Abstract
Plant isoprenoids (also known as terpenes or terpenoids) are a wide family of primary and secondary metabolites with multiple functions. In particular, most photosynthesis-related isoprenoids (including carotenoids and chlorophylls) as well as diterpenes and polyterpenes derive from geranylgeranyl diphosphate (GGPP) produced by GGPP synthase (GGPPS) enzymes in several cell compartments. Plant genomes typically harbor multiple copies of differentially expressed genes encoding GGPPS-like proteins. While sequence comparisons allow to identify potential GGPPS candidates, experimental evidence is required to ascertain their enzymatic activity and biological function. Actually, functional analyses of the full set of potential GGPPS paralogs are only available for a handful of plant species. Here we review our current knowledge on the GGPPS families of the model plant Arabidopsis thaliana and the crop species rice (Oryza sativa), pepper (Capsicum annuum) and tomato (Solanum lycopersicum). The results indicate that a major determinant of the biological role of particular GGPPS paralogs is the expression profile of the corresponding genes even though specific interactions with other proteins (including GGPP-consuming enzymes) might also contribute to subfunctionalization. In some species, however, a single GGPPS isoforms appears to be responsible for the production of most if not all GGPP required for cell functions. Deciphering the mechanisms regulating GGPPS activity in particular cell compartments, tissues, organs and plant species will be very useful for future metabolic engineering approaches aimed to manipulate the accumulation of particular GGPP-derived products of interest without negatively impacting the levels of other isoprenoids required to sustain essential cell functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are eukaryotic organisms with an astounding metabolic diversity mostly derived from their capacity to produce secondary (i.e., non-essential, specialized) metabolites that modulate their environmental interactions. Among the different groups of plant metabolites, isoprenoids (also known as terpenes or terpenoids) are remarkable for their structural and functional variety (Pulido et al. 2012; Tholl 2015). Some plant isoprenoids are present in all plant species and they are considered as primary or essential metabolites due to their irreplaceable functions in vital processes, such as photosynthesis (e.g., carotenoids, chlorophylls, phylloquinones or plastoquinone), respiration (e.g., ubiquinone), membrane dynamics (sterols) or developmental control (e.g., hormones, such as cytokinins, brassinosteroids, gibberellins, abscisic acid and strigolactones). The vast majority of plant isoprenoids, however, are secondary metabolites that are typically confined to particular plant species and/or organs and/or produced in response to environmental challenges. They include monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), sesterterpenes (C25), triterpenes (C30), tetraterpenes (C40) and polyterpenes (> C45) with roles as volatiles, pigments and defense-related molecules in plants and applications as flavors, colorants, drugs, polymers, biofuels or nutraceuticals for the industry.
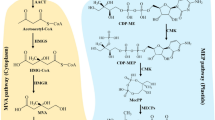
Despite the stunning diversity of chemical structures of isoprenoid end-products, they all derive from the same 5-carbon (C5) building blocks, namely, isopentenyl diphosphate (IPP) and its double-bond isomer dimethylallyl diphosphate (DMAPP). The biosynthesis of these two universal precursors in plant cells involves two independent pathways (Fig. 1), the mevalonic acid (MVA) pathway in the cytosol and the methylerythritol 4-phosphate (MEP) pathway in plastids (Pulido et al. 2012; Rodriguez-Concepcion and Boronat 2015; Tholl 2015). The MVA pathway provides most C5 precursors for the production of sesquiterpenes and triterpenes (including sterols and brassinosteroids), whereas most monoterpenes and tetraterpenes (including chlorophylls, carotenoids and derived hormones, such as abscisic acid and strigolactones) derive from the MEP pathway. Diterpenes, sesterterpenes and polyterpenes (including ubiquinone and plastoquinone) as well as cytokinins can be formed from precursors supplied by the two pathways, depending on their subcellular production site (Fig. 1). Although IPP and DMAPP can move between cytosol and plastid compartments, the exchange rate under normal conditions is not high enough to rescue the lethal genetic or pharmacological blockage of one of the two pathways, thus explaining their coexistence in plants. It is believed that the tight and often antagonist regulation of both pathways at transcriptional, post-transcriptional and post-translational level finely modulates the flux of precursors towards the production of specific groups of final isoprenoid products (Rodriguez-Concepcion and Boronat 2015).
