Abstract
Plenty of pieces of evidence suggest that the resistance to radiotherapy greatly influences the therapeutic effect in cervical cancer (CCa). MicroRNAs (miRNAs) have been reported to regulate cellular processes by acting as tumor suppressors or promoters, thereby driving radioresistance or radiosensitivity. Meanwhile, it has been reported that microRNA-1323 (miR-1323) widely participates in cancer progression and radiotherapy effects. However, the role of miR-1323 is still not clear in CCa. Hence, in this study, we are going to investigate the molecular mechanism of miR-1323 in CCa cells. In the beginning, miR-1323 was found aberrantly upregulated in CCa cells via RT-qPCR assay. Functional assays indicated that miR-1323 was transferred by cancer-associated fibroblasts-secreted (CAFs-secreted) exosomes and miR-1323 downregulation suppressed cell proliferation, migration, invasion, and increased cell radiosensitivity in CCa. Mechanism assays demonstrated that miR-1323 targeted poly(A)-binding protein nuclear 1 (PABPN1). Besides, PABPN1 recruited insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) to regulate glycogen synthase kinase 3 beta (GSK-3β) and influenced Wnt/β-catenin signaling pathway. Therefore, rescue experiments were implemented to validate that PABPN1 overexpression rescued the inhibited cancer development and radioresistance induced by the miR-1323 inhibitor. In conclusion, miR-1323 was involved in CCa progression and radioresistance which might provide a novel insight for CCa treatment.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer is a kind of gynecologic malignant tumor which occurs in the uterovaginal part and cervical canal [1]. Though CCa is one of the best preventable malignancies among all cancers, the occurrence remains high [2]. Surgery, radiotherapy, and chemotherapy are the main treatment options for CCa [3]. Among them, radiotherapy occupies an important position and greatly improves the overall survival rate [4]. Nevertheless, it cannot be ignored that the existence of radioresistance significantly influences the therapeutic effect of radiotherapy since resistance to radiotherapy accounts for most treatment failures in CCa patients who undergo radiotherapy [5, 6]. Based on the literature review, even the aggressive chemoradiation has a 36.7% of local failure rate within the central pelvis. The 5-year recurrence-free survival is about 79% for stage IB, IIA disease and 59% for III/IVA disease[7]. Therefore, identifying valuable factors associated with radioresistance is increasingly urgent.
Cancer-associated exosomal miRNAs have been identified in tumor growth, metastasis, angiogenesis, and drug resistance [8]. In addition, miRNAs have been determined to be important biomarkers in cervical cancer [9, 10]. For instance, Wang et al. have proposed that miR-449a is a tumor suppressor in cervical cancer [11]. Hua et al. have elucidated that miR-338-3p influences cell proliferation in cervical cancer via the MAPK signaling pathway [12]. Meanwhile, miRNAs in cervical cancer have been proven to exert regulatory functions on radioresistance [13]. For example, Zhang et al. have proved that miR-4778-3p is involved in cervical cancer radioresistance by targeting NR2C2 and Med19 [14]. Wei et al. have also disclosed the inhibitory role of miR-9-5p in radioresistance in cervical cancer [15].
As the miRNAs we researched, miR-1323 was reported to regulate tumor progression. For instance, lncRNA GAS5 mediated miR-1323 to enhance the development of tumor via targeting TP53INP1 in hepatocellular carcinoma (HCC); miR-1323 promotes the migration of lung adenocarcinoma (LUAD) cells via inhibiting Cbl-b expression; the knockdown of miR-1323 promotes cell migration and invasion in breast cancer (BC) via targeting tumor protein D52 [16,17,18]. Moreover, the effect of miR-1323 on radioresistant cells has been reported in previous studies. For example, miR-1323 downregulation restores sensitivity to radiation by suppressing PRKDC activity in radiation-resistant lung cancer cells [19]. In this study, we laid emphasis on exploring the function of miR-1323 in modulating radioresistance in CCa and delving into the regulatory mechanism.
Materials and Methods
Cell Culture
CCa cell lines (HeLa, SiHa, CaSki, and C33A) and normal endocervical epithelioid cell lines (End1/E6E7) were all acquired from American Type Culture Collection (ATCC; Manassas, VA, USA). C33A, SiHa, and HeLa cell lines were cultured in EMEM. CaSki cell line was maintained in RPMI-1640 medium, while End1/E6E7 cell line was incubated in K-SFM. HEK-293T cells were acquired from the National Institute for the Control of Pharmaceutical and Biological Products (NICPBP), being maintained in Dulbecco’s modified Eagle medium (DMEM). All mediums were added with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 1% penicillin-streptomycin (Pen-Strep). The whole incubation process was performed at 37 °C in an atmosphere humidified by 5% CO2. Cancer-associated fibroblasts (CAFs) and NFs (normal fibroblasts) were commercially procured from ATCC. To simulate exosome-mediated intercellular communication between CCa cells and CAFs/NFs, coculture experiments were carried out via using Transwell membranes. C33A was seeded in the upper chamber, and CAFs and NFs were plated on the bottom of the 12-well plates. After 72-h incubation, NFs and CAFs were collected for RNA extraction or further experiments. In addition, the passage of fibroblasts used for experiments was twice a week at the logarithmic growth phase. For irradiation experiments, irradiation treatment was performed on the next day. Irradiated C33A cells were harvested for further experiments.
