Abstract
Kallar grass (Leptochloa fusca) is a highly salt-tolerant C4 perennial halophytic forage. The regulation of ion movement across the plasma membrane (PM) to improve salinity tolerance of plant is thought to be accomplished with the aid of the proton electrochemical gradient generated by PM H+-ATPase. In this study, we cloned a partial gene sequence of the Lf PM H+-ATPase and investigated its expression and activity under salt stress. The amino acid sequence of the isolated region of Lf PM H+-ATPase possesses the maximum identity up to 96% to its ortholog in Aeluropus littoralis. The isolated fragment of Lf PM H+-ATPase gene is a member of the subfamily Π of plant PM H+-ATPase and is most closely related to the Oryza sativa gene OSA7. The transcript level and activity of the PM H+-ATPase were increased in roots and shoots in response to NaCl and were peaked at 450 mM NaCl in both tissues. The induction of activity and gene expression of PM H+-ATPase in roots and shoots of Kallar grass under salinity indicate the necessity for this pump in these organs during salinity adaptation to establish and maintain the electrochemical gradient across the PM of the cells for adjusting ion homeostasis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plants are continually exposed to a large variety of biotic and abiotic stresses during their whole life that severely reduces their productivity (Cao et al. 2017). Salinity is a significant abiotic stress affecting plant growth and development and is expected to increase because of global weather changes and as a consequence of many irrigation practices (Reddy et al. 2017). Usually, the high salt concentration can disturb vital physiological processes through inducing low osmotic potential of the soil solution (water deficits), hyperosmotic stress, ion imbalance, nutritional imbalance, metabolic disorders, and even death (Munns and Tester 2008). However, ionic imbalance caused by the accumulation of excessive amounts of Na+ and Cl− ions is a major contributory factor (Acosta-Motos et al. 2017). Under conditions of elevated NaCl levels outside the cell, Na+ excessively accumulates in the cytoplasm, which results in the inhibition of plant growth and development (Janicka-Russak et al. 2013). The reactions of the plant to salinity stress are complex and involving responses to cellular osmotic and ionic stresses, subsequent secondary stresses (e.g., oxidative stress), and whole plant coordination of these responses (Gupta and Huang 2014). The regulation of intracellular ion concentration is an essential strategy for growth in a saline environment. To prevent the accumulation of excessive amounts of Na+ in the cytosol, Na+ is excluded from the cytosol to the apoplast or compartmentalize inside the vacuole utilizing the Na+/H+ antiporters (Park et al. 2016). Elimination of sodium ions out of the cell, catalyzed by the specific PM Na+/H+ antiporter, depends on the membrane proton electrochemical gradient (Blumwald et al. 2000). H+-ATPase is a significant enzyme protein of the plant plasma membrane. It is involved in many physiological processes during plant growth and development, including those related to environmental stress tolerance, intracellular pH regulation, stomatal movements, and cellular expansion (Haruta et al. 2015; Zhu et al. 2015). The PM H+-ATPase uses the energy released upon hydrolysis of ATP to transport proton across the plasma membrane of the cells against chemical and electrical gradients (Sze et al. 1999; Falhof et al. 2015). Using this mechanism, the proton electrochemical gradient, which is generated and maintained across the plasma membrane, provides the driving force for secondary transporters and channel proteins to uptake and efflux of ions and metabolites across the plasma membrane (Palmgren 2001; Almeida et al. 2017). Multiple genes encoding PM H+-ATPase have been cloned in many plants such as Arabidopsis thaliana (Harper et al. 1994), rice (Zhang et al. 1999), tomato (Kalampanayil and Wimmers 2001), Chloris virgata (Zhang et al. 2014), Hevea brasiliensis (Zhu et al. 2015), and potato (Stritzler et al. 2017). The molecular study of plant PM H+-ATPase has shown that this enzyme is encoded by a multigene family organized into five subfamilies according to their amino acid sequence identities and expression profiles (Gaxiola et al. 2007; Zhu et al. 2015). The expression of genes belonging to subfamilies I and П is not restricted to particular organs and highly expressed in all plant tissues. Conversely, expression of genes belonging to the other three groups is more specialized, and their expression is limited to specific organs or cell types and/or to the particular conditions, including salt stress (Pedersen et al. 2012). Several studies have revealed that activity and transcript levels of PM H+-ATPase differentially regulated in response to environmental and hormonal signals, including salinity in both halophytes and glycophytes (Kalampanayil and Wimmers 2001; Janicka-Russak and Kłobus 2007; Sahu and Shaw 2009; Pedersen et al. 2012).
