Abstract
Automated methods using computer-aided decision-making process are effectively used for timely detection of VAs which are the most life-threatening conditions. In this work, we have proposed a decision support system for detection of ventricular arrhythmias (VAs) with a low computational complexity using hybrid features derived from three different transform techniques i.e. DWT, EEMD, and VMD. The methods mainly consist of a windowing technique, signal decomposition, feature extraction, and classification. About 24 time–frequency based features were extracted with 22,721 × 24 and ranked for selection of higher ranked features having maximum information of disease. The reduced feature set consists of only 6 number of highly ranked features which are then classified with SVM and decision tree classifier for efficient recognition of VAs. Aim of the reduction in data size is to reduce the computational time. Our proposed method achieves high classification accuracy in hybrid-based features and by reducing the feature dimension, it reduces the computational complexity significantly. The accuracy of 99.83% and computational time of 4.82 s is achieved when considering all 24 features. In reduced feature set, an accuracy of 99.62% with a very less computational time of 2.71 s was obtained for decision tree classifier which indicates the importance of selecting the important features for classification of VAS. With superior classification accuracy and low computation complexity, this system can be utilized in clinical practice for the recognition of ventricular arrhythmias.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The studies have shown that the dynamics of the cardiac signal are very complex and non-linear in nature, therefore, the non-linear models are suitable for the analysis of ECG signals [1]. Each analysis methods to detect heart disease provides valuable diagnostic information with different conclusions requires further investigation in ECG signal analysis [2]. Hence some non-linear techniques have been developed for efficient detection of life-threatening ventricular arrhythmias such as VT/VF using ECG signal [3]. Sun et al. [4] proposed a non-linear method using the Hurst index to characterize the VT and VF episode in ECG analysis. Both time and frequency-based features improves the classification of ECG beats [5]. Arafat et al. [6] proposed a threshold crossing sample count (TCSC) method which is in time domain analysis for accurate detection of VF episodes. Decomposition of the ECG signal using DWT, EMD, variational mode decomposition with machine learning techniques has been found to be effective in detecting cardiac arrhythmias [7,8,9,10].
The wavelet transform based features is an important approach in classifying the non-stationary ECG signals as it provides both time and frequency information of an ECG signal [11]. Rakshit et al. [12] have proposed an automatic method using wavelet transform and a Hilbert transform for R-peaks detection. The method achieved an average accuracy of 99.83%, with an error rate of 0.17%. Nazarahari et al. [13] presented a new wavelet functions WFs based on the hybrid approach that consists of GA-PSO framework for effective classification of the ECG signals. The discrete wavelet transforms and morphology-based features were successfully used to classify abnormal and normal class ECG signal with neural network classifier [14]. In a linear method, Martis et al. [15] applied the principal component analysis i.e., PCA based feature reduction technique in discrete wavelet transform (DWT) based features to discriminate between normal and arrhythmia classes. They achieved an overall classification accuracy of 98.78% with a neural network (NN) classifier. A novel method was presented by Garcia et al. [16] for automatic detection of AF episodes using the relative wavelet energy (RWE) of the TQ interval in the ECG signal. The method was able to detect the AF condition with an accuracy higher than 90% in less than 7 beats. A novel method based on stationary wavelet transform and SVM classifier was applied successfully for automatic detection of AF and achieves a sensitivity of 97.0% and specificity of 97.1%, respectively [17].