Cell Transfection
Cell transfections were conducted with the employment of Lipofectamine 2000, purchased from Thermo Fisher Scientific (Waltham, IL, USA). The short hairpin RNAs (shRNAs) targeting PABPN1 (CCGGCCTTAGATGAGTCCCTATTTACTCGAGTAAATAGGGACTCATCTAAGGTTTTTG), ELAVL1, CSTF2T, IGF2BP1, IGF2BP2, DDX54, and relative controls were generated by GenePharma (Shanghai, China). In addition, pcDNA3.1 vectors were subjected to insertion with the sequences of PABPN1 for overexpression of PABPN1. miR-1323 inhibitors and NC inhibitors were all bought from Sangon Biotech (Shanghai, China).
Quantitative Real-Time PCR (RT-qPCR) Analysis
According to the manufacturer’s instructions, TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) was utilized to obtain total RNAs. Then RNAs were reversely transcribed into cDNA with the application of the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, IL, USA). The quantification of RNA expressions was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The 2−ΔΔCt method was utilized to calculate, with GAPDH or U6 as the internal control. The experiment was independently conducted at least three times. Besides, alpha-amanitin (α-amanitin) treatment was adopted to evaluate the transference of miR-1323 by exosomes.
Cell Counting Kit-8 (CCK-8) Assay
Cells (2 × 105 cells per well) were seeded in the 24-well plates. The cell viability was determined by measuring the absorbance (450 nm). The experiment was independently performed at least in triplicate.
Colony Formation Assay
The cells were inoculated in the 6-well plates for incubation. After 14 days, cells were treated in ethanol for fixation. The cell colonies were stained by crystal violet, followed by manual counting. The experiment was independently implemented at least in triplicate.
5-Ethynyl-20-Deoxyuridine (EdU) Staining Assay
1 × 104 transfected CCa cells were collected and cultured in the 96-well plates for 2 h of incubation with EdU Staining Kit (Ribobio). After that, cells were treated in DAPI staining solution for 5 min, followed by the observation of a fluorescence microscope from Olympus (Tokyo, Japan). The experiment was independently performed at least in triplicate.
Transwell Assay
A total of 2 × 104 transfected CCa cells were collected, followed by the inoculation in the upper chamber of Transwell chambers (24-well; Corning Incorporated, Corning, NY, USA), filled with serum-free medium for cell migration testing. Cell invasion was determined using Matrigel membrane (BD Biosciences, Franklin Lakes, NJ, USA). Complete culture medium was added into the lower chamber. Twenty-four hours later, migrated or invaded cells were fixed and stained by crystal violet. Finally, visualization was conducted using an optical microscope procured from Olympus. The experiment was independently implemented at least in triplicate.
Exosome Isolation
CAFs or NFs freshly isolated from CCa patients were cultured in DMEM/F12 containing 10% exosome-depleted FBS. Forty-eight hours later, the collected medium was subjected to centrifugation. With the utilization of Total Exosome Isolation reagent (Invitrogen, Carlsbad, CA, USA), exosomes were isolated. Lastly, the isolated exosomes were washed by sterilized PBS and then treated with centrifugation for purification. CAFs-exo represents the exosome derived from CAFs. NFs-exo represents the exosome derived from NFs. The experiment was independently conducted at least three times.
Exosome Tracing
Exosome immunoprecipitation reagent (protein G) purchased from Thermo Fisher Scientific (USA) was used to label the exosomes as per the supplier’s manual. C33A cells were co-incubated with exosomes. The uptake of exosomes by C33A cells was analyzed under a confocal microscope.
Transmission Electron Microscopy (TEM) Observation
Exosomes were subjected to 100 μl of PBS for suspending and fixed with 5% glutaraldehyde at 4 °C until TEM analysis. Then, the specimens were dehydrated by gradient alcohol and immersed in epoxy resin. The ultrathin sections were observed using a transmission electron microscope (Tecnai G2 Spirit Bio TWIN, FEI, USA). The experiment was independently performed at least in triplicate.
Nanoparticle Tracking Analysis (NTA)
NanoSight NS300 Sub-micron particle imaging system (NanoSight Technology, Malvern, UK) was used to analyze the number and size of the exosomes. The samples were diluted with Dulbecco’s phosphate-buffered saline (DPBS) without any nanoparticles at a final concentration of 1–20 × 108 particles per milliliter for analysis. Exosome concentrations and size distribution were calculated with the application of Stokes–Einstein equation. The experiment was independently conducted at least three times.
Western Blot Analysis
Total protein samples were subjected to SDS-PAGE for separation and later transferred onto PVDF membranes. After being blocked in nonfat milk, the membranes were subjected to incubation with the primary antibodies against CD63, CD9, TSG101, HSP90, GM130, PABPN1, GSK-3β, nuclear-β-catenin, p-β-catenin (S33), p-β-catenin (S37), histone H3, and GAPDH. Afterward, the protein samples were incubated with a secondary antibody for 1 h at room temperature. Finally, protein quantification analysis was conducted by ECL Western blotting substrate from Invitrogen (Carlsbad, CA, USA). The experiment was independently conducted at least in triplicate.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
RIP assay was achieved by the use of Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA). Cells were lysed in RIP lysis buffer. After that, cell lysates were cultivated with the magnetic beads conjugated with argonaute-2 (Ago2) antibody (anti-Ago2) or IGF2BP1 antibody. IgG antibody (anti-IgG) was used in NC control. All results were monitored using RT-qPCR. The experiment was independently performed at least in triplicate.