Leptochloa fusca (L.) Kunth, also known as Diplachne fusca, is a C4 perennial halophytic forage plant from the Poaceae family and widely spread in salt-affected regions of Pakistan and India. This forage plant is locally known as Kallar grass. It is an attractive model plant to study the mechanism of salinity tolerance, mainly due to its characteristics as a typical euhalophyte having both accumulating and excreting properties (Adabnejad et al. 2015). In a previous study, we reported the isolation, characterization, and gene expression analysis of PM Na+/H+ antiporter (SOS1) gene from Kallar grass during salinity stress (Taherinia et al. 2015). The extrusion of Na+ through this Na+/H+ antiporter is driven by an inwardly directed proton gradient produced by the PM H+-ATPase (Janicka-Russak and Kabala 2015). However, there are no reports on the isolation and identification of PM H+-ATPase gene from this monocot halophyte. Therefore, to complete the previous study, in the present work, a cDNA fragment representing the partial coding sequence of PM H+-ATPase gene was isolated by a PCR approach, and the effect of NaCl on gene expression and activity of the PM H+-ATPase in the halophyte Leptochloa fusca was studied. The results provide useful information for future studies on the molecular and biochemical basis of the PM H+-ATPase role in salinity tolerance and to gain a better understanding of adaptation to salt stress in this halophyte.
Materials and methods
Plant material, growth conditions, and treatments
The surface-sterilized seeds were planted in 15-cm diameter pots filled with prewashed and nutrient-free sand. Plants were grown in a greenhouse under conditions of a 16:8 h (light:dark) photoperiod at temperature 26 ± 2 ℃ and irrigated daily with half-strength Hoagland’s solution for 60 days before starting salt treatments. After this period, the seedlings were treated with half-strength Hoagland’s solution containing different concentrations of NaCl. To avoid salt shock injuries, salt treatments were gradually stepped up in 75 mM/day increments until final salinity concentrations of 300, 450 and 600 mM were achieved. Control plants were maintained in half-strength Hoagland’s solution without NaCl. For every treatment, three replicates were maintained. Shoots and roots were harvested 3 weeks after final treatment concentrations were achieved, frozen in liquid nitrogen, and kept at − 80 ℃ until enzyme activity and Real-time qRT-PCR analyses.
Determination of Na+ and K+ contents in shoots and roots
After oven-drying, root and shoot samples were ground to a fine powder and burned in 560℃ to obtain ash, which was then digested in 10 mL of 1N HCl. Na and K ion concentrations in the digested samples were determined using a flame photometer (JENWAY, PFP-7, Staffordshire, UK).
Cloning of partial PM H+-ATPase coding sequence
Total RNA was extracted from root tips of Kallar grass by Topazol plus Kit (TopazGene, Iran) according to the manufacturer’s instructions and then was treated with RNase-free DNase I (Thermo Fisher Scientific) to remove DNA contamination. One µg of RNA was reverse-transcribed to its corresponding cDNA according to the instructions of the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The resulted cDNA can finally be used as a template in PCR. The primers were designed using Primer Premier 6 (Table 1) according to conserved regions of PM H+-ATPase orthologs in other members of Poaceae, i.e., Oryza sativa, Triticum aestivum, Zea mays, Hordeum vulgar and Aeluropus littoralis. The PCR was performed in a total volume of 25 μl containing 1 pmol of forward and reverse primer. The PCR condition was 3 min at 94 ℃ (one cycle), 45 Sec at 94 ℃, 45 Sec at 54 ℃, 1 min at 72 ℃ (30 cycles), and 10 min at 72 ℃ (one cycle).
Cloning of PM H+-ATPase amplicon into sequencing vector
To clone the amplicon of PM H+-ATPase into pTZ57R/T vector, InsTAclone™ PCR Cloning Kit (Thermo Fisher Scientific), and competent cells of E. coli strain DH5α were used according to manufacturer’s instructions. In brief, based on blue/white screening, recombinant colonies were selected for DNA extraction by GF-1 Plasmid DNA Extraction Kit (Vivantis, Malaysia). The sequencing of an isolated coding region of the PM H+-ATPase gene was carried out by Faza Pajooh Biotech (Tehran, Iran) using M13 universal primers. The resulted sequence was checked using Chromas Lite 2.01 (Technelysium) after the clipping of vector sequences.
Conserved domains, homology, and phylogenetic analyses
The sequence homology and the deduced amino acid sequence comparisons were carried out using the BLAST algorithm at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast). The conserved domain platform (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and InterProscan (www.ebi.ac.uk/tools/pfa/iprscan), were employed to examine the protein family, domain, or functional site. Gene translation and prediction of the deduced protein were carried out using the EXPASY tool (http://au.expasy.org/translate/). Multiple sequence alignment was performed using BioEdit version 7.1.9. The phylogenetic tree was constructed by the MEGA5 software using the neighbor-joining method with 1000 bootstrap replications.