The empirical mode decomposition (EMD) has been also used for accurate detection of VF. In this method, the ECG signals are decomposed into different intrinsic mode functions (IMFs). Arafat et al. [18] have used EMD technique for decomposition of ECG signals and extracted orthogonality indices from the first three consecutive IMFs and used to detect VF condition. The method was applied for a window with a duration of 3 s and obtained the accuracy of detection as 99.70%. In an efficient process, the EMD method was used to discriminate between ventricular fibrillation and ventricular tachycardia with an accuracy of 91% using ECG signals [19]. Similarly, few studies have been reported the use of a variational mode of decomposition for effective application of detection of ventricular arrhythmias. Xu et al. [20] have proposed an adaptive-VMD process for decomposing the signals into five band-limited intrinsic modes (BLIMs). Then the feature extraction and classification were done using a Boosted-CART classifier to recognize the VT/VF condition and obtained an overall 98.29% of accuracy and 97.32%. of sensitivity. Based on the non-linearity and nonstationary decomposition methods, Abdalla et al. [21] have developed a method called complete ensemble empirical mode decomposition with adaptive noise (CEEMDAN) for classification of arrhythmias using MIT-BIH database. It was used to find intrinsic mode functions (IMFs) and four different parameters were computed to construct the feature vector and classified with ANN classifier with an accuracy of 99.9%.
In this work we have proposed a hybrid feature set approach that combines the features based on DWT, EEMD, and VMD methods. The hybrid features having large dimension have been extracted to make a large dataset in order to achieve higher accuracy. The dimension of the feature set was reduced by selecting the useful informative features using a ranker search method. In order to balance between the reliable recognition system and computational burden, in this work, we have considered a hybrid feature set and ranked them to obtain the most significant information about the VA conditions. The selected informative features are classified using the SVM and the decision tree classifier to recognize the life-threatening ventricular arrhythmias.
Methodology
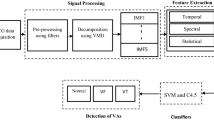
The flow chart for the projected ventricular arrhythmia classification system using hybrid feature has been shown in Fig. 1. The electrocardiogram signals were acquired from the first channel of both CUDB and VFDB databases. The acquired ECG signals are first got pre-processed using a high pass (fc = 1 Hz), low pass (fc = 30 Hz), and a notch filter to eliminate the baseline wander, high-frequency noise and power line interference respectively. Using a window length of 5 s, the acquired electrocardiogram signals have been decomposed using DWT, EEMD and VMD methods. From every mode, the 24 features have been extracted in the temporal, spectral, and statistical domain to make a large dataset having size 22,721 × 24. The dataset is then feed in machine learning approach for classification of the ventricular arrhythmias.
ECG Signal Decomposition
Discrete Wavelet Transform (DWT)
The wavelet transform has been broadly used for signal decomposition and feature extraction for the analysis of nonstationary signals like ECG [22]. In DWT approach, the signal is decomposed into the approximate coefficients as LPF and detail coefficients as HPF. The signal is then down sampled to half in each step. In this work, the ECG signal is decomposed into 8 levels by using Daubechies db6 mother wavelet transform. The decomposed signal of detail coefficients at D2-D8 and approximation coefficient A8 is consider for processing. In case of VT condition, the signal consists of detail coefficients (D1–D3) and approximation coefficient A3.
Let X → be the original signal
-
n → be the no. of decomposition levels
-
A (n) → be the approximation coefficients
-
D (n) → be the detail coefficients
$${\text{S}}\left[ {\text{n}} \right] = \left( {\text{X*A}} \right)\left[ {\text{n}} \right] = \mathop \sum \limits_{{\text{k}} = - \infty }^\infty {\text{X}}\left[ {\text{k}} \right]{\text{A}}\left[ {{\text{n}} - {\text{k}}} \right]$$(1)$${\text{S}}_{{\text{low}}} \left[ {\text{n}} \right] = \mathop \sum \limits_{{\text{k}} = - \infty }^\infty {\text{X}}\left[ {\text{k}} \right]{\text{ A}}\left[ {2{\text{n}} - {\text{k}}} \right]$$(2)$${\text{S}}_{{\text{high}}} \left[ {\text{n}} \right] = \mathop \sum \limits_{{\text{k}} = - \infty }^\infty {\text{X}}\left[ {\text{k}} \right]{\text{ D}}\left[ {2{\text{n}} - {\text{k}}} \right]$$(3)
Ensemble Empirical Mode Decomposition (EEMD)
The effective time–frequency analysis of nonlinear and non-stationary signals, such as electrogram signals, can be done be done using the EEMD technique [23]. Due to the signal intermittency, often the mode mixing problem arise which is the main bottleneck of the earlier EMD techniques. The EEMD technique provides a solution to the mode mixing problem by adding a finite noise to the original signal, while also retaining the decomposition information.