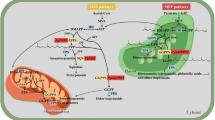
Subcellular compartmentation of isoprenoid biosynthetic pathways and GGPPS-like proteins. Dotted arrows represent multiple steps and open arrowheads represent transport. Hormones are abbreviated in italics: CKs cytokinins, BRs brassinosteroids, GAs gibberellins, SLs strigolactones, ABA abscisic acid. Colors and first two letters of the proteins identify the species: blue for Arabidopsis (At), green for rice (Os), orange for pepper (Ca), and red for tomato (Sl). The truncated form of AtGGPPS11 lacking the N-terminal plastid-targeting peptide is represented with a blunt end. Subcellular localization of the proteins is based on the analysis of GFP-tagged versions. AtGGPPS1, At1g49530; AtGGPPS2, At2g18620; AtGGPPS3, At2g18640; AtGGPPS4, At2g23800; AtGGPPS11, At4g36810; CaGGPPS1, Capana04g000412; CaPTP1, Capana05g000800; CaPTP2, Capana00g002450; CaPTP3, Capana00g002451; CaPTP4, Capana00g002452; OsGGPPS1, Os07g39270; OsGPPS, Os01g14630; SlGGPPS1, Solyc11g011240; SlGGPPS2, Solyc04g079960; SlGGPPS3, Solyc02g085700; SlTPT1, Solyc02g085710; SlTPT2, Solyc02g085720
The sequential condensation of IPP units to one molecule of DMAPP results in linear isoprenyl diphosphate intermediates, such as C10 geranyl diphosphate (GPP), C15 farnesyl diphosphate (FPP), C20 geranylgeranyl diphosphate (GGPP), C25 geranylfarnesyl diphosphate (GFPP), and so on. These intermediates, of up to thousands of carbon atoms, represent the starting precursors that a series of enzymatic modifications, such as self-condensation, cyclization, isomerization, conjugation and redox reactions eventually convert into the known diversity of isoprenoids (Tholl 2015). GPP is produced almost exclusively from MEP-derived IPP and DMAPP in plastids and used for the biosynthesis of monoterpenes, while FPP is mostly produced in the cytosol and mitochondria from MVA-derived precursors and used for the production of sesqui- and triterpenes. By contrast, GGPP is synthesized in several cell compartments from precursors supplied by the MEP or the MVA pathways and used itself as a precursor of di-, tetra- and polyterpenes (Fig. 1). In particular, MEP-derived GGPP is required to synthesize gibberellins and isoprenoids essential for plant photosynthesis, including photosynthetic pigments (carotenoids and chlorophylls) but also components of the electron transport chain, such as phylloquinones and plastoquinone and photoprotectants, such as tocopherols. Besides their fundamental roles to ensure plant fitness, some of these metabolites are also of nutritional interest for humans, including pro-vitamin A carotenoids, tocopherols (vitamin E) and phylloquinones (vitamin K). Here we will review some of the recent advances in our understanding of how different plant species regulate the production of GGPP and its channeling into downstream isoprenoid pathways. This knowledge is of fundamental importance for the rational design of biotechnological approaches aimed to improve the contents of these metabolites in crops.
GGPP synthases
The synthesis of GGPP and other branch-point precursors, such as GPP and FPP, is mediated by isoprenyl diphosphate synthases or prenyltransferases (PTs). These enzymes catalyze the elimination of the diphosphate moiety from the allylic substrate that remains as an allylic cation prone to be attacked by an IPP molecule. The addition of the IPP unit generates a new 1’-4 double-bond in the product. Depending on the stereochemical conformation of the double bonds formed during the elongation of the isoprenyl diphosphate, PTs are classified as trans or cis PTs. Despite sharing the same substrates and carrying out similar enzymatic reactions, trans and cis PTs form genetically unrelated protein families with completely different protein sequences and different catalytic and substrate binding mechanisms. GPP, FPP and GGPP are normally produced by short-chain trans PTs, which mediate sequential head-to-tail condensation of IPP with allylic substrates including DMAPP, GPP or FPP (Tholl 2015; Vandermoten et al. 2009).
GPP, FPP and GGPP synthases are short-chain trans PTs that contain highly conserved domains essential for their catalytic activity referred to as FARM (First Aspartate-Rich Motif; DDX2-4D, where ‘X’ is any amino acid) and SARM (Second Aspartate-Rich Motif; DDX2D). The length of the final product appears to be regulated by the amino acid identity of the chain-length determination motif (CLD), located upstream to the FARM. In this elongation pocket, residues with large side chains at the fourth and/or the fifth position before the FARM form a “floor” to block the stretching of the product into the pocket, thereby determining the chain length of the final product (Nagel et al. 2015; Vandermoten et al. 2009). A recent study proposed a “three floors” model that suggests that the molecular weight of the residues at different floors would be critical for determining the chain length (Wang et al. 2016). According to this model, floor 1 determines the product FPP versus GGPP. When floor 1 contains a large side-chain residue (such as Y or F) FPP cannot stretch into the chain elongation pocket and it is thus released. However, when small side-chain residues are present in this position, FPP can pass through floor 1 and reach floor 2, leading to the production of GGPP. If floor 1 contains residues with a side chain of medium length (such as M), the effect will be intermediate: the allyl substrate will be able to pass the floor 1 but it will become blocked at floor 2 and it will be released as GGPP. Similar to floor 1, the size of the residues at floor 2 would influence the production of GGPP vs. GFPP, whereas the size of the floor 3 residues would determine whether GFPP or longer products are formed (Wang et al. 2016). Despite this model, it is still difficult to predict the specific product of PT enzymes with high confidence relying solely on sequence homology.