Gene expression analysis
RNA extraction and cDNA synthesis
Total RNA was isolated from shoots and roots of the control and treated seedlings using Topazol plus Kit (TopazGene, Iran) following the manufacturer’s instructions. RNA quantification was carried out using a Thermo Scientific™ NanoDrop™ OneC Spectrophotometer and gel electrophoresis to test the RNA quality and purity, as described by Sambrook et al. (1989). Next to DNaseI treatment of RNA samples, 1 μg of total RNA was reverse-transcribed to corresponding cDNAs using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific).
Real-time qRT-PCR analysis
Real-Time Quantitative Reverse Transcription PCR was performed on a Bio-RadiQ5 Multicolor Real-Time PCR detection system (BioRad, Hercules, CA, USA) following the manufacturer’s protocol using the SYBR Green PCR Master Mix. The primer pairs used for Real-time qRT-PCR are listed in Table 1. 18SrRNA was used as an internal reference gene (Nicot et al. 2005). Relative expression software tool (REST) analysis provided ratios of expression, statistical analysis, and confidence intervals for the results. The reaction conditions included an initial denaturing step of 95 ℃ for 10 min followed by 40 cycles of 95 ℃ for 15 s, 58 ℃ for 15 s, and 72 ℃ for 30 s, after which a melt curve was produced at 60–95 ℃. Each Real-time qRT-PCR analysis was carried out in triplicates.
PM H+-ATPase activity assay
PM H+-ATPase activity was determined in purified membranes isolated by a two-step aqueous two-phase partitioning system as described by Buckhout et al. (1989) from shoots and roots of Kallar grass seedlings with or without salt treatment. The protein content of the plasma membrane was calculated from a BSA standard curve, as described by Bradford (1976). The hydrolytic activity of PM H+-ATPase was assayed according to the method of Blumwald and Poole (1987) by adding 50 μL (approximately 5 µg) plasma membrane vesicles into 500 μL reaction medium containing 30 mM Hepes–Tris (pH 6.5), 50 mM KNO3, 3 mM MgSO4, 0.1 mM (NH4)2MoO4, 0.1 mM NaN3 and 0.01% (w/v) Triton X-100. The reaction was initiated by adding 3 mM ATP-Na2. After 20 min of reaction at 37 ℃, the reaction was stopped by adding 50 μL 55% (v/v) trichloroacetic acid. Inorganic phosphate hydrolyzed by the plasma membrane vesicles was determined according to Ohnishi et al. (1975), and the activity was expressed as µmol Pi/mg protein/h.
Statistical analysis
Statistical analysis of all parameters was performed using SAS 9.1 software. Averages of three replicates for all data in this study were used. Standard deviations are given as bars. One-way between-subjects ANOVA was conducted to compare the effect of different levels of salinity on Na+ and K+ contents, gene expression, and enzyme activity of Lf PM-H+-ATPase in roots and shoots of Kallar grass. Post-hoc comparisons using Duncan’s multiple range tests (p = 0.05) were used to compare means when ANOVA was significant.
Results
The effect of salt stress on Na+ and K+ accumulation
The amount of Na+ and K+ accumulated in shoots and roots and the Na+/K+ ratio is shown in Table 2. In general, Na+ accumulation exhibited a continuous increase in the shoots and roots of Kallar grass in response to increasing salt stress. In both tissues, the K+ content increased with rising salinity and reached the highest value at 300 mM NaCl and then decreased gradually. The Na+/K+ ratio of Kallar grass gradually increased in both tissues following treatment with increasingly higher concentrations of NaCl. Under the extreme salt stress (600 mM NaCl), Na+ content in both tissues was peaked, thereby resulting in an increase in Na+/K+ ratio by 6.5- and 6.3-fold in shoots and roots, respectively (Table 2).
Conserved domains, homology, and phylogenetic analyses of Lf PM H+-ATPase
Using conserved sequences of PM H+-ATPase, Lf PM H+-ATPase partial sequence (660 bp) was cloned from Leptochloa fusca and deposited in GenBank with accession number KM234610. As prophesied with the ORF finder, the greatest length of the Lf PM H+-ATPase ORF is 615 bp, which encodes a 204 amino acid peptide. According to the results of the conserved domain analysis, the isolated region of Kallar grass PM H+-ATPase is related to HAD_like superfamily (cl21460).