The decomposed data Y(t) has been given as the intrinsic mode functions (IMFs) as:
where the number of IMFs is given by n.
Then for successive observations, the modified data Yi(t) after adding random white noise has been given as:
where N indicates the added white noise.
Based upon the decomposition, the required steps for the EEMD technique are given as follows:
-
Initially, a random white noise has been added to the given data.
-
Then the IMFs have been generated after decomposing the given data set.
-
Repeat step 1 and step 2 by adding white noise series every time.
-
Finally, obtain the mean of the signal i.e. ensemble of respective IMFs of the decomposition as the absolute result.
In the final mean of the corresponding IMFs, the added white noise series cancel each other. The average IMF resides within the filter windows and thus reduces the occurrence of mode mixing.
Variational Mode Decomposition (VMD)
The EMD approach is extensively used for recursive decomposition of a signal into various modes but has few limitations such as sampling and sensitivity to noise. Hence, to overcome such limitations, a non-recursive VMD model was developed which has the ability of concurrent extractions of modes. A signal, where the difference between the number of local extrema and zero-crossings is at most one, is defined as a mode [24, 25]. The IMFs of VMD are given as:
where N is the number of modes.
\(\emptyset_{\text{k}} \left( {\text{t}} \right){ }\) is the phase \((\emptyset_{\text{k}}^{^{\prime} } ({\text{t}}) > 0\)) and \({\text{E}}_{\text{k}} \left( {\text{t}} \right){ }\) is the envelope \({\text{(E}}_{\text{k}} \left( {\text{t}} \right) \ge 0).\)
The variational substitute to the empirical mode decomposition or the empirical wavelet transform is given by VMD. This approach decomposes an input signal into different sub-signals i.e. IMFs which have the ability to reproduce the input with their sparsity properties.
Basically, 3 parameters are needed for VMD decomposition technique i.e. data fidelity constraint parameter (α), number of modes (N) and Lagrangian multiplier (λ). The parameter α is used to determine the frequency ranges in different modes. The parameter N has to be predefined for a signal in order to determine the number of decomposed modes. The parameter λ is used for quality check of reconstruction of an original signal from its IMFs.
The decomposition algorithm VMD is given as follows:
-
Step I. At count n = 1, the Lagrangian multiplier \({{(\uplambda }}_1 )\) is initiated and the center frequency \(({\text{w}}_{\text{k}} )\) and the spectrum \(({\text{I}}_{\text{k}} )\) of the kth mode are generated.