GGPP synthase (GGPPS) enzymes generate GGPP by three sequential IPP condensation steps to DMAPP, via GPP and finally FPP. Consistent with the requirement of GGPP in multiple locations and growth stages to ensure proper development and response to environmental challenges, plant genomes have retained families of differentially expressed genes encoding GGPPS isoforms that interact with specific protein partners and localize to particular cell compartments, including cytosol, plastids, mitochondria, and endoplasmic reticulum (Barja et al. 2021; Beck et al. 2013; Coman et al. 2014; Ruiz-Sola et al. 2016b; Wang and Dixon 2009; Wang et al. 2018; You et al. 2020; Zhou and Pichersky 2020; Zhou et al. 2017). Multiple copies of phylogenetically related (i.e., homologous) GGPPS-encoding genes that result from duplication events involving individual genes, chromosomal segments, or entire genomes are referred to as paralogs (in the same species) and orthologs (in different species). Paralogs tend to acquire different roles, while orthologs tend to share similar functions, (Studer and Robinson-Rechavi 2009). Following duplication, GGPPS paralogs can accumulate inactivating mutations and become a pseudogene or, alternatively, they can be preserved in the genome if they confer selective advantages. Retention of multiple paralogs may involve one of them retaining the ancestral function and the other(s) acquiring new functions (neofunctionalization), but it may also partition the ancestral function between the paralogs for them to specialize and retain only distinct subsets of the ancestral function (subfunctionalization). With the increasing availability of whole genome information for more and more plants, we now have access to the full set of potential GGPPS paralogs in many species. A functional characterization of the complete family, however, is only available for a handful of plants. The next sections will review our current knowledge on the GGPPS families of the model plant Arabidopsis thaliana and the crop species rice (Oryza sativa), pepper (Capsicum annuum) and tomato (Solanum lycopersicum).
Arabidopsis
The first genome-wide list of plant GGPPS candidates was reported in Arabidopsis (Lange and Ghassemian 2003). Initially, twelve Arabidopsis genes (named AtGGPPS1 to 12) were predicted to encode GGPPS homologs. However, subsequent characterization removed many candidates from the list. AtGGPPS5 was described as a pseudogene (Beck et al. 2013). AtGGPPS12 was found to lack GGPPS activity likely due to absence of a functional SARM (Okada et al. 2000) but to heterodimerize with true GGPPS isoforms to produce GPP at much higher levels than homodimeric GGPPS enzymes (Chen et al. 2015; Wang and Dixon 2009). Heterodimeric GPP synthase enzymes are formed by a large and a small subunit (LSU and SSU, respectively) in many plant species. LSUs are typically active or inactive GGPPS-like enzymes, whereas SSUs show lower similarity to GGPPS and only display functional activity when physically interacting with the catalytic LSU monomer (Chen et al. 2015; Tholl 2015; Wang and Dixon 2009). In mint (Mentha piperita), this interaction forms a heterotetrameric (LSU/SSU)2 GPP synthase enzyme (Chang et al. 2010). Two types of SSUs exist in plants. Type I SSUs (SSU-I) are associated with the production of GPP and monoterpenes, whereas interaction of type II SSUs (SSU-II) with LSU monomers can yield GPP but also enhance the production of GGPP (Wang and Dixon 2009). Binding of both SSU-I and SSU-II isoforms to LSUs/GGPPS requires the presence of CxxxC domains (‘x’ is any hydrophobic residue) in both partners: typically two are present in SSUs and one in LSUs (Wang and Dixon 2009). Based on the described features, AtGGPPS12 (At4g38460) was renamed as AtSSU-II.
The rest of the Arabidopsis GGPPS candidates were found to produce GGPP in vitro or in Escherichia coli strains engineered to produce carotenoids (Beck et al. 2013; Okada et al. 2000; Wang and Dixon 2009). However, the use of highly sensitive analytical methods for isoprenyl diphosphate identification later showed that six of the remaining ten isoforms (AtGGPPS1, and AtGGPPS6–10) produced GFPP or even longer isoprenyl diphosphates as major product in vitro (Nagel et al. 2015; Wang et al. 2016). In the case of AtGGPPS1, recombinant His-tagged versions produced mostly GGPP in vitro (Wang et al. 2016; Zhu et al. 1997), whereas a version with a N-terminal GST fusion was found to synthesize 72% of GFPP, 25% of GGPP and 2% of GPP (Nagel et al. 2015). The product mixture of recombinant AtGGPPS6–10 enzymes also contained minor but detectable amounts of GGPP, which can explain the GGPPS activity deduced for these isoforms in previous in vitro and in vivo assays. For the rest of candidates, AtGGPPS2–4 and AtGGPPS11, GGPP was confirmed as their primary product. After these findings, the Arabidopsis GGPPS family was concluded to contain five members: AtGGPPS1 (At1g49530), AtGGPPS2 (At2g18620), AtGGPPS3 (At2g18640), AtGGPPS4 (At2g23800) and AtGGPPS11 (At4g36810). AtGGPPS8 was considered a polyprenyl diphosphate synthase and renamed AtPPPS2 (Wang et al. 2016). AtGGPPS6, 7, 9 and 10 were considered GFPP synthases and renamed AtGFPPS1, 2, 3 and 4, respectively. These GFPPS-encoding Arabidopsis genes occur in tandem with genes encoding sesterterpene synthases that use GFPP as a substrate (Chen et al. 2019; Huang et al. 2017; Shao et al. 2017; Wang et al. 2016).