RNA Pull-Down Assays
For RNA-RNA pull-down assay, biotin-labeled miR-1323 was treated with structure buffer and then denatured by heating at 95 °C for 2 min and subjected to ice bath for 3 min, and finally left at room temperature for 30 min. Streptavidin beads and biotin-labeled miR-1323-WT/miR-1323-Mut/NC (Bio-miR-1323-WT/Bio-miR-1323-Mut/Bio-NC) were incubated at 4 °C for 2 h. For RNA-protein pull-down assay, Pierce magnetic RNA-protein pull-down kit (Thermo Fisher, IL, USA) was used following the manufacturer’s protocol. Biotinylated GSK-3β sense and antisense probes (Bio-GSK-3β S and Bio-GSK-3β AS) were incubated with streptavidin beads. Cells were lysed and cell lysates were mixed with beads-probe complexes, followed by overnight incubation and centrifugation. RNAs were extracted using Trizol and subjected to RT-qPCR analysis in the RNA-RNA pull-down assay. The enriched proteins were measured by western blot with IGF2BP1 antibody in the RNA-protein pull-down assay. The experiment was independently conducted three times.
Dual-Luciferase Reporter Assay
The wild-type (WT) or mutant (Mut) of PABPN1 3′-UTR fragments were cloned into a luciferase vector from Promega (Madison, WI, USA) to construct PABPN1 reporter vector (Wt-Luc/Mut-Luc). The predicted binding sites of PABPN1 were mutated to synthesize PABPN1-Mut1 and PABPN1-Mut2, which were then used to construct PABPN1-Mut reporter vectors (Mut1-Luc and Mut2-Luc). Subsequently, an LF-3000 transfection reagent was applied to transfect the cells with the above composite plasmids and NC inhibitor or miR-1323 inhibitor. Similarly, WT or Mut β-catenin promoter containing the binding sites between the β-catenin promoter and PABPN1 was subjected to insertion into the pGL3 vector obtained from Promega (Madison, WI, USA) to construct pGL3-β-catenin promoter vector (pGL3-β-catenin promoter WT or pGL3-β-catenin promoter Mut), which was co-transfected along with PABPN1 or the empty vector into HEK-293T and C33A cells. After co-transfection for 2 days, cells were collected for subsequent luciferase activity analysis. The experiment was independently carried out three times.
TOP/FOP Luciferase Reporter Assay
TOP/FOP luciferase reporter assay was applied to examine the effects of miR-1323 and PABPN1 on the activity of the Wnt/β-catenin pathway in C33A cells transfected with NC inhibitor, miR-1323 inhibitor, or sh-PABPN1 + miR-1323 inhibitor. Also, the TOP/FOP reporter system was adopted to evaluate the transcriptional activity of β-catenin in C33A cells added with exosomes after being co-transfected with miR-1323 inhibitor and PABPN1 or empty vector. In brief, the cells were transfected with 1 μg of Renilla luciferase vector from Promega (Madison, WI, USA) and 1 μg of TOPFlash vector (Millipore, Bedford, MA, USA) or the NC FOPFlash (Millipore, Bedford, MA, USA). After being cultured for 24 h, the cells were harvested. The experiment was independently carried out three times.
Immunohistochemistry (IHC) Assay
First, paraformaldehyde was used to fix fresh tissues obtained from the in vivo experiment. Afterward, graded ethanol solutions were adopted to dehydrate the fixed specimens. Fixed specimens after dehydration were then inserted into the paraffin and cut into 4-μm sections, followed by dewaxing and antigen retrieval. The sections were cultivated with primary antibodies against KLHDC1, Ki-67, and PCNA at 4 °C overnight and then cultivated with horseradish peroxidase-conjugated (HRP-conjugated) secondary antibodies. The visualization of all sections was performed by Olympus BX-41 microscope (Olympus). The assay was independently carried out three times. Tissues from the mice injected with empty vector-transfected C33A cells were used as the negative control.
In Vivo Experiments
After transfection of PABPN1 or empty vector, C33A cells added with exosomes and miR-1323 antagomir were subcutaneously injected into 4-week-old nude mice. The mice were euthanized after about 1 month. Tumor size was measured every 4 days, and the tumor weight was also recorded. The study was approved by the ethics committee of Renmin Hospital of Wuhan University.
Statistical Analysis
Experimental data were analyzed by SPSS 22.0 statistical software package. All data were expressed as mean ± standard deviation (SD). The differences between two groups or more groups were analyzed with the employment of Student’s t-test or ANOVA. All the experimental results are representative of at least three independent experiments. The values of P < 0.05 were regarded to be statistically significant.