-
Step II. For all modes i.e. for k = 1 to N,
-
(a)
the spectrum of the kth mode in the next iteration i.e. count = n + 1 is updated as:
$${\text{ I}}_{\text{k}}^{{\text{n}} + 1} \left( {\text{t}} \right) = \frac{{{\text{I}}\left( {\text{w}} \right) - \sum_{{\text{i}} < k} {\text{I}}_{\text{i}}^{{\text{n}} + 1} \left( {\text{w}} \right) - \sum_{{\text{i}} > k} {\text{I}}_{\text{i}}^{\text{n}} \left( {\text{w}} \right) + \left( {\left( {{\uplambda }^{\text{n}} ({\text{w}})/2} \right)} \right)}}{{1 + 2{{ \upalpha }}\left( {{\text{w}} - {\text{w}}_{\text{k}}^{\text{n}} } \right)^2 }}$$(7) -
(b)
For each mode, the center frequency is updated as follows:
$${\text{ w}}_{\text{k}}^{{\text{n}} + 1} = \frac{{\smallint_0^\infty {\text{w}}\left| {{\text{I}}_{\text{k}}^{{\text{n}} + 1} ({\text{w}})} \right|^{2{ }} {\text{dw}}}}{{\smallint_0^\infty \left| {{\text{I}}_{\text{k}}^{{\text{n}} + 1} ({\text{w}})} \right|^{2{ }} {\text{dw}}}}$$(8) -
(c)
The Lagrangian multiplier is updated as follows:
$${{ \uplambda }}^{{\text{n}} + 1} = {\uplambda }^{\text{n}} + {\uptau }\left( {\text{I}}\left( {\text{w}} \right) - \sum \limits_{\text{k}}\, {\text{I}}_{\text{k}}^{{\text{n}} + 1} ({\text{w}}) \right)$$(9)
-
(a)
-
Step III. Repeat step 2 until convergence occurs. The criterion for convergence is given as:
$$\sum\limits_{\text{k}} \frac{{\left\| {{\text{I}}_{\text{k}}^{{\text{m}} + 1} - {\text{I}}_{\text{k}}^{\text{m}}} \right\|_{2}^{2} }}{{\left\| {{\text{I}}_{\text{k}}^{\text{m}} } \right\|_{2}^{2}}} < \in$$(10)
Feature extraction
A total of 24 number of features having spectral, statistical and, temporal information conte have been extracted in order to classify the ventricular arrhythmia. The following features have been extracted to provide the precise classification results: Leakage Measure (LKG) [21, 26], Normalized spectral moment (FSMN, A1, A2, and A3), Auxiliary counts and Filter coefficient (C1, C2) [21, 26], Frequency Measure (FB) [21, 26], Covariance (CO) [21, 26], Area Measure (AB) [21, 26], Exponential Parameter (CROSS) [21, 26], Modified Exponential (MEA) [21, 26], Threshold crossing interval (TCI) [21, 26], Threshold crossing sample count (TCSC) [21, 26], Hurst Parameter (Hurst) [27], Mean Absolute Value (VAL) [21, 26], Permutation Entropy (Perm) [28], Non-linear Features such as Shannon entropy (Shan_ent), Norm Entropy (Norm_ent), Log Entropy (Log_ent), Threshold Entropy (Th_ent) and, Sure Entropy (Sure_ent) [29], Kurtosis (Kur) [30], Skewness (Skw) [30]. Each and individual features are evaluated by the correlation attribute evaluation method and then ranked by a ranker search algorithm to improve the classification accuracy.
Feature Selection
The time–frequency domain features gather the relevant information in the signal for distinguishing the classes. Due to this reason, the feature selection and ranking play a key responsibility in the success of classification accuracy. To compute the order of variables according to their weightage value, a feature selection procedure called the Gain Ration Attribute Evaluation method was used.
Gain Ratio Attribute Evaluation [31]: The information gain to the intrinsic information ratio is termed as information gain ratio i.e. IGR. Assuming Tr be the training examples set, A be the attributes set, and H be the entropy. The value (q,p) with q ∈ Tr describes the worth of an accurate pattern q for attribute p ∈ A. The information gain for an attribute p ∈ A is given as:
The intrinsic information is given as:
The information gain ratio is specified as:
Classification and Validation
Since last decades, a number of approaches have been reported in the literature for the classification of cardiac arrhythmias using machine learning techniques. In proposed work, the extracted feature set was fed to the SVM and C4.5 classifiers for classification of VF, VT, and NSR arrhythmia conditions.
Support Vector Machine
Support vector machines are the supervised algorithms used in machine learning for signal classification [17]. The decision planes are used to outline the decision having dissimilar class memberships. The hyperplane classifiers draw a line to discriminate between objects based on memberships functions. A hyperplane separates the variables into either class 0 or class 1.