GFP fusions of individual members of the Arabidopsis GGPPS family were found to localize in mitochondria (AtGGPPS1), plastids (AtGGPPS2 and 11) and endoplasmic reticulum (AtGGPPS3 and 4) (Fig. 1) (Beck et al. 2013; Bick and Lange 2003; Okada et al. 2000). Genes encoding the organellar GGPPS enzymes (AtGGPPS1, 2 and 11) are constitutively expressed, with AtGGPPS11 showing the highest mRNA levels, particularly in photosynthetic tissues. By contrast, the expression of the genes encoding AtGGPPS3 and 4 is restricted to siliques, flowers and roots (Beck et al. 2013). A deeper study of the genes encoding the two plastidial isoforms showed that the AtGGPPS2 paralog was mostly co-expressed with genes encoding enzymes of the gibberellin biosynthetic pathway, whereas the AtGGPP11 gene was co-regulated with genes involved in the MEP pathway and in the production of photosynthesis-related isoprenoids (Ruiz-Sola et al. 2016b). Consistent with the described gene expression pattern and co-regulation network, the AtGGPPS11 protein was found to physically interact with phytoene synthase (PSY), solanesyl diphosphate synthase 2 (SPS2), and geranylgeranyl reductase (GGR), which are GGPP-consuming enzymes involved in the synthesis of carotenoids, plastoquinone, and the phytol chain of chlorophylls, tocopherols and phylloquinone (Figs. 1, 2) (Ruiz-Sola et al. 2016b). Later results confirmed that AtGGPPS11 and PSY closely interact to efficiently channel GGPP into the carotenoid pathway in Arabidopsis (Camagna et al. 2019). Direct interactions with these GGPP-using enzymes might allow specifically diverting GGPP to particular downstream pathways in the same subcellular compartment when needed. The control of this GGPP allocation mechanism (e.g., how protein–protein interactions are regulated) remains unknown. AtGGPPS11 was also found to physically interact with AtSSU-II, resulting in a change in the main product from GGPP to GPP (Fig. 2) (Wang and Dixon 2009). These findings illustrate how the formation of specific AtGGPPS11 protein complexes can contribute to the production of particular classes of plastidial isoprenoids in Arabidopsis. AtSSU-II can also interact with AtGGPPS2 but not with AtGGPPS6/AtGFPPS1 despite they are all plastid-targeted enzymes (Beck et al. 2013; Wang and Dixon 2009).
Genetic approaches are key to determine the in vivo role of plant proteins. In the case of Arabidopsis GGPPS paralogs, analysis of individual mutants defective in individual GGPPS enzymes did not show any developmental defects with the only exception of those defective in AtGGPPS11 (Ruiz-Sola et al. 2016a, b). The deleterious phenotypes observed in different mutant alleles of AtGGPPS11 indicated that the second plastidial isoform, AtGGPPS2, was unable to complement the loss of AtGGPPS11 function, perhaps because it is expressed at much lower levels. The AtGGPPS11-defective mutant allele with the weakest reported phenotype is ggpps11-1, which harbors a point mutation in a conserved residue (Ruppel et al. 2013). This mutant shows a temperature-dependent variegated phenotype similar to that of the chs5 line, which also harbors a point mutation in the gene encoding the first enzyme of the MEP pathway (Araki et al. 2000). This is consistent with a role of AtGGPPS11 in the production of MEP-derived isoprenoids and hence in photosynthesis. In addition, consistently, decreased AtGGPPS11 expression due to a T-DNA insertion upstream of the predicted ATG translation start codon in the ggpps11-5 allele caused a pale phenotype and a developmental delay (Ruiz-Sola et al. 2016b). These alterations were accompanied by a significant reduction in the levels of plastidial GGPP-derived isoprenoids (carotenoids, chlorophylls, tocopherols and plastoquinone) in the mutant, which were restored to wild-type levels after complementing the ggpps11-5 allele with the native gene (Ruiz-Sola et al. 2016b). Further supporting the conclusion that AtGGPPS11 is the major isoform transforming MEP-derived precursors into GGPP to produce photosynthesis-related isoprenoid products, a seedling-lethal albino phenotype visually identical to that of knockout MEP pathway mutants (Phillips et al. 2008) was observed in the case of the ggpps11-2 allele, which harbors a T-DNA insertion in the N-terminal end of the protein coding region. In this allele, the insertion disrupts the plastid-targeting signal but does not prevent the production of a shorter protein with GGPPS activity which localizes in the cytosol (Ruiz-Sola et al. 2016a; Ruppel et al. 2013). Strikingly, T-DNA insertions in the C-terminal end of the protein resulted in non-functional enzymes and blocked embryo development in alleles ggpps11-3 and ggpps11-4 (Ruiz-Sola et al. 2016a, b; Ruppel et al. 2013). This embryo lethal phenotype has never been observed in MEP pathway mutants (Phillips et al. 2008) but it is common in mutants defective in MVA-derived isoprenoids (Okada et al. 2004; Schrick et al. 2002). Further experiments demonstrated that embryo development required the presence of AtGGPPS11 activity in the cytosol (despite the existence of two other GGPPS enzymes in Arabidopsis associated to the endoplasmic reticulum), whereas photosynthetic development required the activity of AtGGPPS11 in the plastid (despite a second plastidial enzyme with the same activity exists in Arabidopsis) (Ruiz-Sola et al. 2016a). At the mechanistic level, it was shown that the AtGGPPS11 gene can produce two types of enzyme isoforms: those translated from the first ATG codon carry a plastid-targeting peptide and localize in plastids, whereas shorter versions translated from a downstream ATG codon lack the plastid-targeting peptide and remain in the cytosol (Ruiz-Sola et al. 2016a). The production of these two differentially targeted isoforms may rely on the use of alternative transcription start sites, the use of alternative translation start sites, or both. Together, these findings confirmed that AtGGPPS11 is a hub GGPPS isozyme for the production of most photosynthesis-related isoprenoids and other unidentified products required for embryo development in the cytosol. Other GGPPS isoforms might participate in more specialized developmental and/or condition-specific functional processes yet to be identified. Indeed, a combination of computational analyses and integration with meta-analysis of existing data sets led to propose that the Arabidopsis GGPPS paralogs are not redundant and that their persistence in the genome is likely attributed to subfunctionalization in terms of differential gene expression (Coman et al. 2014).