The aa sequence of the isolated Lf PM H+-ATPase fragment comprising 288 aa (GenBank: AIV00616) was used as an initial query to search, using the protein–protein BLAST tool, against the non-redundant protein sequences. As a result, the identity of the deduced amino acid sequence of isolated Lf PM H+-ATPase gene to that of Aeluropus littoralis, Zea mays/Sorghum bicolor, Oryza sativa, Setaria italica, and Triticum aestivum/Hordeum vulgare was 96%, 93%, 91%, 90%, and 84%, respectively (Fig. 1).
Multiple alignment of the deduced amino acid sequence of Kallar grass PM H+-ATPase with other related orthologs of different species using BioEdit version 7.1.9 (Hall 1999). Black blocks indicate highly conserved residues. Accession numbers are Leptochloa fusca (AIV00616), Aeluropus littoralis (AEO22063), Triticum aestivum (AAV71150), Oryza sativa Japonica group (CAD29313), Setaria italica (XP_004960134), Sorghum bicolor (XP_002447249), Arabidopsis thaliana (Q9SJB3), Nicotiana plumbaginifolia (Q03194), Hevea brasiliensis (XP_021660005), and Panicum miliaceum (RLM65155). The red lines show the sequences of the regions which used to design the primers for isolation of partial LfPM H+-ATPase coding sequence
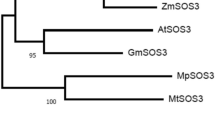
To investigate the relationship between the Kallar grass PM H+-ATPase gene and other P-type ATPase genes, a phylogenetic tree was created using the MEGA 5.1 software by comparing the amino acid sequences of Lf PM H-+ATPase, Nicotiana plumbaginifolia, Oryza sativa, and Arabidopsis thaliana. The results showed that PM H+-ATPase could be divided into five distinct subgroups as described previously (Pedersen et al. 2012; Falhof et al. 2015), in which Lf PM H+-ATPase was classified as a subfamily including OSA7, PMA4, AHA3, AHA1, AHA2, AHA5 and most closely related to the Oryza sativa gene OSA7 (Fig. 2).
Phylogenetic analysis of Leptochloa fusca, Arabidopsis thaliana, Nicotiana plumbaginifolia, and Oryza sativa PM H+-ATPases. To generate the phylogenetic tree, the amino acid sequences of PM H+-ATPase from Kallar grass (KHA), O. sativa (OSA1, OSA2 and OSA4-OSA10), N. plumbaginifolia (PMA1-PMA6, PMA8 and PMA9), and A. thaliana (AHA1-AHA11) was compared using MEGA5.1 software (http://www.megasoftware.net/) and Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is indicated next to the branches. GenBank accession numbers are O. sativa OSA1 (Q43001), OSA2 (Q43002), OSA4 (AJ440002), OSA6 (AJ440217), OSA7 (AJ440218), OSA8 (AJ440219), OSA9 (AJ440220) and OSA10 (AJ440221); N. plumbaginifolia PMA1 (Q08435), PMA2 (Q42932), PMA3 (Q08436), PMA4 (Q03194), PMA5 (AAV49160), PMA6 (Q9SWH2), PMA8 (Q9SWH1), PMA9 (Q9SWH0); and A. thaliana AHA1 (P20649), AHA2 (P19456), AHA3 (P20431), AHA4 (Q9SU58), AHA5 (Q9SJB3), AHA6 (Q9SH76), AHA7 (Q9LY32), AHA8 (Q9M2A0), AHA9 (Q42556), AHA10 (Q43128), AHA11 (Q9LV11); Accession numbers for L. fusca PM H+-ATPase (AIV00616)
Effects of salinity stress on PM H+-ATPase gene expression and activity
The results showed that NaCl significantly induced LfPM H+-ATPase gene expression in shoots [F (3, 8) = 7.27, p = 0.0113], and roots [F (3, 8) = 27.29, p = 0.0001] of Kallar grass at p < 0.05 level (Fig. 3). In shoots, the expression of PM H+-ATPase was increased by 1.15- and 1.4-fold at 300 and 450 mM NaCl, relative to control, respectively. In roots, exposing the plants to 300 and 450 mM NaCl significantly increased the expression levels of PM H+-ATPase by 1.6- and 1.9-fold higher than control, respectively. By increasing the salinity level to 600 mM, the mRNA levels of the PM H+-ATPase down-regulated in both tissues and reached the control level. The NaCl treatments significantly changed the activity of the PM H+-ATPase in shoots [F (3, 8) = 206.32, p ≤ 0.0001] and roots [F (3, 8) = 233.45, p ≤ 0.0001] of Kallar grass at p < 0.05 level (Fig. 4). When seedlings were treated with 300 mM or 450 mM NaCl, the activity of the PM H+-ATPase significantly increased in shoots (+50% and +108% at 300 mM and 450 mM compared to control, respectively) and roots (+75% and +119% at 300 mM and 450 mM compared to control, respectively) tissues. In contrast, the activity of the PM H + -ATPase dramatically decreased at 600 mM NaCl and reached a level below control in shoots and roots. The PM H+-ATPase activity was closely related to the concentrations of NaCl (Fig. 4).
