Abstract
Endometriosis, as a prevalent gynecological disease, is characterized by the presence of endometrial-like tissue outside the uterus, causing infertility and considerable pain and affecting the quality of life of women. The pathogenic mechanism has not been fully elucidated, and there are no effective biomarkers for endometriosis. In our study, microRNA (miRNA) expression profiling of 10 ectopic endometrial plasma from patients with ovarian endometriosis and 10 normal plasma from healthy controls was analyzed using a microarray. As a result, 114 differentially expressed miRNAs were identified. Among them, 14 miRNAs were significantly downregulated in patients with ovarian endometriosis, which matched the microarray results. The diagnostic value of the 14 downregulated miRNAs in ovarian endometriosis was evaluated by receiver operating characteristic (ROC) curve analysis, and hsa-let-7i-5p showed the highest area under the ROC curve (AUC) with a value of 0.900. The target genes of the 14 miRNAs were predicted by miRWalk2.0, and the genes that were targeted by at least 2 of the 14 miRNAs were analyzed by function enrichment. The target genes were significantly enriched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, such as microRNAs in cancer, bladder cancer, and endocrine resistance pathways, and the Gene Ontology (GO) terms such as nucleobase-containing compound metabolic process, cellular nitrogen compound biosynthetic process, and heterocycle metabolic process. The identified 14 differentially expressed miRNAs could be potential biomarkers and therapeutic targets for the diagnosis and treatment of endometriosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endometriosis is characterized by the presence of endometrial-like tissue, such as endometrial glands and stroma tissue outside the uterus, inducing an inflammatory reaction along with adhesions, fibrosis, scarring, and anatomical distortion [1, 2]. It mainly consists of three types: ovarian endometriosis, peritoneal endometriosis, and deep infiltrating endometriosis [3]. The incidence rate of endometriosis is 6–10% among women of reproductive age [4]. In the USA, endometriosis is the third major cause of gynecologic hospitalization [2]. Approximately 40% of women with endometriosis are infertile [5]. Endometriosis often appears as lesions or cysts at several parts of the body, such as pelvic organs, peritoneum and lung, which affect the physical and mental wellbeing and quality of life of women [6, 7].
Currently, the primary diagnostic approaches for endometriosis, such as laparoscopy, ultrasound, magnetic resonance imaging (MRI), and serum markers, have some limitations in terms of their accuracy and specificity [8]. Therapeutic strategies, which mainly consist of medical therapy and surgical treatment, have been proved to be non-cytoreductive with a high recurrence rate [9,10,11]. However, the underlying molecular mechanism and etiology of endometriosis remain unknown, and no theory has interpreted its origin and progression comprehensively. Therefore, the introduction and investigation of novel specific biomarkers will facilitate the early diagnosis, effective management, and successful treatment of endometriosis.
MicroRNAs (miRNAs), as single-stranded noncoding RNA molecules, are about 22 nucleotides in length and may be associated with endometriosis [12, 13]. They have attracted much attention for their diagnostic value in endometriosis [14]. Aberrant expressions of miR-135a and miR-135b were involved in endometriosis by regulating HOXA10 expression and inhibiting implantation-related genes [15]. Elevated miR-210 expression contributed to endometriosis pathogenesis by targeting activator of transcription 3 and signal transducer [16]. It was revealed that miR-126 was related to the progression of endometriosis by modulating the expression of Crk [17]. MiR-191 was shown to be associated with the ovarian endometriosis response and its malignant transformation [18]. Although numerous miRNAs have been proved to be related to endometriosis, the lack of clinically effective miRNAs makes a precise diagnosis of endometriosis a significant challenge.
In this study, we analyzed the miRNA expressions in patients with ovarian endometriosis and healthy controls with microarray analysis to identify differentially expressed miRNAs. The differentially expressed miRNAs were further validated in additional patients with ovarian endometriosis and healthy controls, and the diagnostic value of these miRNAs was further assessed. The relationships between the miRNAs and target genes were analyzed to screen for significant miRNAs that might be associated with the etiology and progression of endometriosis. Our research provides potential biomarkers for the diagnosis and treatment of endometriosis.