Rice
The rice genome harbors several genes with homology to PTs, of which only two clustered with GGPPS-like enzymes and one with SSU-II proteins (You et al. 2020; Zhou et al. 2017). Similar to Arabidopsis, no gene encoding SSU-I was found in the rice genome. One of the two GGPPS-like proteins produced GGPP as that the main product from IPP and DMAPP in vitro and it was designated OsGGPPS1 (Os07g39270). The other candidate produced GPP as the only product from IPP and DMAPP but it was also able to produce GGPP when fed with IPP and FPP. This second GGPPS paralog was hence named as OsGPPS (Os01g14630). The SSU-II homolog (Os02g44780) was confirmed to lack enzymatic activity and to interact with OsGGPPS1, resulting in an improved catalytic efficiency and product specificity towards GGPP (Zhou et al. 2017). GFP fusions of OsGGPPS1, OsGPPS and OsSSU-II proteins were targeted to chloroplasts (Fig. 1) (You et al. 2020; Zhou et al. 2017). Interestingly, OsSSU-II was found to recruit OsGGPPS1 from the chloroplast stroma to the thylakoid membranes, directing it to a large protein complex containing GGR and other components of the chlorophyll biosynthetic pathway, such as the light-harvesting-like protein LIL3 and the chlorophyll synthase CHLG (Fig. 2). Based on this property, OsSSU-II was referred to as GGPPS Recruiting Protein (OsGRP) (Zhou et al. 2017). It remains unknown whether OsSSU-II/OsGRP could also bind OsGPPS and the consequences of this interaction in terms of enzymatic activity or/and subplastidial localization.
Because the formation of heterodimers of OsGGPPS1 and OsGRP was more favorable than that of OsGGPPS1 homodimers, it was proposed that the abundance of OsGRP might determine OsGGPPS1 allocation in chloroplasts and hence influence the metabolic flux from GGPP towards downstream pathways located in the stroma (via OsGGPPS1 homodimers) or associated to thylakoid membranes (via heterodimers). In agreement, overexpression of OsGRP led to an increase in the amount of chlorophylls but reduced levels of carotenoids and gibberellins (Zhou et al. 2017), presumably because enhanced thylakoid allocation of GGPP-producing heterodimers increased flux toward chlorophyll biosynthesis but reduced flux toward competing isoprenoid pathways. Although interaction of OsGGPPS1 with PSY was not observed, a T-DNA mutant with reduced levels of OsGGPPS1 transcripts showed decreased levels of both chlorophylls and carotenoids (Zhou et al. 2017). In summary, the available data suggest that, similar to Arabidopsis, rice contains a hub GGPPS enzyme (OsGGPPS1) and an SSU-II protein (OsSSU-II/OsGRP) but no SSU-I homolog. The biological function of the SSU-II orthologs and the mechanisms for GGPP allocation in chloroplasts, however, appear to differ between the two species. Strikingly, no GGPPS isoforms could be found associated to other cell compartments besides chloroplasts, suggesting that other PTs might be producing GGPP in these extraplastidial locations. Alternatively, shorter versions of OsGGPPS1 or/and OsGPPS lacking their plastid-targeting peptides might be produced as described for Arabidopsis AtGGPPS11 (Ruiz-Sola et al. 2016a).
Pepper
Another plant species with a relatively well characterized GGPPS family is pepper (Wang et al. 2018). Of the eight genes found in the pepper genome with homology to PTs, one was likely a pseudogene, five clustered with confirmed GGPPS enzymes from Arabidopsis (AtGGPPS11) and rice (OsGGPPS1), and two showed homology to SSU proteins: one to SSU-I and another one to SSU-II. Localization of GFP-fused proteins in transfected protoplasts showed that the two SSU-like proteins, Capana00g004199 (CaSSU-I) and Capana09g002331 (CaSSU-II), as well as two of the GGPPS-like proteins, Capana04g000412 (CaGGPPS1) and Capana05g000800 (here referred to as CaPTP1) were targeted to chloroplasts. The other three GGPPS-like proteins, which were encoded by genes clustered together in the genome, had either cytosolic (CaPTP2, Capana00g002450 and CaPTP3, Capana00g002451) or mitochondrial (CaPTP4, Capana00g002452) localizations (Fig. 1) (Wang et al. 2018). CaGGPPS1 was found to be the only one displaying GGPPS activity in E. coli strains engineered with a heterologous carotenoid biosynthetic pathway. In vitro activity assays confirmed that this paralog produced GGPP as that the main product from IPP and DMAPP. While these experiments may lead to conclude that CaGGPPS1 is the only functionally active GGPPS enzyme in pepper, extending the in vitro assays to all the family members with several substrate combinations (including DMAPP, GPP and FPP as allylic substrates) and/or genetic evidence (e.g., pepper mutants) would be necessary to support such a conclusion.