Effects of NaCl stress on the relative mRNA level of LfPM H+-ATPase in the shoots and roots of Kallar grass seedlings after 21 days of treatments. The values are represented as mean ± SD of three replicates. Different letters indicate significant differences at p < 0.05 according to the Duncan’s test
Discussion
To barricade the accumulation of toxic Na+ in the cytosol, active Na+ efflux into the apoplast is one of the main strategies applied by plants to improve salt tolerance (Blumwald et al. 2000). The Na+ efflux from the cytosol can be fulfilled with the Na+/H+ antiporter activity in the plasma membrane (SOS1). The driving force, for Na+ exclusion by this antiporter, was created by the membrane H+-ATPase pump (Shi et al. 2002). In the present study, we for the first time, isolated and characterized a partial cDNA clone of PM H+-ATPase gene from the L. fusca shoots using RT-PCR method and the validated sequence was registered in the GenBank (Accession No. KM234610). The obtained cDNA clone and sequence data of Lf PM H+-ATPase will promote the detection of its full-length. The conserved domain analysis revealed that putative Lf PM H+-ATPase protein is related to haloacid dehydrogenase (HAD) superfamily (cl21460). This superfamily comprises different enzymes, including P-type ATPases and can be involved in a variety of cellular processes ranging from amino acid biosynthesis to detoxification (Marchler-Bauer et al. 2011). The P-type ATPases are a large and vital protein family that pumps lipids and ions across cellular membranes, and can be classified into five subfamilies (P1-P5) (Thever and Saier 2009). P3A-ATPases are PM H+ pumps that transport H (+) from the cytosol to the extracellular space, thus energizing the plasma membrane for the uptake of nutrients and ions and are expressed in plants and fungi (Palmgren 2001; Pedersen et al. 2012). From the homology point of view, the aa sequence of Lf PM H + -ATPase (GenBank: AIV00616) retains considerable identities to its orthologues in other plant species, especially Poaceae family (Fig. 1). Kallar grass is a member of the Poaceae family to which many crops, like wheat, barley, rice, millet, and maize as well belong. Hence, Kallar grass has many characteristics in common with these major crop plants. Determining the mechanisms for salinity tolerance in Kallar grass may lead to the breeding of salt tolerance into cereals and other crop plants in the future. Multiple alignments of the protein sequences of A. thaliana (AHA), N. plumbaginifolia (PMA), O. sativa (OSA), and L. fusca (KHA) PM H+-ATPases were carried out to generate a phylogenetic tree (Fig. 2). The results indicated that PM H+-ATPase could be divided into five distinct subfamilies as previously defined (Zhu et al. 2015; Stritzler et al. 2017), in which KHA was classified as subfamilies II. The continuous accumulation of Na+ observed in L. fusca with increasing external NaCl concentration is a hallmark of halophytes and indicates the active sodium efflux into the apoplast and its compartmentalization inside the vacuole to adapt to saline conditions (Munns and Tester 2008). These results conform with previous findings in other salt-tolerant species such as Salicornia persica (Tale Ahmad et al. 2013) and Halogeton glomeratus (Wang et al. 2015). Halophytic plants, which live in saline environments typically, accumulate high concentrations of inorganic ions (especially Na+) as an osmolyte with low energy cost to lower their osmotic potential in response to increasing salinity (Shabala 2013). The ability of plants to prevent the detrimental effects of salt stress strongly depends on their potassium nutrition and maintenance of a high cytosolic K+ concentration (Almeida et al. 2017). The accumulation of sodium and the concomitant decrease of potassium levels appear to be one of the general characteristics of halophytes (Shabala 2013). Under salinity, the Na+ can compete with K+ for the same binding sites and therefore interferes with potassium transport into the cell using its physiological transport systems (Orlovsky et al. 2016). However, in both tissues of Kallar grass, the K+ content at 300 mM NaCl was increased compared with the control, suggesting