Materials and Methods
Study Population
Written informed consent forms were obtained from all participants prior to surgery, and our study was approved by the Ethics Committee of the Chinese PLA General Hospital. Sixty participants were recruited for the study, including the experimental group (n = 29) and the control group (n = 31). Blood samples from ectopic endometrial patients were collected in the morning (~5:30–6:00 a.m.) in the scheduled preoperative examination. For paired healthy control, the peripheral blood samples were collected into EDTA collection tubes in the morning (~5:30–6:00 a.m.). After collection, the plasma samples were processed within 1 hour and stored at –80 °C for further use.
The inclusion criteria for the experimental group were as follows: women of childbearing age with regular menstrual cycles, patients who were diagnosed with ovarian endometriosis by laparoscopy and had their samples collected at the secretory phase, and patients who were diagnosed with ovarian endometriosis according to pathology reports after surgery. Patients who had taken hormonal drugs within 6 months prior to their surgery or had malignancy or other diseases such as endometrial hyperplasia, submucous myoma, adenomyosis, or chronic pelvic inflammation were excluded from the study.
RNA Isolation and miRNA Microarray Analysis
Total RNA was extracted from plasma using the mirVana™ RNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). The RNA quantity and integrity were determined by the NanoDrop ND-2000 (Thermo Fisher Scientific, Wilmington, DE, USA) and Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA) respectively.
The miRNA expressions were analyzed by Agilent Human miRNA microarray Release 21.0. Feature Extraction software (version10.7.1.1, Agilent Technology, Santa Clara, CA, USA) was used to process the acquired images. Genespring software (version13.1, Agilent Technology, Santa Clara, CA, USA) was used for quantile normalization and processing of the subsequent data.
Analysis of the Differentially Expressed miRNAs
The data were normalized using the RMA algorithm. The Limma package was used to identify the differentially expressed miRNAs between patients with ovarian endometriosis and the healthy controls with |log2FC| >1 (FC, fold change) and P < 0.05 as the thresholds.
Prediction of the miRNAs Target Genes
The target genes of the miRNAs were analyzed by miRWalk2.0. The experimentally validated miRNA-target interaction database was used. The genes that were targeted by at least three of the differentially expressed miRNAs were retained for further analysis.
Function Enrichment Analysis
Genes that were targeted by at least two of the differentially expressed miRNAs were screened for function enrichment analysis with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). With a P value of < 0.05 as the threshold, the significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms were selected.
Real-time Quantitative Polymerase Chain Reaction
Total RNA was isolated from plasma using Trizol reagent. Then, RNA was reverse transcribed into cDNA using the TIANGEN® miRcute Kit (kr211, TIANGEN Biotech Co., Ltd., Beijing, China). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed according to the instructions of the TIANGEN® miRcute Plus miRNA qPCR Kit (FP411, TIANGEN Biotech Co., Ltd., Beijing, China) on the StepOnePlus™ real-time PCR system (Applied Biosystems, Foster City, CA, USA). The experimental data were analyzed using the 2-ΔΔCt method. The t test was used to compare the difference in expression levels between the experimental and control groups, and P < 0.001 was considered statistically significant.
Receiver Operating Characteristic Analysis
The receiver operating characteristic (ROC) curves were plotted using package Epi (https://cran.r-project.org/web/packages/Epi/index.html) to evaluate the diagnostic value of the miRNAs. The area under the ROC curve (AUC) was considered the diagnostic index for distinguishing ovarian endometriosis patients from healthy controls. An AUC value of 0.5 reflected classifications assigned randomly.
Results
Differentially Expressed miRNAs in Ovarian Endometriosis
The miRNA microarray was performed on ovarian plasma with endometriosis and healthy control groups to identify the differentially expressed miRNAs between them. The miRNA expressions in the two groups are shown in Fig. 1. Compared with the control group, 69 miRNAs were significantly downregulated and 45 miRNAs were significantly upregulated in patients with ovarian endometriosis (Fig. 2).