Let the training data with N pairs i.e. \(( x_{1}, y_{1}),( x_{2,} y_{2 }), \ldots \ldots \ldots ( x_{N,} y_{N })\) with \(x_{i} \in {\Re }^{p}\) and \(y_{i} \in\{ { - 1, 1} \}.\)
Let a hyper-plane be \(\left\{ {x:f\left( x \right) = x^T \beta + \beta_0 = 0} \right\}\), where β is a unit vector,
Using WEKA software tool, the tuning parameters of the SVM classifier chosen as:
-
(a)
Batch size is the preferred number of instances to process may be set to 100.
-
(b)
C: It is the complexity parameter. This value has been set to 1.0.
-
(c)
Epsilon: It is used for round-off error. This value has been set to 1.0E−12.
-
(d)
Kernel: It indicates which kernel to use. We have chosen polykernel.
Decision Tree (C4.5) Algorithm
C4.5 is an algorithm is a class of decision tree was developed by Ross Quinlan which is an improved version of the ID3 algorithm [32]. It uses the perception of information entropy to construct decision trees the same as ID3. The classification begins at the root node. At each node, it selects the attribute that divides the data samples into two subsets. The attribute having peak normalized information gain is considered for making decisions. At each node, the information gain for an attribute Z, is estimated as:
where I be the set of instances and |I| be its cardinality, Iv is the subset of I for which attribute Z has value v. The entropy of I is estimated as:
where pi is the amount of instances in I that have the ith class value as output attribute.
The tuning parameters of the C4.5 classifier using WEKA software tool have been chosen by batch size of 100, confidence factor of 0.5 and the minimum instances per leaf is of 2.
Performance Measures
The classification parameters such as sensitivity (Se), specificity (Sp), precision (Pr) and accuracy (Acc) are is calculated using standard equations as follows:
where T_P denotes true positive, \({\text{F}}\_{ }_{\text{N}}\) denotes false negative, \({\text{T}}\_{ }_{\text{N}}\) denotes true negative and \({\text{F}}\_{ }_{\text{P}}\) denotes false positive.
Results
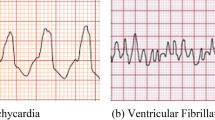
The analysis on ventricular arrhythmias identification and classification was carried out by using all the available ECG signals in CUDB and VFDB databases of PhysioNet repository. The acquired signals are often contaminated with noises and interference; hence a pre-processing stage has been carried out for removal of unwanted artifacts and de-noising of the ECG signals. The de-noised ECG signals are decomposed into different levels using three different decomposition techniques (DWT, EEMD and VMD) and the results are as shown in Figs. 2, 3 and 4 respectively. A set of 24 features has been extracted from the decomposed signals of all the three methods to make a hybrid feature set with large size data of 22,721 × 24. The hybrid feature set is then evaluated with correlation attribute evaluation process along with ranker search method as shown in Table 1. All 24 hybrid features are assigned with a rank according to the weightage of respective features. By taking a threshold value of 0.2, a set of only first 6 features having highest rank has been selected which is as shown in Table 2.