at least this concentration of salt probably does not hurt K+ uptake by Kallgrass. The high concentration of Na+ (600 mM) in the growth environment negatively affects the intracellular K+ influx in Kallar grass and decreased K+ content inside the plant and resulted in increased Na+/K+ ratio in shoots and roots compared with control (Table 2). Salt stress significantly increases PM H+-ATPase expression and activity in roots and shoots of Kallar grass, suggesting a requirement for this pump in these organs during salt adaptation. PM H+-ATPase plays a principal role in physiological functions and adaptation of plants to changing conditions, specifically stress. Thus, PM H+-ATPase may be generally substantial to environmental stress resistance (Palmgren and Nissen 2011). Our results were consistent with previous reports that salinity stimulates the gene expression or hydrolytic activity of PM H+-ATPase in roots and shoots of other halophytes (Niu et al. 1993; Sibole et al. 2005; Chen et al. 2010; Olfatmiri et al. 2014). The increased activity of the PM H+-ATPase in roots and shoots of Kallar grass, followed an increase in the transcript levels of the enzyme, may reflect that the stimulation of PM H+-ATPase activity in roots and shoots could partially have emanated from increased transcript levels. Moreover, in the current study, at all salinity levels, the expression levels and activity of PM H+-ATPase were higher in roots than in shoots. The high degree of NaCl induction of the PM H+-ATPase expression/activity in roots compared to shoots could contribute to restriction of the toxic ions’ transport to the photosynthesizing tissues and adjustment of ionic homeostasis. Under salinity stress, the ion homeostasis was disturbed in root tissues. Increasing pump activity due to the accumulation of gene transcripts was required for the establishment and maintenance of the electrochemical potential gradient across the plasma membrane of epidermal, cortical, and endodermal cells. This electrochemical gradient prevented further apoplast transport of Na+ and Cl− into the xylem, through which the ion will be translocated into the shoots (Falhof et al. 2015). The gene expression level and enzyme activity of Lf PM H+-ATPase were gradually increased with the increased levels of salinity, peaked in 450 mM NaCl, and after that reduced at 600 mM NaCl. It appears that the activity of this protein is required for ion homeostasis in L. fusca under 450 mM NaCl. However, at the higher salt concentration (600 mM), the plant requires more energy to cope with the existing condition, so the amount of protein reduces to minimize the cost of energy consumed by proton pumps, thus enhancing the overall suitability of the plant. Besides, high salt concentrations may disrupt the cell membrane and hence reduce the need for PM H+-ATPase activity and its expression. Our previous studies showed that the transcript level of the PM Na+/H+ antiporter (SOS1) was up-regulated in shoots and roots of L. fusca in response to salt stress (Mohammadi et al. 2019). The coordinate induction of SOS1 and PM H+-ATPase supports this idea that Kallar grass has a regulated mechanism that controls Na+ influx and efflux across the plasma membrane and explains the ability of this halophyte to survive and keep growth under high salt concentrations.
Conclusion for future biology
To avoid the toxic effects of salt, Na+ ions are compartmentalized into vacuoles, or extruded to the apoplast, and remain at low concentrations in the cytosol. The PM Na+/H+ antiporter and the PM H+-ATPase pump can mediate Na+ efflux from the cytosol and contribute to Na+ tolerance. Salt stress increased Lf PM H+-ATPase expression and activity, which suggests that PM H+-ATPase could play an essential role in maintaining ion homeostasis and salt tolerance of this monocot halophyte. Since most of the agriculturally important plants are monocot glycophytes, studying a closely related halophyte might be an effective way to unravel and improve salt tolerance in crops. Lf PM H+-ATPase gene isolated from this grass could be a potential candidate gene for enhancing salt tolerance in various crops through genetic engineering in the future.