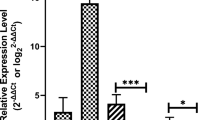
To further validate the change in the miRNA expression in the microarray analysis, the expression values of 20 miRNAs, which were selected based on previous studies and their fold changes, were determined by RT-qPCR in additional patients with ovarian endometriosis (n = 19) and healthy controls (n = 21). As shown in Fig. 3, 14 miRNAs (hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-let-7g-5p, hsa-let-7i-5p, hsa-miR-199a-3p, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-328-3p, hsa-miR-331-3p, hsa-miR-320e) matched the results of the microarray and were clearly downregulated in patients with ovarian endometriosis compared with the healthy controls.
Evaluation of the Diagnostic Value of the 14 miRNAs in Ovarian Endometriosis
To assess the diagnostic value of the 14 downregulated miRNAs that were validated by RT-qPCR, ROC curve analysis was performed. Their AUC values are shown in Fig. 4. It was revealed that miRNA hsa-let-7i-5p had the highest AUC of 0.900, and the other miRNAs also had relatively high AUC values (hsa-let-7a-5p, 0.884; hsa-let-7b-5p, 0.868; hsa-let-7d-5p, 0.869; hsa-let-7f-5p, 0.856; hsa-let-7g-5p, 0.894; hsa-miR-199a-3p, 0.841; hsa-miR-320a, 0.845; hsa-miR-320b, 0.853; hsa-miR-320c, 0.857; hsa-miR-320d, 0.832; hsa-miR-328-3p, 0.891; hsa-miR-331-3p, 0.870; hsa-miR-320e, 0.823).
Receiver operating characteristic (ROC) curve analysis of miRNAs hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-let-7g-5p, hsa-let-7i-5p, hsa-miR-199a-3p, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-328-3p, hsa-miR-331-3p, and hsa-miR-320e. The miRNA hsa-let-7i-5p presented the highest area under the ROC curve (AUC) of 0.900
hsa-miR-320a, hsa-miR-320d, hsa-let-7b-5p, and hsa-let-7a-5p Had Multiple Target Genes
The miRNA-target gene regulatory network is shown in Fig. 5. To simplify the network, only the genes that were targeted by at least three of the differentially expressed miRNAs were retained. miRNAs hsa-miR-320a, hsa-miR-320d, hsa-let-7b-5p, and hsa-let-7a-5p could target several genes simultaneously and may be significant biomarkers in ovarian endometriosis. There was no target gene for hsa-miR-320e, and it was not shown in this figure. Genes ANKRD52, AGO1, QKI, CCND2, and SP1 were targeted by several miRNAs simultaneously.
Network diagram between miRNAs and corresponding target genes. The triangles represented miRNAs, and the dots represented target genes. Only the genes that were targeted by at least three of the differentially expressed miRNAs were shown in this figure. There was no target gene for hsa-miR-320e, and it was not shown
Target Genes Were Significantly Enriched in MicroRNAs in the Cancer Pathway and Nucleobase-Containing Compound Metabolic Process
Genes that were targeted by at least two of the 14 miRNAs were selected for functional enrichment analysis. The top 30 significantly enriched KEGG pathways and GO terms are shown in Fig. 6. It revealed that the target genes were significantly enriched in KEGG pathways such as microRNAs in cancer, bladder cancer, and endocrine resistance pathways and GO terms such as the nucleobase-containing compound metabolic process, cellular nitrogen compound biosynthetic process, and heterocycle metabolic process.