In most pepper cultivars, fruit ripening involves degradation of chlorophylls and a boost in the production of carotenoids, changing fruit color from green to yellow, orange or red colors when ripe. Besides providing attractive colors, carotenoid accumulation involves nutritional benefits as these plastidial isoprenoids are pro-vitamin A and health-promoting metabolites in humans (Rodriguez-Concepcion et al. 2018). In agreement with the expected requirement of an increased supply of GGPP precursors for carotenoid biosynthesis, GGPPS protein levels substantially increase during pepper fruit ripening (Rodiger et al. 2021). Proteomic analysis identified CaGGPPS1 in ripe fruit chromoplasts (Siddique et al. 2006), and the gene encoding this paralog but also that for CaSSUII were highly induced during ripening (Wang et al. 2018). Both yeast two hybrid and bimolecular fluorescence complementation assays failed to clearly show CaGGPPS1 homodimerization but demonstrated a clear heterodimerization with CaSSU-II, resulting in a higher catalytic efficiency of CaGGPPS1 without altering its product specificity (Fig. 2) (Wang et al. 2018). It is unknown whether CaGGPPS1 could also bind CaSSU-I and the consequences of this interaction. Possible interactions of CaPTP1 (the second plastid-targeted pepper GGPPS-like protein) with CaSSU-I and CaSSU-II also remain to be tested. As to interaction with other proteins, a GGPPS was found associated with PSY in pepper chromoplasts (Dogbo and Camara 1987). Yeast two-hybrid assays confirmed interaction of CaGGPPS1 but also of CaSSU-II with the main PSY paralog present in pepper fruit (Wang et al. 2018). All these data together suggest that CaSSU-II might not only stimulate GGPP production upon heterodimerization with the hub GGPPS enzyme CaGGPPS1 but it might also help to bridge the interaction with PSY for carotenoid biosynthesis, similar to that reported in rice for OsGGPPS1/OsSSU-II and GGR for chlorophyll biosynthesis (Fig. 2). Whether other GGPPS isoforms provide GGPP in pepper for other metabolic pathways remains to be tested.
Tomato
A recent analysis of the updated reference tomato genome found five genes encoding GGPPS-like enzymes and two showing homology to SSU proteins (Zhou and Pichersky 2020). From them, paralogs SlGGPPS1 (Solyc11g011240), SlGGPPS2 (Solyc04g079960) and SlGGPPS3 (Solyc02g085700) were experimentally shown to be targeted to plastids and to produce GGPP as the main product when fed with IPP and DMAPP in vitro (Barja et al. 2021; Zhou and Pichersky 2020). The other two GGPPS-like proteins, encoded by genes located close together with SlGGPPS3 in the genome (SlTPT1, Solyc02g085710 and SlTPT2, Solyc02g085720), encoded proteins that lacked GGPPS or GFPPS activity and localized in mitochondria (Fig. 1) (Zhou and Pichersky 2020). As to the SSU proteins, one showed homology to SSU-I (SlSSU-I, Solyc07g064660) and the other one to SSU-II (SlSSU-II, Solyc09g008920). Both proteins localized in plastids and lacked catalytic activity in vitro in the presence of IPP and DMAPP. While SlSSU-I was found to change the product specificity of the true GGPPS enzymes SlGGPPS1, SlGGPPS2 and SlGGPPS3 from GGPP to GPP, heterodimerization with the SlSSU-II protein resulted in an improved specificity towards GGPP (Zhou and Pichersky 2020). As a summary, the tomato GGPPS family was concluded to contain three members located in plastids, where they may interact with SSU proteins to modulate their enzymatic activity (Fig. 2).
The possible subfunctionalization of the SlGGPPS1–3 enzymes was initially addressed by analyzing their gene expression patterns (Barja et al. 2021). It was concluded that SlGGPPS1 might be the main GGPPS paralog providing GGPP in roots, in agreement with other studies (Stauder et al. 2018). The much more highly expressed SlGGPPS2 and SlGGPPS3 genes would be in charge of supplying GGPP to produce carotenoids and other plastidial isoprenoids in leaves and fruits. Protein complexes containing unidentified isoforms of GGPPS and PSY enzymes have been found in tomato chloroplasts but also in the chromoplasts of ripe fruit, which accumulate very high levels of carotenoids (Fraser et al. 2000; Maudinas et al. 1977). Interestingly, co-immunoprecipitation experiments have shown that SlGGPPS2 but not SlGGPPS3 can interact with the PSY paralogs present in tomato leaves and fruits (Barja et al. 2021). It is possible, however, that SlGGPPS3 can also directly provide GGPP for PSY enzymes to produce phytoene and downstream carotenoids by either heterodimerization with PSY-interacting SlGGPPS2 proteins or through interaction with SlSSU-II, similar to that proposed for CaSSU-II and CaGGPPS1 (Fig. 2). Further experiments would be needed to test this possibility.