References
Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco MJ, Hernandez JA (2017) Plant responses to salt stress: adaptive mechanisms. Agronomy 7:1–38. https://doi.org/10.3390/agronomy7010018
Adabnejad H, Kavousi HR, Hamidi Ravari H, Tavassolian I (2015) Assessment of the vacuolar Na+/H+ antiporter (NHX1) transcriptional changes in Leptochloa fusca L. in response to salt and cadmium stresses. Mol Biol Res Commun 4:133–142. https://doi.org/10.22099/mbrc.2015.3115
Almeida DM, Oliveira MM, Saibo NJM (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40:326–345. https://doi.org/10.1590/1678-4685-GMB-2016-0106
Blumwald E, Poole RJ (1987) Salt Tolerance in suspension cultures of sugar beet: induction of Na+/H+ antiport activity at the tonoplast by growth in salt. Plant Physiol 83:884–887. https://doi.org/10.1104/pp.83.4.884
Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochim Biophys Acta 1465:140–151. https://doi.org/10.1016/s0005-2736(00)00135-8
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Buckhout TJ, Bell PF, Luster DG, Chaney RL (1989) Iron-stress induced redox activity in tomato (Lycopersicum esculentum Mill.) is localized on the plasma membrane. Plant Physiol 90:151–156. https://doi.org/10.1104/pp.90.1.151
Cao XQ, Jiang ZH, Yi YY, Yang Y, Ke LP, Pei ZM, Zhu S (2017) Biotic and abiotic stresses activate different Ca2+ permeable channels in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00083
Chen M, Song J, Wang BS (2010) NaCl increases the activity of the plasma membrane H+-ATPase in C 3 halophyte Suaeda salsa callus. Acta Physiol Plant 32:27–36. https://doi.org/10.1007/s11738-009-0371-7
Falhof J, Pedersen T, Fuglsang AT, Palmgren M (2015) Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol Plant 9:323–337. https://doi.org/10.1016/j.molp.2015.11.002
Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581:2204–2214. https://doi.org/10.1016/j.febslet.2007.03.050
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. https://doi.org/10.1186/s12864-020-6524-1(Article ID 701596)
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. https://doi.org/10.14601/Phytopathol_Mediterr-14998u1.29
Harper JF, Manney L, Sussman MR (1994) The plasma membrane H+-ATPase gene family in Arabidopsis: genomic sequence of AHA10 which is expressed primarily in developing seeds. Mol Gen Genet 244:572–587. https://doi.org/10.1007/bf00282747
Haruta M, Gray WM, Sussman MR (2015) Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Curr Opin Plant Biol 28:68–75. https://doi.org/10.1016/j.pbi.2015.09.005
Janicka-Russak M, Kabala K (2015) The role of plasma membrane H+-ATPase in salinity stress of plants. In: Luttage U, Beyschlag W (eds) Progress in Botany 76. Springer International Publishing, Switzerland, pp 77–92
Janicka-Russak M, Kłobus G (2007) Modification of plasma membrane and vacuolar H+-ATPases in response to NaCl and ABA. J Plant Physiol 164:295–302. https://doi.org/10.1016/j.jplph.2006.01.014
Janicka-Russak M, Kabała K, Wdowikowska A, Klobus G (2013) Modification of plasma membrane proton pumps in cucumber roots as an adaptation mechanism to salt stress. J Plant Physiol 170:915–922. https://doi.org/10.1016/j.jplph.2013.02.002
Kalampanayil BD, Wimmers LE (2001) Identification and characterization of a salt stressed-induced plasma membrane H+-ATPase in tomato. Plant Cell Environ 24:999–1005. https://doi.org/10.1046/j.1365-3040.2001.00743.x
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:225–229. https://doi.org/10.1093/nar/gkq1189
Mohammadi F, Kavousi HR, Mansouri M (2019) Effects of salt stress on physio biochemical characters and gene expressions in halophyte grass Leptochloa fusca (L.) Kunth. Acta Physiol Plant 41:143. https://doi.org/10.1007/s11738-019-2935-5
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914. https://doi.org/10.1093/jxb/eri285
Niu X, Narasimhan ML, Salzman RA, Bressan RA, Hasegawa PM (1993) NaCl regulation of plasma membrane H+-ATPase gene expression in a glycophyte and a halophyte. Plant Physiol 103:713–718. https://doi.org/10.1104/pp.103.3.713
Ohnishi T, Gall RS, Mayer ML (1975) An improved assay of inorganic phosphate in the presence of extralabile phosphate compounds: application to the ATPase assay in the presence of phosphocratine. Anal Biochem 69:261–267. https://doi.org/10.1016/0003-2697(75)90585-0
Olfatmiri H, Alemzadeh A, Zakipour Z (2014) Up-regulation of plasma membrane H+-ATPase under salt stress may enable Aeluropus littoralis to cope with stress. Mol Biol Res Commun 3:67–75. https://doi.org/10.1016/j.sjbs.2014.05.001