Function enrichment of the genes that were targeted by at least two of the differentially expressed miRNAs. These target genes were significantly enriched in the KEGG pathways such as MicroRNAs in cancer, Bladder cancer, Endocrine resistance pathways (a) and the GO terms such as nucleobase-containing compound metabolic process, cellular nitrogen compound biosynthetic process, and heterocycle metabolic process (b). KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology
Discussion
As an estrogen-dependent disease, endometriosis often presents in the peritoneal cavity and ovary, leading to dysmenorrhea, pelvic pain, and infertility [15, 19]. In our study, by microarray analysis, the miRNA expression profiling of 10 ectopic endometrial plasma and 10 normal plasma was done. There were 114 differentially expressed miRNAs between the ectopic endometrial plasma and normal controls, among which 69 miRNAs were significantly downregulated, and 45 miRNAs were significantly upregulated. According to previous studies and the fold changes, 20 differentially expressed miRNAs were selected for validation by RT-qPCR in additional patients with ovarian endometriosis and healthy controls. Among them, 14 miRNAs (hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-let-7g-5p, hsa-let-7i-5p, hsa-miR-199a-3p, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-328-3p, hsa-miR-331-3p, and hsa-miR-320e) matched the microarray analysis and were downregulated in patients with ovarian endometriosis.
Hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7d-5p, and hsa-let-7f-5p have been proved to be downregulated in humans with endometriosis [20, 21]. There have not been reports on the involvement of the other 10 miRNAs in endometriosis. To our knowledge, this is the first report on the dysregulation of the 10 miRNAs (hsa-let-7g-5p, hsa-let-7i-5p, hsa-miR-199a-3p, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-328-3p, hsa-miR-331-3p, and hsa-miR-320e) in endometriosis. To evaluate the diagnostic value of the 14 downregulated miRNAs, we conducted an ROC curve analysis and found that hsa-let-7i-5p had the highest AUC value of 0.900 among the 14 miRNAs. The result indicates that hsa-let-7i-5p is a potential diagnostic biomarker for ovarian endometriosis. The let-7 family, the first discovered miRNAs in nematode Caenorhabditis elegans and humans, has a conserved sequence and functions, and their dysregulation is associated with cell differentiation and cell-related diseases [22]. The let-7 family can target KRAS and the polymorphism of the let-7 binding site in KRAS, which is involved in the pathogenesis of endometriosis [23]. As a member of the let-7 family, aberrant hsa-let-7i-5p expression has been shown to participate in multiple biological processes, such as decidua, brite adipocyte function, maintenance of cancer stem cells in cervical cancer, and colon cancer development [24,25,26,27]. However, the effects of hsa-let-7i-5p in ovarian endometriosis have not been studied. Our research showed its downregulation as well as the diagnostic value in ovarian endometriosis.
The network diagram of the miRNA-gene interaction revealed that hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-320a, and hsa-miR-320d had multiple target genes and might have a hub role in the regulation of endometriosis. Besides, they had relatively high AUC values (0.884, 0.868, 0.845, and 0.832, respectively), indicating that the 4 miRNAs were also potential diagnostic biomarkers for ovarian endometriosis. It was noted that the downregulation of hsa-let-7a-5p and hsa-let-7b-5p should be involved in complicated regulations and several organs in endometriosis [20]. Inflammation is one of the main symptoms of endometriosis [28]. It was reported that in the serum of patients with endometriosis, hsa-let-7b-5p expression was decreased, and a negative correlation between hsa-let-7b-5p and TNF-α was observed. However, hsa-let-7b-5p might contribute to endometriosis by regulating the inflammatory response and inducing the production of macrophage inflammatory cytokines [29]. Furthermore, the let-7 family is associated with the migration of endometrial cells in endometriosis by targeting aromatase [30]. Therefore, we hypothesized that hsa-let-7a-5p and hsa-let-7b-5p might affect the pathogenesis and progression of endometriosis by targeting multiple pathways and genes.
Downregulation of another hub miRNA, hsa-miR-320a, in the miRNA-gene interaction was shown to lead to overexpression of pro-inflammatory cytokines by promoting COX-2 expression, which was regulated by ERK/NF-κB pathways [31]. In patients with inflammatory bowel disease, hsa-miR-320a modulated NOD2 expression, which was induced by inflammation [32]. Besides, it was estimated that hsa-miR-320a could influence approximately 145 pathways and 1500 target genes [33, 34]. hsa-miR-320d is a cancer suppressor. Downregulated expression of hsa-miR-320d occurs in breast cancer, and its overexpression can suppress the proliferation, migration, and invasion of cancer cells [35]. The decreased expression of hsa-miR-320d was also observed in aortic dissection, which would affect staurosporine and tumor necrosis factor-α-induced apoptosis and the expressions of multiple genes [36]. Herein, we speculated that hsa-miR-320a and hsa-miR-320d might be involved in endometriosis by regulating the production of inflammatory cytokines or influencing endometrial cells through several pathways and target genes.