The features set is then classified with SVM and C4.5 classifier for comparative analysis. The best result was obtained with CP = 1 for SVM algorithm for all 24 hybrid features. The results for the evaluation of different feature combinations are as shown in Table 3, which confirms that the combination of all 24 hybrid features taken together gives the highest accuracy rate of 86.69% with a sensitivity of 74.63%. Taking the reduced feature set, the accuracy rate was found to be 80.73% along with the sensitivity of 63.28% as shown in Table 4. The performance analysis for feature set has been done by C4.5 algorithm with different confidence factors (CF) and the best result was obtained with CF = 0.5. Then all 24 features were evaluated for different combinations using a C4.5 algorithm at CF = 0.5 and the evaluated results are as shown in Table 5. It confirms that the larger the number of features higher the classification accuracy but the computational time also increases. By taking all twenty-four features give a high accuracy rate of 99.83% with a sensitivity of 99.75% with a computational time of 4.82 s. In a reduced feature set, the best accuracy rate was found to be 99.62% along with the sensitivity of 99.41% as shown in Table 6. A comparative analysis has been done for both the classifiers by taking all 24 hybrid features and selected high ranked 6 features and the results are as shown in Table 7. By considering all 24 features into account, the hybrid feature set was evaluated by both SVM and C4.5 classifiers. The accuracy rate was found to be 99.83% and 86.99% respectively for C4.5 and SVM classifiers. Figures 5 and 6 have been plotted taking accuracy and sensitivity values for both classifiers using all 24 and 6 selected features respectively. Figure 7 has been plotted for comparative analysis of evaluated parameters of C4.5 classifier using all 24 feature and a reduced set of 6 features. The mean absolute error (MAE) value was found to be 27.48% and 0.31% for SVM and decision tree (C4.5) algorithm respectively. The computational time for both classifiers was also measured and it is found to be 85.9 s and 4.82 s for SVM and C4.5 classifiers respectively for all 24 hybrid features. From the results, it has been observed that the SVM classifier has a very high computational time of 85.9 s for all 24 hybrid features and computational time of 9.35 s for the selected feature set. Figure 8 shows the plot for computational times of both classifiers with all 24 and 6 number of reduced features. Similar observations have also been made that the classification using a reduced feature set gives an accuracy of 99.62% for C4.5 classifier which is lesser than the feature set having all 24 features only by 0.21% with respect to accuracy value, but the computational time was reduced significantly to 2.71 s which earlier was a very high value of 4.82 s in all 24 hybrid features. The computational time is reduced by 43.77% using the selected features. As the computational time is an important parameter in detecting the VAS, therefore the reduced feature sets having important information is a useful approach for diagnosis of heart condition. Therefore, the reduce feature approach is appropriate in detecting VA conditions with faster rate and comparable accuracy.
Discussion
Precise recognition and classification of ventricular arrhythmias are very much crucial for the application of automatic external defibrillation in case of extreme emergency. The main aim of this work was to determine the accuracy to detect VT/ VF suing hybrid dataset and a decision tree classifier. Ciaccio et al. [33] have proposed a quantitative method for atrial fibrillation and ventricular tachycardia analyses using MEDLINE search tool. A convolutional neural network approach has been used classification and achieves an accuracy of 93.18% and sensitivity of 91.04% [34]. A raised cosine wavelet transform (RCWT) has been used for the recognition of VAs which is found to be useful for distinguishing between VF, VT, and atrial fibrillation [35]. Precise discrimination of VT and VF rhythms was done with an accuracy of 94.74% [36]. Balasundaram et al. [29] discriminated the VT, Organized VF and disorganized VF with an accuracy of 93.7% using wavelet analysis and binary classifier. Anas et al. [30] suggested a novel method based on EMD that significantly improved the recognition of the life-threatening cardiac arrhythmias. Tripathy et al. [24] used ECG signals to detect the shockable/non-shockable ventricular arrhythmias by using VMD and random forest (RF) classifier and they achieved an accuracy of 97.23%. In the proposed method, a set of all 24 hybrid features gives the highest accuracy rate of 99.83% with a sensitivity of 99.75% in C4.5 classifier. By taking a threshold value of 0.2, a set of only first 6 high ranked features was evaluated by decision tree classifier. The accuracy rate was of 99.62% along with the sensitivity of 99.41% as described in Table 7. It has been observed that in the reduced feature set the classification accuracy is lesser by only 0.21%, but the computational time has been reduced significantly by 43.77%. Hence, the method with a few selected important features can be a solution in the fastest detection of cardiac arrhythmias. It has been proven that the proposed hybrid method has high detection accuracy with less computational time. Furthermore, this method has significantly low computational complexity and achieves the best performance so far (Table 8).