Analysis of tomato mutants defective in SlGGPPS2 or/and SlGGPPS3 confirmed that these two isoforms coordinately supply GGPP in shoot tissues. Decreased levels of carotenoids and chlorophylls in tomato lines lacking SlGGPPS3 resulted in pale young leaves with a decreased photosynthetic activity, whereas the reduction of GGPP supply in fruits led to pigmentation and ripening defects in both slggpps2 and slggpps3 mutants (Barja et al. 2021). Partial loss of SlGGPPS3 activity in slggpps2 mutants or SlGGPPS2 activity in slggpps3 mutants exacerbated their phenotypes, consistent with the conclusion that these are functionally exchangeable isoforms that participate in the same biological processes. Subfunctionalization, however, can be explained due to distinct expression profiles. SlGGPPS3 expression is fairly high and constitutive, suggesting a house-keeping role for the encoded paralog to maintain a continuous supply of GGPP. By contrast, expression of the SlGGPPS2 gene is more responsive to peak demands of GGPP to boost the production of carotenoids and other plastidial isoprenoids (e.g., during deetiolation and fruit ripening). The very low and restricted expression level of the SlGGPPS1 gene further suggests a localized and specialized role for this paralog. Similar to that observed in rice and pepper, extraplastidial GGPPS paralogs appear to be missing in tomato. Interestingly, complete loss of both SlGGPPS2 and SlGGPPS3 function in double mutant plants resulted in an embryo lethal phenotype that resembles that described in Arabidopsis mutant alleles lacking AtGGPPS11 activity in the cytosol (Barja et al. 2021; Ruiz-Sola et al. 2016a). It is, therefore, possible that short extraplastidial versions of the tomato SlGGPPS1–3 enzymes could be produced in vivo.
Concluding remarks
While gene families encoding GGPPS paralogs are typically present in plant genomes, single isoforms appear to be responsible in some plants (e.g., rice and pepper) for the production of most if not all GGPP for cell functions. Such isoforms are localized in plastids when fused to GFP, consistent with these compartments being the main site for the production and accumulation of GGPP-derived isoprenoids (including carotenoids and chlorophylls). In cases when more than one GGPPS enzyme is present (e.g., in Arabidopsis and tomato), the predominant role of particular paralogs appears to mainly rely on their spatio-temporal gene expression pattern. In Arabidopsis, genetic evidence shows that only one of the two plastid-targeted isoforms (AtGGPPS11) is essential to produce both plastidial and cytosolic GGPP, whereas in tomato at least two plastidial isoforms (SlGGPPS2 and SlGGPPS3) coordinately supply GGPP to produce carotenoids and other isoprenoids essential for photosynthesis, fruit pigmentation, and seed viability. Furthermore, specific interactions with other proteins (including PSY and other GGPP-consuming enzymes but also non-catalytic SSU proteins) might be relevant for subfunctionalization. A conclusion based on the available knowledge is that what we learn in one species might not be directly applied to others. For example, the cucumber (Cucumis sativus) plastid-targeted isoforms CsaGGPPS1 (Csa2G296070) and CsaGGPPS2 (Csa6G487640) interact with CsaSSU-I (Csa7G211090) and CsaSSU-II (Csa7G017680), as expected, but only the CsaGGPPS1 / CsaSSU-I heterodimer produces GPP as the main product (more than 70%) in vitro (Wei et al. 2016). Future work should provide additional clues to understand the mechanisms governing the supply of plastidial GGPP for the synthesis of isoprenoids with different biological functions in particular tissues, organs and plant species. Deciphering such mechanisms will be very useful for future metabolic engineering approaches aimed to manipulate the accumulation of particular GGPP-derived products of interest without negatively impacting the levels of other isoprenoids required to sustain essential cell functions.