In conclusion, we identified 14 miRNAs that might be involved in the pathogenesis of ovarian endometriosis. Among them, 10 miRNAs were reported for the first time to be associated with endometriosis. hsa-let-7i-5p and hub miRNAs including hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-320a, and hsa-miR-320d might be potential diagnostic biomarkers. Our study provides potential targets for the specific diagnosis and effective treatment of endometriosis.
References
Leyland N, Casper R, Laberge P, Singh SS, SOGC. Endometriosis: diagnosis and management. J Obstet Gynaecol Can. 2010;32:S1–32.
McLeod BS, Retzloff MG. Epidemiology of endometriosis: an assessment of risk factors. Clin Obstet Gynecol. 2010;53:389–96.
Basta A, Brucka A, Górski J, et al. The statement of Polish Society’s Experts Group concerning diagnostics and methods of endometriosis treatment. Ginekol Pol. 2012;83:871–6.
Giudice LC. Endometriosis. N Engl J Med. 2010;362:2389–98.
Carvalho LF, Rossener R, Azeem A, Malvezzi H, Simões Abrão M, Agarwal A. From conception to birth - how endometriosis affects the development of each stage of reproductive life. Minerva Ginecol. 2013;65:181–98.
Kennedy S, Bergqvist A, Chapron C, et al. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod. 2005;20:2698–704.
De Graaff AA, D'Hooghe TM, Dunselman GA, et al. The significant effect of endometriosis on physical, mental and social wellbeing: results from an international cross-sectional survey. Hum Reprod. 2013;28:2677–85.
Hsu AL, Khachikyan I, Stratton P. Invasive and noninvasive methods for the diagnosis of endometriosis. Clin Obstet Gynecol. 2010;53:413–9.
Valle RF, Sciarra JJ. Endometriosis: treatment strategies. Ann N Y Acad Sci. 2003;997:229–39.
Kennedy S, Bergqvist A, Chapron C, D’Hooghe T, Dunselman G, Greb R, et al. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod. 2005;20:2698–704.
Vercellini P, Viganò P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10:261–75.
Du T, Zamore PD. Micro Primer: the biogenesis and function of microRNA. Development. 2005;132:4645–52.
Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, et al. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011;25:821–32.
Braicu OL, Budisan L, Buiga R, et al. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225–38.
Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96:E1925–33.
Okamoto M, Nasu K, Abe W, Aoyagi Y, Kawano Y, Kai K, et al. Enhanced miR-210 expression promotes the pathogenesis of endometriosis through activation of signal transducer and activator of transcription 3. Hum Reprod. 2015;30:632–41.
Liu S, Gao S, Wang XY, Wang DB. Expression of miR-126 and Crk in endometriosis: miR-126 may affect the progression of endometriosis by regulating Crk expression. Arch Gynecol Obstet. 2012;285(4):1065–72.
Tian X, Xu L, Wang P. MiR-191 inhibits TNF-α induced apoptosis of ovarian endometriosis and endometrioid carcinoma cells by targeting DAPK1. Int J Clin Exp Pathol. 2015;8(5):4933–42.
Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–79.
Seifer BJ, Su D, Taylor HS. Circulating miRNAs in murine experimental endometriosis. Reprod Sci. 2017;24(3):376–81.
Cho S, Mutlu L, Grechukhina O, Taylor HS. Circulating microRNAs as potential biomarkers for endometriosis. Fertil Steril. 2015;103(5):1252–60.
Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16.
Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012;4(3):206–17.