Orlovsky N, Japakova U, Zhang H, Volis S (2016) Effect of salinity on seed germination, growth and ion content in dimorphic seeds of Salicornia europeae L. (Chenopodiaceae). Plant Diversity 38:83–189. https://doi.org/10.1016/j.pld.2016.06.005
Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52:817–845. https://doi.org/10.1146/annurev.arplant.52.1.817
Palmgren MG, Nissen P (2011) P-Type ATPases. Annu Rev Biophys 40:243–266. https://doi.org/10.1146/annurev.biophys.093008.131331
Park HJ, Kim WY, Yun DJ (2016) A new insight of salt stress signaling in plant. Mol Cells 39:447–459. https://doi.org/10.14348/molcells.2016.0083
Pedersen CNS, Axelsen KB, Harper JF, Palmgren MG (2012) Evolution of plant P-type ATPases. Front Plant Sci 3:31. https://doi.org/10.3389/fpls.2012.00031
Reddy INBL, Kim BK, Yoon KK, Kim KH (2017) Salt tolerance in rice: focus on mechanisms and approaches. Rice Sci 24:123–144. https://doi.org/10.1016/j.rsci.2016.09.004
Sahu B, Shaw B (2009) Salt-inducible isoform of plasma membrane H+-ATPase gene in rice remains constitutively expressed in natural halophyte, Suaeda maritima. J Plant Physiol 166:1077–1089. https://doi.org/10.1016/j.jplph.2008.12.001
Sambrook J, Fritsch EF, Maniatist T (1989) Molecular cloning: a laboratory manual, vol 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor,NY, p 626
Shabala S (2013) Learning from halophytes: physiological basis and strategies to improve stress tolerance in crops. Ann Bot 112:1209–1221. https://doi.org/10.1093/aob/mct205
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477. https://doi.org/10.1105/tpc.010371
Sibole VS, Cabot C, Michalke W, Poschenrieder C, Barcel OJ (2005) Relationship between expression of the PM H+-ATPase, growth and ion partitioning in the leaves of salt-treated Medicago species. Planta 221:557–566. https://doi.org/10.1007/s00425-004-1456-6
Stritzler M, Muñiz García MN, Schlesinger M, Cortelezzi JI, Capiati DA (2017) The plasma membrane H+-ATPase gene family in Solanum tuberosum L. Role of PHA1 in tuberization. J Exp Bot 68:4821–4837. https://doi.org/10.1093/jxb/erx284
Sze H, Li X, Palmgren MG (1999) Energization of plant cell membranes by H+-pumping ATPases: regulation and biosynthesis. Plant Cell 11:677–689. https://doi.org/10.1105/tpc.11.4.677
Taherinia B, Kavousi HR, Dehghan S (2015) Isolation and characterization of plasma membrane Na+/H+ antiporter (SOS1) gene during salinity stress in Kallar grass (Leptochloa fusca). Eurasia J Biosci 9:12–20. https://doi.org/10.5053/ejobios.2015.9.0.2
Tale Ahmad S, Kosh Kholgh Sima NA, Mirzaei HH (2013) Effect of sodium chloride on physiological aspects of Salicornia persica. J Plant Nutr 36:401–414. https://doi.org/10.1080/01904167.2012.746366
Thever MD, Saier MH Jr (2009) Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. J Membr Biol 229:115–130. https://doi.org/10.1007/s00232-009-9176-2
Wang J, Meng Y, Li B, Ma X, Lai Y, Si E, Yang K, Xu X, Shang X (2015) Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ 38:655–669. https://doi.org/10.1111/pce.12428
Zhang JS, Xie C, Li Z, Chen SY (1999) Expression of the plasma membrane H+-ATPase gene in response to salt stress in a rice salt-tolerant mutant and its original variety. Theor Appl Genet 99:1006–1011. https://doi.org/10.1007/s001220051408
Zhang X, Wang H, Liu S, Takano T (2014) Cloning and characterization of a plasma membrane H+-ATPase (PMA) gene from a salt-tolerant plant Chloris virgata. Mol Soil Biol 5:16–22. https://doi.org/10.5376/msb.2014.05.0003
Zhu J, Xu J, Chang W, Zhang Z (2015) Isolation and expression analysis of four members of the plasma membrane H+-ATPase gene family in Hevea brasiliensis. Trees 29:1355–1363. https://doi.org/10.1007/s00468-015-1213-4
Acknowledgements
The authors are grateful to the Shahid Bahonar University of Kerman, Kerman, Iran for financially supporting this research.
Funding
None.
Author information
Authors and Affiliations
Contributions
HHR and FM performed most of the experiments. HRK supervised the experimental design and wrote the manuscript. SP did some of the experimentation and provided reagents and materials. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflict of interests associated with the manuscript.
Rights and permissions
About this article
Cite this article
Ravari, H.H., Kavousi, H.R., Mohammadi, F. et al. Partial cloning, characterization, and analysis of expression and activity of plasma membrane H+-ATPase in Kallar grass [Leptochloa fusca (L.) Kunth] under salt stress. BIOLOGIA FUTURA 71, 231–240 (2020). https://doi.org/10.1007/s42977-020-00019-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-020-00019-3