Results
miR-1323 Downregulation Dramatically Inhibits Cell Radioresistance, Proliferation, Migration, and Invasion in CCa
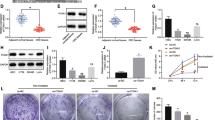
To assess the correlation between miR-1323 and cervical cancer radioresistance, we firstly detected its expression in CCa cells. In comparison with the normal cell line (End1/E6E7), we found that miR-1323 was obviously upregulated in CCa cells (HeLa, SiHa, CaSki, and C33A) (Fig. 1A). Due to its higher expression of miR-1323, we selected C33A for the following experiments. Then miR-1323 expression was knocked down in C33A cells after transfection of miR-1323 inhibitor for the subsequent loss-of-function assays (Fig. 1B). CCK-8 assay demonstrated that, compared with a miR-1323 inhibitor in the no-irradiated group, the miR-1323 inhibitor significantly inhibited cell viability in the irradiated group (Fig. 1C). At the same time, colony formation and EdU assays also proved that downregulation of miR-1323 markedly hindered cell proliferation under the condition of irradiation as evidenced by the decline of colonies and EdU-positive cells (Fig. 1D–E). Similarly, Transwell assays were carried out to observe cell migration and invasion. The experimental results indicated that upon exposure to irradiation, miR-1323 inhibitor prominently weakened cell migratory and invasive capacities (Fig. 1F–G). To sum up, miR-1323 promotes cell proliferation, migration, and invasion and enhances radioresistance in CCa.
miR-1323 downregulation dramatically inhibits cell radioresistance, proliferation, migration, and invasion in CCa. A RT-qPCR examined miR-1323 expression in CCa cell lines and normal cell line. B The expression of miR-1323 was reduced in C33A cells. C CCK-8 assay assessed the function of miR-1323 inhibitor on cell viability in the nonirradiated group and irradiated group. D–E Colony formation and EdU assays observed cell proliferation after miR-1323 was downregulated with the presence or absence of irradiation. F–G Cell migratory and invasive capacities were tested upon exposure to irradiation when miR-1323 was silenced. *P < 0.05, **P < 0.01
Characterization of Exosomes Secreted by Cancer-Associated Fibroblasts
Previous studies have shown that miRNAs have been identified in exosomes, and exosomal miRNAs due to the secretion from CAFs, NFs as well as cancer cells [20, 21]. Therefore, we analyzed the characterization of exosomes which were secreted from CAFs and NFs. Through transmission electron microscopy (TEM), the presented circular or elliptical membranous vesicle-like vesicles featuring a diameter of approximately 30–60 nm were detected in NFs and CAFs, which were in accordance with the morphological characteristics of the exosomes (Fig. 2A). The particle diameters of exosomes were about 100 nm (Fig. 2B). Western blot analyzed the protein levels of exosome markers (CD63, CD9, TSG101, HSP90, and GM130) in cell lysates with exosomes (cell group) and purified exosomes (exosome group), which confirmed the isolation of exosomes (Fig. 2C). From the results of the laser confocal microscope, we noticed the existence of exosomes in NFs and CAFs (Fig. 2D). RT-qPCR analyzed that in comparison with exosomes which were secreted by NFs, miR-1323 expression was much higher in exosomes which were derived from CAFs (Fig. 2E). A similar result could also be observed in the CAFs-exo group in C33A cells (Fig. 2F). Colony formation and EdU assays indicated that CAFs-exo contributed to cell proliferation. After the addition of GW4869, an inhibitor of exosome biogenesis/release, the proliferative capacity promoted by CAFs-exo in the nonirradiated group and irradiated group was inhibited (Fig. 2G, S1A). In the same way, the results of the Transwell assay demonstrated that after GW4869 was added, the promoted cell migration and invasion on account of CAFs-exo was suppressed (Fig. 2H–I). All in all, the abovementioned results indicated that miR-1323 is transferred by CAF-secreted exosomes.
Characterization of exosomes secreted by cancer-associated fibroblasts. A Transmission electron microscopy images of exosomes were isolated from CM-CAFs and CM-NFs. B Nanoparticle tracking analysis of NFs-exo and CAFs-exo. C The protein levels of exosomal markers (CD63, CD9, TSG101, HSP90, and GM130) in NFs-exos, CAFs-exos, and the corresponding supernatant of NFs-CM and CAFs-CM obtained through ultracentrifugation were analyzed by western blot (cell group represents cell lysates with exosomes, exosome group represents purified exosomes). D Laser scanning confocal microscope examined the internalization of exosomes by C33A cells. E Relative expression of miR-1323 in NFs, CAFs, and exosomes derived from NFs and exosomes derived from CAFs. F RT-qPCR analysis of miR-1323 expression in NFs-exo and CAFs-exo. G Cell proliferation was examined in C33A cells treated with PBS, NFs-exo, CAFs-exo, and CAFs-exo + GW4869. H–I Transwell assay detected cell migration and invasion. *P < 0.05, **P < 0.01
Exosome-Mediated Transfer of miR-1323 Promotes CCa Progression and Radioresistance
After the treatment with α-amanitin, an inhibitor of RNA transcription, we found that exosomes derived from CAFs exerted no influence on the expression of miR-1323. This result suggested that the upregulation of miR-1323 in recipient cells was induced by exosomes secreted by CAFs, as the transcription of miR-1323 was inhibited by α-amanitin (Fig. 3A). Colony formation assay testified that with the existence of irradiation, exo-miR-1323 inhibitor greatly repressed cell proliferation in C33A cells (Fig. 3B). Likewise, the experimental results of Transwell assays also attested that the migratory and invasive capacities of C33A cells were also reduced in the exo-miR-1323 inhibitor group (Fig. 3C–D). Taken together, miR-1323 transferred by exosomes promotes CCa progression and radioresistance.