Conclusions
The automatic arrhythmia classification system is of great significance for the diagnosis of the cardiac diseases. 24 parameters are taken to form a hybrid feature set that includes time–frequency, and statistical domain for classification of normal, VF and VT arrhythmias. The pre-processed ECG signals are decomposed into different levels to extract features using three different transform techniques i.e. wavelet transform, ensemble EMD, and VMD. The proposed system achieves a high classification accuracy of 99.83% in decision tree classifier a short duration (5 s) of ECG data. In the selected high ranked features, the classification accuracy was of 99.62% which is less by only 0.21% with respect to all features. However, this compromise in the accuracy value has been achieved with lesser computational time. The reduced feature set has the computational time about 2.71 s, in case of all 24 parameters based feature was 4.82 s which is reduced by 43.77%. Therefore the proposed method standouts in terms of computational time, as well as in detection success for recognizing the ventricular arrhythmias. The main limitation of this study is the application of deep learning techniques for prediction of VAs which is the future scope of the work using image datasets [37].
References
Dirk H, Bernd P, Hanspeter H, et al. Non-linear coordination of cardiovascular automatic control. IEEE Eng Med Biol. 1998;17(6):17–21.
Leisy PJ, Coeytaux RR, Wagner GS, et al. ECG-based signal analysis technologies for evaluating patients with acute coronary syndrome: a systematic review. J Electrocardiol. 2013;46:92–7.
Brugada J. Relevance of atrial fibrillation classification in clinical practice. J Cardiovasc Electrophysiol. 2002;20:S27–30.
Sun Y, Chan KL, Krishnan SM. Life-threatening ventricular arrhythmia recognition by nonlinear descriptor. Biomed Eng Online. 2005;4:6.
Mathivanan P, Ganesh AB. ECG steganography based on tunable Q-factor wavelet transform and singular value decomposition. Int J Imaging Syst Technol. 2020;31(1):270–87.
Arafat MA, Chowdhury AW, Hasan MK. A simple time domain algorithm for the detection of ventricular fibrillation in electrocardiogram. SIViP. 2011;5:1–10.
Wasan PS, Uttamchandani M, Moochhala S, et al. Application of statistics and machine learning for risk stratification of heritable cardiac arrhythmias. Expert Syst Appl. 2013;40(7):2476–86.
Miquel Alfaras M, Soriano MC, Ortín S. A fast machine learning model for ECG-based heartbeat classification and arrhythmia detection. Front Phys. 2019;7:1–11.
Sahoo S, Das T, Sabut S. Adaptive thresholding based EMD for delineation of QRS complex in ECG signal analysis. In: Int. Conf. Wireless Comm. Sig. Proc. and Network. 2016.
Maji U, Mitra M, Pal S. Characterization of cardiac arrhythmias by variational mode decomposition technique. Biocybern Biomed Eng. 2017;37:578–89.
Rai HM, Trivedi A, Shukla S. ECG signal processing for abnormalities detection using multi-resolution wavelet transform and artificial neural network classifier. Measurement. 2013;46(9):3238–46.
Rakshit M, Das S. An efficient wavelet-based automated R-peaks detection method using Hilbert transform. Biocybern Biomed Eng. 2017;37:566–77.
Nazarahari M, Namin SG, Hossein A, et al. A multi-wavelet optimization approach using similarity measures for electrocardiogram signal classification. Biomed Signal Process Control. 2015;20:142–51.
Zhang K, Aleexenko V, Jeevaratnam K. Computational approaches for detection of cardiac rhythm abnormalities: are we there yet? J Electrocardiol. 2020;59:28–34.
Martis RJ, Acharya UR, et al. Computer aided diagnosis of atrial arrhythmia using dimensionality reduction methods on transform domain representation. Biomed Signal Process Control. 2014;13:295–305.
Garcia M, Rodenas J, Alcaraz R, Rieta JJ. Application of the relative wavelet energy to heart rate independent detection of atrial fibrillation. Comput Methods Programs Biomed. 2016;131:157–68.
Asgari S, Mehrnia A, Moussavi M. Automatic detection of atrial fibrillation using stationary wavelet transform and support vector machine. Comput Biol Med. 2015;60:132–42.