References
Araki N, Kusumi K, Masamoto K, Niwa Y, Iba K (2000) Temperature -sensitive Arabidopsis mutant defective in 1-deoxy-d-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiol Plant 108:19–24
Barja MV et al (2021) Several geranylgeranyl diphosphate synthase isoforms supply metabolic substrates for carotenoid biosynthesis in tomato. New Phytol. https://doi.org/10.1111/nph.17283
Beck G, Coman D, Herren E, Ruiz-Sola MA, Rodriguez-Concepcion M, Gruissem W, Vranova E (2013) Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol Biol 82:393–416
Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415:146–154
Camagna M, Grundmann A, Bar C, Koschmieder J, Beyer P, Welsch R (2019) Enzyme fusion removes competition for geranylgeranyl diphosphate in carotenogenesis. Plant Physiol 179:1013–1027
Chang TH, Hsieh FL, Ko TP, Teng KH, Liang PH, Wang AH (2010) Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. Plant Cell 22:454–467
Chen Q, Fan D, Wang G (2015) Heteromeric Geranyl(geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol Plant 8:1434–1437
Chen Q et al (2019) Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci China Life Sci 62:947–958
Coman D, Altenhoff A, Zoller S, Gruissem W, Vranova E (2014) Distinct evolutionary strategies in the GGPPS family from plants. Front Plant Sci 5:230
Dogbo O, Camara B (1987) Purification of isopentenyl pyrophosphate isomerase and geranylgeranyl pyrophosphate synthase from Capsicum chromoplasts by affinity-chromatography. Biochim Biophys Acta Lipids Lipid Metab 920:140–148
Fraser PD, Schuch W, Bramley PM (2000) Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts–partial purification and biochemical properties. Planta 211:361–369
Huang AC, Kautsar SA, Hong YJ, Medema MH, Bond AD, Tantillo DJ, Osbourn A (2017) Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc Natl Acad Sci USA 114:E6005–E6014
Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51:925–948
Maudinas B, Bucholtz ML, Papastephanou C, Katiyar SS, Briedis AV, Porter JW (1977) The partial purification and properties of a phytoene synthesizing enzyme system. Arch Biochem Biophys 180:354–362
Nagel R et al (2015) Arabidopsis thaliana isoprenyl diphosphate synthases produce the C intermediate, geranylfarnesyl diphosphate. Plant J 84:847–859
Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122:1045–1056
Okada K et al (2004) The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol Biol 55:567–577
Phillips MA, Leon P, Boronat A, Rodriguez-Concepcion M (2008) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci 13:619–623
Pulido P, Perello C, Rodriguez-Concepcion M (2012) New insights into plant isoprenoid metabolism. Mol Plant 5:964–967
Rodiger A, Agne B, Dobritzsch D, Helm S, Muller F, Potzsch N, Baginsky S (2021) Chromoplast differentiation in bell pepper (Capsicum annuum) fruits. Plant J 105:1431–1442
Rodriguez-Concepcion M, Boronat A (2015) Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr Opin Plant Biol 25:17–22
Rodriguez-Concepcion M et al (2018) A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Progr Lipid Res 70:62–93
Ruiz-Sola MA, Barja MV, Manzano D, Llorente B, Schipper B, Beekwilder J, Rodriguez-Concepcion M (2016a) A single Arabidopsis gene encodes two differentially targeted geranylgeranyl diphosphate synthase isoforms. Plant Physiol 172:1393–1402
Ruiz-Sola MA et al (2016b) Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol 209:252–264
Ruppel NJ, Kropp KN, Davis PA, Martin AE, Luesse DR, Hangarter RP (2013) Mutations in GERANYLGERANYL DIPHOSPHATE SYNTHASE 1 affect chloroplast development in Arabidopsis thaliana (Brassicaceae). Am J Bot 100:2074–2084
Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jurgens G (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31:61–73
Shao J et al (2017) (+)-Thalianatriene and (−)-retigeranin B catalyzed by sesterterpene synthases from Arabidopsis thaliana. Org Lett 19:1816–1819
Siddique MA, Grossmann J, Gruissem W, Baginsky S (2006) Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant Cell Physiol 47:1663–1673
Stauder R, Welsch R, Camagna M, Kohlen W, Balcke GU, Tissier A, Walter MH (2018) Strigolactone levels in dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family front. Plant Sci 9:255
Studer RA, Robinson-Rechavi M (2009) How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25:210–216
Tholl D (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148:63–106
Vandermoten S, Haubruge É, Cusson M (2009) New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cell Mol Life Sci 66:3685–3695
Wang G, Dixon RA (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA 106:9914–9919
Wang C, Chen Q, Fan D, Li J, Wang G, Zhang P (2016) Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae. Mol Plant 9:195–204
Wang Q, Huang XQ, Cao TJ, Zhuang Z, Wang R, Lu S (2018) Heteromeric geranylgeranyl diphosphate synthase contributes to carotenoid biosynthesis in ripening fruits of red pepper ( Capsicum annuum var. conoides). J Agric Food Chem 66:11691–11700
Wei G et al (2016) Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular insight into voc diversity in cucumber plants (Cucumis sativus). Plant Physiol 172:603–618
You MK, Lee YJ, Yu JS, Ha SH (2020) The predicted functional compartmentation of rice terpenoid metabolism by trans-prenyltransferase structural analysis, expression and localization. Intl J Mol Sci 21:8927
Zhou F, Pichersky E (2020) The complete functional characterisation of the terpene synthase family in tomato. New Phytol 226:1341–1360
Zhou F et al (2017) A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc Natl Acad Sci USA 114:6866–6871
Zhu XF, Suzuki K, Okada K, Tanaka K, Nakagawa T, Kawamukai M, Matsuda K (1997) Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant Cell Physiol 38:357–361
Funding
Work in our lab is funded by grants BIO2017-84041-P from the Spanish Agencia Estatal de Investigacion (AEI) and 2017SGR-710 from Generalitat de Catalunya to MRC. MVB was supported by PhD fellowship FPU14/05142 from the Spanish Ministerio de Educacion y Cultura.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest or competing interest.
Rights and permissions
About this article
Cite this article
Barja, M.V., Rodriguez-Concepcion, M. Plant geranylgeranyl diphosphate synthases: every (gene) family has a story. aBIOTECH 2, 289–298 (2021). https://doi.org/10.1007/s42994-021-00050-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-021-00050-5