Exosome-mediated transfer of miR-1323 promotes CCa progression and radioresistance. A After α-amanitin was added, miR-1323 expression was detected in the control group and CAFs-exo group. B The proliferative capacity was examined in C33A cells transfected with exo, exo-NC inhibitor and exo-miR-1323 inhibitor in the nonirradiated group and irradiated group. C–D Cell migration and invasion in exo, exo-NC inhibitor, and exo-miR-1323 inhibitor group with the presence or absence of irradiation. *P < 0.05, **P < 0.01
PABPN1 Is a Target Gene of miR-1323
Through starBase (http://starbase.sysu.edu.cn/index.php), we predicted 3 mRNAs (MARCKS, MTPN, and PABPN1) based on miRmap and PicTar (CLIP data ≥ 5, Degradome data ≥ 3). Among these 3 mRNAs, we found that MARCKS and PABPN1 were obviously downregulated in CCa cells (Fig. 4A). Ago2 RIP assay uncovered that miR-1323 had a strong affinity with PABPN1 instead of MARCKS (Fig. 4B). Next, the results of the western blot showed that PABPN1 was underexpressed at the protein level in CCa cells (Fig. S1B). Thereby, PABPN1 was chosen for the following studies. Two predicted binding sequences between miR-1323 and PABPN1 were demonstrated in Fig. 4C. Luciferase reporter assay revealed that miR-1323 inhibitor enhanced the luciferase activity in the Wt-Luc group and Mut1-Luc group, while no apparent changes were observed in the Mut2-Luc group, which further confirmed that the definite binding region between miR-1323 and PABPN1 was the second one (Fig. 4D). RNA pull-down assay further verified the interaction between miR-1323 and PABPN1 (Fig. 4E). More intriguingly, RT-qPCR and western blot analysis exposed that miR-1323 downregulation enhanced the expression of PABPN1 at mRNA and protein level (Fig. 4F–G). Taken together, miR-1232 directly targets PABPN1.
PABPN1 is a target gene of miR-1323. A Expression of MARCKS, MTPN, and PABPN1 in CCa cell lines and normal cell line. B RIP assay demonstrated the affinity between miR-1323 and MARCKS/PABPN1. C Predicted binding sequences between miR-1323 and PABPN1. D The binding between miR-1323 and PABPN1 was verified by luciferase reporter assay. E RNA pull-down assay validated the interaction between miR-1323 and PABPN1. F–G The expression of PABPN1 at mRNA level and protein level was decreased in C33A cells transfected with miR-1323 inhibitor. *P < 0.05, **P < 0.01
miR-1323 Targets PABPN1 and Activates Wnt/β-catenin Signaling Pathway
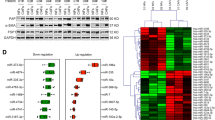
Firstly, at the mRNA and protein levels, PABPN1 expression was reduced in C33A cells (Fig. 5A, S1C). TOP/FOPFlash luciferase reporter assay was implemented to examine the activity of the Wnt/β-catenin signaling pathway. It turned out that PABPN1 knockdown partly rescued the decreased activation of the Wnt signaling pathway caused by a miR-1323 inhibitor (Fig. 5B). Western blot analysis manifested that PABPN1 upregulation lowered the protein levels of nuclear-β-catenin, while PABPN1 downregulation exhibited the opposite effect (Fig. 5C, S1D). As the activation of the Wnt/β-catenin signaling pathway features enrichment of β-catenin in the nucleus, PABPN1 overexpression suppressed the activity of this signaling pathway. The binding between PABPN1 and β-catenin promoter was excluded by luciferase reporter assay in HEK-293T (a widely adopted cell in luciferase reporter assay) and C33A cells, indicating that PABPN1 cannot directly regulate the expression of β-catenin (Fig. 5D). Through western blot analysis, we noticed that PABPN1 promoted the protein levels of GSK-3β, p-β-catenin (S33), and p-β-catenin (S37), while the protein levels of nuclear-β-catenin were observably reduced, which further verified the effect of PABPN1 on the Wnt/β-catenin signaling pathway (Fig. 5E). To further prove the effect of PABPN1, we knocked down the PABPN1 expression and found that the protein levels of GSK-3β, p-β-catenin (S33), and p-β-catenin (S37) were reduced, while the protein levels of nuclear-β-catenin were upregulated (Fig. S1E). Via western blot analysis, we then found that miR-1323 inhibitor facilitated the protein levels of GSK-3β, p-β-catenin (S33), and p-β-catenin (S37) and lowered the levels of nuclear-β-catenin (Fig. S1F). Overall, miR-1323 targets PABPN1 and regulates GSK-3β expression to affect β-catenin phosphorylation, thereby motivating the Wnt/β-catenin signaling pathway.