Arafat MA, Sieed J, Hasan MK. Detection of ventricular fibrillation using empirical mode decomposition and Bayes decision theory. Comput Biol Med. 2009;39:1051–7.
Kaur L, Singh V. Ventricular fibrillation detection using empirical mode decomposition and approximate entropy. Int J Emerg Technol Adv Eng. 2013;3:260–8.
Xu Y, Wang D, Zhang W, et al. Detection of ventricular tachycardia and fibrillation using adaptive variational mode decomposition and boosted-CART classifier. Biomed Signal Process Control. 2018;39:219–29.
Abdalla FO, Wu L, Ullah H, et al. ECG arrhythmia classification using artificial intelligence and nonlinear and nonstationary decomposition. Signal Image Video Process. 2019;13:1283–91.
Acharya UR, Fujita H, Sudarshan VK, et al. An integrated index for detection of sudden cardiac death using discrete wavelet transform and nonlinear features. Knowl-Based Syst. 2015;83:149–58.
Zhaohua W, Huang NE. Ensemble empirical mode decomposition: a noise-assisted data analysis method. Adv Adapt Data Anal. 2009;1:1–41.
Tripathy RK, Sharma LN, Dandapat S. Detection of shockable ventricular arrhythmia using variational mode decomposition. J Med Syst. 2016;40:40–79.
Rajesh PS, Dhuli R. Classification of ECG heartbeats using nonlinear decomposition methods and support vector machine. Comput Biol Med. 2017;87:271–84.
Nguyen MT, Shahzad A, Nguyen BV, Kim K. Diagnosis of shockable rhythms for automated external defibrillators using a reliable support vector machine classifier. Biomed Signal Process Control. 2018;44:258–69.
Maji U, Mitra M, Pal S. Automatic detection of atrial fibrillation using empirical mode decomposition and statistical approach. Procedia Technol. 2013;10:45–52.
Acharya UR, Oh SL, Hagiwara Y, et al. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017;89:389–96.
Balasundaram K, Masse S, Nair K, Umapathy K. A classification scheme for ventricular arrhythmias using wavelets analysis. Med Biol Eng Comput. 2013;51:153–64.
Anas E, Lee S, Hasan M. Sequential algorithm for life-threatening cardiac pathologies detection based on mean signal strength and emd functions. Biomed Eng Online. 2010;9(1):43–64.
Novakovic J. Using information gain attribute evaluation to classify sonar targets. In: Telecom. forum TELFOR. 2009. p. 1351–4.
Mantas CJ, Abellán J. Credal-C4.5: Decision tree based on imprecise probabilities to classify noisy data. Expert Syst Appl. 2014;41:4625–37.
Ciaccio EJ, Biviano AB, Iyer V, Garan H. Trends in quantitative methods used for atrial fibrillation and ventricular tachycardia analyses. Inform Med Unlocked. 2017;6:12–27.
Tripathy RK, Zamora-Mendez A, Serna JA, et al. Detection of life threatening ventricular arrhythmia using digital taylor fourier transform. Front Physiol. 2018;9:1–12.
Khadra L, Al-Fahoum AS, Al-Nashash H. Detection of life-threatening cardiac arrhythmias using the wavelet transformation. Med Biol Eng Comput. 1997;35:626–32.
de Gauna SR, Lazkano A, Ruiz J, Aramendi E. Discrimination between ventricular tachycardia and ventricular fibrillation using the continuous wavelet transform. Comput Cardiol. 2004;31:21–4.
Xiao W, Gao Q, Kumar R, et al. Implementation of convolutional neural network categorizers in coronary ischemia detection. Int J Imaging Syst Technol. 2021;31:313–26.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
The paper does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohanty, M., Dash, P. & Sabut, S. Decision Support System for Predicting Ventricular Arrhythmias Using Non-linear Features of ECG Signals. SN COMPUT. SCI. 5, 357 (2024). https://doi.org/10.1007/s42979-024-02718-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02718-3