlcNAcylation to regulate DNA demethylation on UTX-dependent enhancers and CpG islands. Life Sci Alliance. 2021. https://doi.org/10.26508/lsa.202101228
Hamada Y, Hiroe T, Suzuki Y, Oda M, Tsujimoto Y, Coleman JR, Tanaka S. Notch2 is required for formation of the placental circulatory system, but not for cell-type specification in the developing mouse placenta. Differentiation. 2007. https://doi.org/10.1111/j.1432-0436.2006.00137.x.
Pieters T, Sanders E, Tian H, van Hengel J, van Roy F. Neural defects caused by total and Wnt1-Cre mediated ablation of p120ctn in mice. BMC Dev Biol. 2020. https://doi.org/10.1186/s12861-020-00222-4.
Johnson J, Espinoza T, McGaughey RW, Rawls A, Wilson-Rawls J. Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev. 2001. https://doi.org/10.1016/s0925-4773(01)00523-8.
Hernandez-Martinez R, Ramkumar N, Anderson KV. p120-catenin regulates WNT signaling and EMT in the mouse embryo. Proc Natl Acad Sci USA. 2019. https://doi.org/10.1073/pnas.1902843116.
Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013. https://doi.org/10.1016/j.devcel.2012.12.015.
Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, Kamiunten A, Sekine M, Hidaka T, Katayose K, Kubuki Y, Yamamoto S, Miike T, et al. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia. 2012. https://doi.org/10.1038/leu.2012.94.
Arand J, Chiang HR, Martin D, Snyder MP, Sage J, Reijo Pera RA, Wossidlo M. Tet enzymes are essential for early embryogenesis and completion of embryonic genome activation. EMBO Rep. 2022. https://doi.org/10.15252/embr.202153968
Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006. https://doi.org/10.1016/j.neuron.2006.05.018.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003. https://doi.org/10.1101/gr.1239303.
Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009. https://doi.org/10.1038/nature08315.
Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012. https://doi.org/10.1371/journal.pgen.1002440.
Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, Kono T, Sasaki H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013. https://doi.org/10.1371/journal.pgen.1003439.
Kobayashi H, Nagao K, Nakajima K. Focus issue on male infertility. Adv Urol. 2012;2012:823582.
Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, Poizat C, Alkuraya FS. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015. https://doi.org/10.1186/s13059-015-0792-0.
Kuchmiy AA, D’Hont J, Hochepied T, Lamkanfi M. NLRP2 controls age-associated maternal fertility. J Exp Med. 2016. https://doi.org/10.1084/jem.20160900.
Yan R, Cheng X, Gu C, Xu Y, Long X, Zhai J, Sun F, Qian J, Du Y, Wang H, Guo F. Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development. Nat Genet. 2023. https://doi.org/10.1038/s41588-022-01258-x.
Sun X, Song X, Zhang L, Sun J, Wei X, Meng L, An J. NLRP2 is highly expressed in a mouse model of ischemic stroke. Biochem Biophys Res Commun. 2016. https://doi.org/10.1016/j.bbrc.2016.09.157.
Minkiewicz J, de Rivero Vaccari JP, Keane RW. Human astrocytes express a novel NLRP2 inflammasome. Glia. 2013. https://doi.org/10.1002/glia.22499.
Israel S, Ernst M, Psathaki OE, Drexler HCA, Casser E, Suzuki Y, Makalowski W, Boiani M, Fuellen G, Taher L. An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-49817-3.
Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol. 2011. https://doi.org/10.1016/j.ydbio.2010.11.033.
Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, Smallwood SA, Chen T, Kelsey G. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015. https://doi.org/10.1101/gad.271353.115.
Uysal F, Ozturk S, Akkoyunlu G. Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J Assist Reprod Genet. 2018. https://doi.org/10.1007/s10815-017-1087-z.
Huo Y, Yan ZQ, Yuan P, Qin M, Kuo Y, Li R, Yan LY, Feng HL, Qiao J. Single-cell DNA methylation sequencing reveals epigenetic alterations in mouse oocytes superovulated with different dosages of gonadotropins. Clin Epigenetics. 2020. https://doi.org/10.1186/s13148-020-00866-w.
Lin J, Xu H, Chen B, Wang W, Wang L, Sun X, Sang Q. Expanding the genetic and phenotypic spectrum of female infertility caused by TLE6 mutations. J Assist Reprod Genet. 2020. https://doi.org/10.1007/s10815-019-01653-0.
Ertzeid G, Storeng R. The impact of ovarian stimulation on implantation and fetal development in mice. Hum Reprod. 2001. https://doi.org/10.1093/humrep/16.2.221.
Funding
This work was supported by grants R01HD092746 and P50HD103555 (for use of the administrative core), from the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health. M.S. is supported by postdoctoral fellowship award from The Lalor Foundation, INC and by T32HD098068. This project was supported in part by the Genomic and RNA Profiling Core at Baylor College of Medicine with funding from the NIH NCI (P30CA125123) and CPRIT (RP200504) grants. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
SM, YWW, and IV conceived the experiments; ZA, SM, IC, and ETN performed experiments; YWW, ZL, IC, LS, and MS analyzed data. ZA and IV wrote the manuscript. All authors contributed to manuscript editing. IV acquired funding and was responsible for overall oversight and final editing.
Corresponding authors
Ethics declarations
Ethics Approval
All experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (protocol AN-2035). Animal facilities were accredited by the Association for Assessment and Accreditation for Laboratory Animal Care International (AAALAC).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Anvar, Z., Chakchouk, I., Sharif, M. et al. Loss of the Maternal Effect Gene Nlrp2 Alters the Transcriptome of Ovulated Mouse Oocytes and Impacts Expression of Histone Demethylase KDM1B. Reprod. Sci. 30, 2780–2793 (2023). https://doi.org/10.1007/s43032-023-01218-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-023-01218-8