miR-1323 targets PABPN1 and activates the Wnt/β-catenin signaling pathway. A PABPN1 was knocked down in C33A cells. B TOP/FOP flash assay examined the activity of the Wnt/β-catenin signaling pathway. C Protein levels of nuclear-β-catenin when PABPN1 was overexpressed. D The luciferase activity of β-catenin promoter in HEK-293T and C33A cells after PABPN1 expression was enhanced. E Protein levels of GSK-3β, nuclear-β-catenin, p-β-catenin (S33), and p-β-catenin (S37) after PABPN1 was overexpressed. *P <0.05, **P < 0.01
PABPN1 Recruits IGF2BP1 and Regulates GSK3B Expression to Motivate Wnt/β-Catenin Signaling Pathway
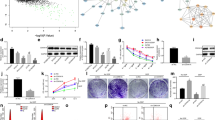
To deeply explore the relationship between PABPN1 and GSK3B (mRNA of GSK3β), we raised a hypothesis that RNA-binding protein (RBP) might play a role in their mechanism. Through the Venn diagram, we selected 5 potential RBPs (ELAVL1, CSTF2T, IGF2BP1, IGF2BP2, and DDX54) (Fig. 6A). Before the selection of certain RBP, we knocked down the expression of the 5 RBPs in C33A cells (Fig. 6B). Furthermore, we used RT-qPCR to detect the level and found that only IGF2BP1 inhibition could decrease the expression of GSK3B (Fig. 6C). Hence, IGF2BP1 was chosen. Western blot also unmasked that depletion of IGF2BP1 led to the decreased protein levels of GSK-3β (Fig. 6D). The relationship among PABPN1, IGF2BP1, and GSK3B was demonstrated by RNA pull-down and RIP assays (Fig. 6E–F). The stability of GSK3B mRNA was tested in C33A cells after the treatment of actinomycin D (Act D). The results suggested that the silencing of IGF2BP1 inhibited the stabilization of GSK3B mRNA (Fig. 6G). Collectively, IGF2BP1 protein, recruited by PABPN1, interacts with and stabilizes GSK3B mRNA.
PABPN1 recruits IGF2BP1 and regulates GSK3B expression to motivate the Wnt/β-catenin signaling pathway. A Venn diagram screened out 5 RBPs. B ELAVL1, CSTF2T, IGF2BP1, IGF2BP2, and DDX54 were all knocked down in C33A cells. C GSK3B expression after depletion of ELAVL1, CSTF2T, IGF2BP1, IGF2BP2, and DDX54. D Protein levels of GSK-3β after IGF2BP1 expression were downregulated. E RNA pull-down assay testified the affinity of IGF2BP1 and GSK3B. F RIP assay certified the relationship among PABPN1, IGF2BP1, and GSK3B. G The stability of GSK3B mRNA was shown after IGF2BP1 interference. **P < 0.01
Exosomal miR-1323 Participates in CCa Progression and Radioresistance in Vitro and in Vivo by Directly Targeting PABPN1
To determine the miR-1323/PABPN1 axis in CCa progression and radioresistance, firstly, C33A cells were treated with exo-miR-1323 inhibitor. The subsequent experiments were conducted in two groups with the absence or presence of irradiation. Then, PABPN1 expression was enhanced in C33A cells (Fig. 7A). TOP/FOPFlash luciferase reporter assay proved that PABPN1 decreased the activity of the Wnt signaling pathway, especially in the group exposed to irradiation (Fig. 7B). The experimental results of CCK8, colony formation, and EdU assays elucidated that overexpression of PABPN1 attenuated the proliferative capacity in C33A cells (Fig. 7C–E). As demonstrated in Fig. 7F–G, Transwell assays also disclosed that PABPN1 upregulation limited cell migration and invasion. Animal experiments validated that PABPN1 hindered tumor growth in vivo, and this effect became more obvious after irradiation (Fig. 7H–I). The results of the IHC assay uncovered that KLHDC1, Ki-67, and PCNA in the nonirradiated group and irradiated group were all less expressed when PABPN1 was upregulated, further proving the inhibition of proliferation (Fig. 7J). In conclusion, miR-1323 targets PABPN1 to influence CCa progression and radioresistance in vitro and in vivo.
Exosomal miR-1323 participates in CCa progression and radioresistance in vitro and in vivo by directly targeting PABPN1. A PABPN1 expression was enhanced in the nonirradiated group and irradiated group in C33A cells treated with exo-miR-1323 inhibitor. B Wnt signaling pathway activation in C33A cells treated with exo-miR-1323 inhibitor was checked after overexpression of PABPN1 in the presence or absence of irradiation. C–E) Cell proliferation in C33A cells treated with exo-miR-1323 inhibitor after PABPN1 was overexpressed with or without the exposure of irradiation. F–G Cell migration and invasion in C33A cells treated with exo-miR-1323 inhibitor with or without the exposure of irradiation. H–I Tumor weight and tumor growth were detected. J Immunohistochemistry of expression of KLHDC1, Ki-67, and PCNA. *P < 0.05, **P < 0.01
Discussion
Cervical cancer ranks the third most common cancer among women across the globe. Nowadays, radiotherapy is considered to be one of the most effective therapies for CCa treatment for that it targets tumors more precisely [22]. However, the occurrence and development of radioresistance hinder the therapeutic effect of radiotherapy. CAFs have been reported to influence the growth and radiation survival of CCa cells [23]. Meanwhile, the dysregulation of exosomal miRNAs plays an important role in CAFs [21]. miR-1323 has been determined to be associated with radiosensitivity in lung cancer cells [19]. In our study, we found that miR-1323 was upregulated in CCa cells. Moreover, miR-1323 transferred by CAF-secreted exosomes promoted cell proliferation, migration, and invasion and enhanced radioresistance in CCa cells.