Lv Y, Gao S, Zhang Y, Wang L, Chen X, Wang Y. miRNA and target gene expression in menstrual endometria and early pregnancy decidua. Eur J Obstet Gynecol Reprod Biol. 2016;197:27–30.
Giroud M, Karbiener M, Pisani DF, et al. Let-7i-5p represses brite adipocyte function in mice and humans. Sci Rep. 2016;6:28613.
Chhabra R. let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis coordinate to maintain cancer stem cell population in cervical cancer. Sci Rep. 2018;8(1):7840.
Song J, Wang L, Ma Q, Yang Y, Yang Z, Wang B, et al. Let-7i-5p inhibits the proliferation and metastasis of colon cancer cells by targeting kallikrein-related peptidase 6. Oncol Rep. 2018;40(3):1459–66.
Morotti M, Vincent K, Becker CM. Mechanisms of pain in endometriosis. Eur J Obstet Gynecol Reprod Biol. 2017;209:8–13.
Nematian SE, Mamillapalli R, Kadakia TS, Majidi Zolbin M, Moustafa S, Taylor HS. Systemic inflammation induced by microRNAs: endometriosis-derived alterations in circulating microRNA 125b-5p and Let-7b-5p regulate macrophage cytokine production. J Clin Endocrinol Metab. 2018;103(1):64–74.
Cho S, Mutlu L, Zhou Y, Taylor HS. Aromatase inhibitor regulates let-7 expression and let-7f-induced cell migration in endometrial cells from women with endometriosis. Fertil Steril. 2016;106(3):673–80.
Cheng Z, Qiu S, Jiang L, Zhang A, Bao W, Liu P, et al. MiR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J Clin Immunol. 2013;33(3):567–76.
Stronati L, Pierdomenico M, Cesi V, et al. MIR320 family regulates NOD2/CARD15: a new mechanism for controlling inflammation? Dig Liver Dis. 2014;46:e82.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Shi S, Hu X, Xu J, et al. MiR-320d suppresses the progression of breast cancer via LncRNA HNF1A-AS1 regulation and SOX4 inhibition. RSC Adv. 2018;8(34):19196–207.
Gokul S, Sivachitra M, Vijayachitra S. Parkinson’s disease prediction using machine learning approaches. Fifth International Conference on Advanced Computing (ICoAC). IEEE. 2013; 246–252.
Shen H, Lu S, Dong L, Xue Y, Yao C, Tong C, et al. hsa-miR-320d and hsa-miR-582, miRNA biomarkers of aortic dissection, regulate apoptosis of vascular smooth muscle cells. J Cardiovasc Pharmacol. 2018;71(5):275–82.
Funding
Grants from National Natural Science Foundation of China: High-throughput sequencing identifying pathogenic genes of endometriosis (Grant No. 81571411).
Author information
Authors and Affiliations
Contributions
Chenglei Gu and Zhe Zhang contributed to the conception of the study; Wensheng Fan and Li Lian performed the experiments; Mingxia Ye, Qian Zhang, and Nina Zhang helped for acquisition of data and analysis and interpretation of data; Yuanguang Meng has been involved in drafting the manuscript or revising it critically for important intellectual content; Zhen Li provided valuable instructions and suggestions for this paper and helped for revising the manuscript; all authors have read and approved the final version of the manuscript for publication.
Corresponding author
Ethics declarations
Written informed consent forms were obtained from all participants prior to surgery, and our study was approved by the Ethics Committee of the Chinese PLA General Hospital.
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Highlights
•Microarray analysis identified 114 DEmiRNAs in endometriosis patients.
•Fourteen miRNAs were validated to be downregulated by RT-qPCR.
•The 14 DEmiRNAs may be potential biomarkers for endometriosis.
Rights and permissions
About this article
Cite this article
Gu, Cl., Zhang, Z., Fan, Ws. et al. Identification of MicroRNAs as Potential Biomarkers in Ovarian Endometriosis. Reprod. Sci. 27, 1715–1723 (2020). https://doi.org/10.1007/s43032-020-00148-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-020-00148-z