It has been reported that the miRNA-mRNA regulatory network is of great importance in the improvement of CCa prognosis and treatment [24]. The activation of the Wnt/β-catenin signaling pathway has been validated to be implicated in the carcinogenesis of CCa [25]. Besides, the Wnt/β-catenin signaling pathway has been considered to be correlated with cancer radioresistance [26]. Consistent with this finding, we discovered that miR-1323 directly targeted PABPN1 and modulated PABPN1 expression to stimulate the Wnt/β-catenin signaling pathway. Rescue assays also determined the miR-1323/PABPN1 axis in CCa development and radioresistance in vitro and in vivo.
RNA binding protein (RBP) mechanism has been regarded to be important in posttranscriptional events in cancers [27]. IGF2BP1 has been defined to be a potential therapeutic target for cancers [28]. In our study, RIP, RNA pull-down, and luciferase reporter assay attested that PABPN1 recruited IGF2BP1 to modulate GSK-3β expression, thereby influencing the phosphorylation of β-catenin and the activation of the Wnt/β-catenin signaling pathway.
In brief, exosomal miR-1323, derived from CAFs, was determined to be upregulated in CCa cells and contributed to CCa progression and radioresistance. From the perspective of the mechanism, miR-1323 targets PABPN1. PABPN1 recruits IGF2BP1 and stabilizes GSK-3β mRNA so as to modulate the activation of the Wnt/β-catenin signaling pathway through β-catenin phosphorylation. Finally, rescue experiments testified that miR-1323 participates in CCa radioresistance and development via targeting PABPN1. In conclusion, exosome-mediated transfer of miR-1323 from CAFs confers radioresistance of C33A cells by targeting PABPN1 and activating the Wnt/β-catenin signaling pathway in CCa. Our study demonstrated the underlying mechanisms of radioresistance in CCa, and miR-1323 might serve as a radiosensitizer to improve the radiotherapy for CCa. However, we have to admit that this investigation has its limitations, which requires further explorations. For instance, the effect of miR-1323 on cell apoptosis in CCa has not been explored. Upstream mechanisms of miR-1323 existing in CCa also remain obscure. In order to ensure the preciseness and reliability of the experimental data, we only conducted animal experiments concerning C33A-exo-miR-1323 inhibitor cells and PABPN1. Extra animal experiments should be performed to evaluate the effects of miR-1323 in vivo. Furthermore, clinicopathological analyses should be utilized to further prove the association between miR-1323 and the radioresistance of CCa. In the future, we will conduct the studies to further explore the mechanisms and clinical value of miR-1323 in CCa.
Data Availability
Research data are not shared.
References
Tsikouras P, Zervoudis S, Manav B, Tomara E, Iatrakis G, Romanidis C, et al. Cervical cancer: screening, diagnosis and staging. Journal of BUON : official journal of the Balkan Union of Oncology. 2016;21(2):320–5.
Petry KU. HPV and cervical cancer. Scandinavian journal of clinical and laboratory investigation Supplementum. 2014;244:59-62; discussion. 2014;10(3109/00365513):936683.
Willmott LJ, Monk BJ. Cervical cancer therapy: current, future and anti-angiogensis targeted treatment. Expert review of anticancer therapy. 2009;9(7):895–903. https://doi.org/10.1586/era.09.58.
Powell ME. Modern radiotherapy and cervical cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2010;20(11 Suppl 2):S49–51. https://doi.org/10.1111/igc.0b013e3181f7b241.
Huang C, Lu H, Li J, Xie X, Fan L, Wang D, et al. SOX2 regulates radioresistance in cervical cancer via the hedgehog signaling pathway. Gynecologic oncology. 2018;151(3):533–41. https://doi.org/10.1016/j.ygyno.2018.10.005.
Li Q, Zhang Y, Jiang Q. SETD3 reduces KLC4 expression to improve the sensitization of cervical cancer cell to radiotherapy. Biochemical and biophysical research communications. 2019;516(3):619–25. https://doi.org/10.1016/j.bbrc.2019.06.058.
Bandyopadhyay A, Mukherjee U, Ghosh S, Ghosh S, Sarkar SK. Pattern of failure with locally advanced cervical cancer– a retrospective audit and analysis of contributory factors. Asian Pacific Journal of Cancer Prevention : APJCP. 2018;19(1):73–9. https://doi.org/10.22034/apjcp.2018.19.1.73.
Sun FD, Wang PC, Luan RL, Zou SH, Du X. MicroRNA-574 enhances doxorubicin resistance through down-regulating SMAD4 in breast cancer cells. European Review for Medical and Pharmacological Sciences. 2018;22(5):1342–50. https://doi.org/10.26355/eurrev_201803_14476.
Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, Borran S, Pourhanifeh MH, Moghoofei M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. Journal of cellular physiology. 2019;234(10):17064–99. https://doi.org/10.1002/jcp.28457.
Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical cancer markers: epigenetics and microRNAs. Laboratory medicine. 2018;49(2):97–111. https://doi.org/10.1093/labmed/lmx080.
Wang L, Zhao Y, Xiong W, Ye W, Zhao W, Hua Y. MicroRNA-449a Is downregulated in cervical cancer and inhibits proliferation, migration, and invasion. Oncology research and treatment. 2019;42(11):564–71. https://doi.org/10.1159/000502122.
Hua FF, Liu SS, Zhu LH, Wang YH, Liang X, Ma N, et al. MiRNA-338-3p regulates cervical cancer cells proliferation by targeting MACC1 through MAPK signaling pathway. European Review for Medical and Pharmacological Sciences. 2017;21(23):5342–52. https://doi.org/10.26355/eurrev_201712_13919.
Pedroza-Torres A, López-Urrutia E, García-Castillo V, Jacobo-Herrera N, Herrera LA, Peralta-Zaragoza O, et al. MicroRNAs in cervical cancer: evidences for a miRNA profile deregulated by HPV and its impact on radio-resistance. Molecules (Basel, Switzerland). 2014;19(5):6263–81. https://doi.org/10.3390/molecules19056263.
Zhang Y, Li P, Hu J, Zhao LN, Li JP, Ma R, et al. Role and mechanism of miR-4778-3p and its targets NR2C2 and Med19 in cervical cancer radioresistance. Biochemical and biophysical research communications. 2019;508(1):210–6. https://doi.org/10.1016/j.bbrc.2018.11.110.
Wei YQ, Jiao XL, Zhang SY, Xu Y, Li S, Kong BH. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. European Review for Medical and Pharmacological Sciences. 2019;23(17):7314–26. https://doi.org/10.26355/eurrev_201909_18837.
Zhang F, Yang C, Xing Z, Liu P, Zhang B, Ma X, et al. LncRNA GAS5-mediated miR-1323 promotes tumor progression by targeting TP53INP1 in hepatocellular carcinoma. OncoTargets and therapy. 2019;12:4013–23. https://doi.org/10.2147/ott.S209439.
Zhao H, Zheng C, Wang Y, Hou K, Yang X, Cheng Y, et al. miR-1323 Promotes cell migration in lung adenocarcinoma by targeting Cbl-b and is an early prognostic biomarker. Frontiers in oncology. 2020;10:181. https://doi.org/10.3389/fonc.2020.00181.
Xu Y, Liu M. MicroRNA-1323 downregulation promotes migration and invasion of breast cancer cells by targeting tumour protein D52. Journal of biochemistry. 2020;168(1):83–91. https://doi.org/10.1093/jb/mvaa035.
Li Y, Han W, Ni TT, Lu L, Huang M, Zhang Y, et al. Knockdown of microRNA-1323 restores sensitivity to radiation by suppression of PRKDC activity in radiation-resistant lung cancer cells. Oncology reports. 2015;33(6):2821–8. https://doi.org/10.3892/or.2015.3884.
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. Journal of medical genetics. 2015;52(10):710–8. https://doi.org/10.1136/jmedgenet-2015-103334.
Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Molecular cancer. 2017;16(1):148. https://doi.org/10.1186/s12943-017-0718-4.
Walsh L, Morgia M, Fyles A, Milosevic M. Technological advances in radiotherapy for cervical cancer. Current opinion in oncology. 2011;23(5):512–8. https://doi.org/10.1097/CCO.0b013e3283499d93.
Chu TY, Yang JT, Huang TH, Liu HW. Crosstalk with cancer-associated fibroblasts increases the growth and radiation survival of cervical cancer cells. Radiation research. 2014;181(5):540–7. https://doi.org/10.1667/rr13583.1.
Mei Y, Jiang P, Shen N, Fu S, Zhang J. Identification of miRNA-mRNA regulatory network and construction of prognostic signature in cervical cancer. DNA and cell biology. 2020;39(6):1023–40. https://doi.org/10.1089/dna.2020.5452.
Bahrami A, Hasanzadeh M, ShahidSales S, Yousefi Z, Kadkhodayan S, Farazestanian M, et al. Clinical significance and prognosis value of Wnt signaling pathway in cervical cancer. Journal of cellular biochemistry. 2017;118(10):3028–33. https://doi.org/10.1002/jcb.25992.
Yang Y, Zhou H, Zhang G, Xue X. Targeting the canonical Wnt/β-catenin pathway in cancer radioresistance: updates on the molecular mechanisms. Journal of cancer research and therapeutics. 2019;15(2):272–7. https://doi.org/10.4103/jcrt.JCRT_421_18.
Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. trends in cancer. 2017;3(7):506–28. https://doi.org/10.1016/j.trecan.2017.05.003.
Huang X, Zhang H, Guo X, Zhu Z, Cai H, Kong X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. Journal of hematology & oncology. 2018;11(1):88. https://doi.org/10.1186/s13045-018-0628-y.
Acknowledgements
Contributions from all participants are appreciated.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
The study was approved by the ethics committee of Renmin Hospital of Wuhan University.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Figure S1
(A) EdU assay detected the proliferation of C33A cells treated with PBS, NFs-exo, CAFs-exo and CAFs-exo+GW4869. (B) Western blot was conducted to examine PABPN1 protein level in CCa cell lines and normal cell line. (C) The protein level of PABPN1 was knocked down in C33A cells. (D) Protein levels of nuclear-β-catenin when PABPN1 was depleted. (E-F) Protein levels of GSK-3β, nuclear-β-catenin, p-β-catenin (S33) and p-β-catenin (S37) after PABPN1 or miR-1323 was inhibited. *P <0.05, **P < 0.01 (PNG 1176 kb)
Rights and permissions
About this article
Cite this article
Fang, F., Guo, C., Zheng, W. et al. Exosome-Mediated Transfer of miR-1323 from Cancer-Associated Fibroblasts Confers Radioresistance of C33A Cells by Targeting PABPN1 and Activating Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Reprod. Sci. 29, 1809–1821 (2022). https://doi.org/10.1007/s43032-021-00820-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-021-00820-y