Abstract
Salinity tolerance has been documented as one of the main abiotic stresses, disapprovingly impeding the growth and fertility of core food crops including rice in the ecosphere. We have evaluated 24 varieties exposed to salt stress (150–300 mM NaCl; for 48–72 h) at the vegetative stage. The cellular-level physiological response was observed by assessing the morphophysiological parameters and significant differences were observed in the rice varieties of the Gujarat region of the Indian continent. Additionally, we have performed a comparative transcriptome analysis of the shoots and roots of rice varieties at the vegetative stage (Dandi, salinity-tolerant; and GR-11, Salinity-sensitive) in response to salinity. Based on transcriptome analysis, the genes specifically activated and differentially over-expressed during salinity stress belong to different functional categories mostly located on chromosomes 1, 2, 3, 4, 8, and 11 in both cultivars. A total of 617 and 676 significant DEGs with log2-fold change in the range of ≥ + 2.0 to ≤ − 2.0 were identified from the comparative analysis of the two data sets DS (Dandi Shoot) and GS (GR-11Shoot) (Dandi vs. Control, GR-11 vs. Control) in the shoot sample. Similarly, a total of 510 and 508 significant DEGs DR (Dandi Root) and GR (GR-11 Root) (Dandi vs. Control, GR-11 vs. Control) in the root samples. The DEGs were examined against the KEGG database to explore the biological pathways of Dandi (226) and GR-11 (397) in the shoot as well as root (218,285) samples. Further PageMan analysis showed that there was a significantly higher number of DEGs (p < 0.05, ≥ ± 2.0-fold) involved in signalling pathways including secondary metabolism, hormone metabolism, abiotic stresses, and development which have significant regulatory functions in shoot and root of Dandi and GR-11 variety of rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
By generating yield loss in cereal crops, salinity has become one of the key stress factors posing a serious threat to long-term agricultural productivity and global food security. By the year 2050, scientists predict that half of all cultivable lands would be under serious threat. In regards to this major issue in cereal crops, rice is more sensitive to salt stress at the early seedling and vegetative stage as compared with the tillering stage (Grattan et al. 2002; Shereen et al. 2005). Vegetative growth and fresh weight decreased with increased salt stress from 5 to 7.5 dS m−1 (Kazemi and Eskandari 2011). Various greenhouse and field trials also revealed that salt stress decreases rice stand density and vegetative biomass production (Zeng and Shannon 2000). Oryza sativa. L (rice), one of the most cultivated crops (especially in Asia), and a staple crop in South India are sensitive to salt stress (Challabathula et al. 2022). Although its sensitivity has limited its productivity to a certain rate, it has led to the breeding of several salt-tolerant cultivars (Ganie et al. 2021; Khan et al. 2021). Salt-induced osmotic stress is the initial effect of salinity in rice. Later, it is followed by ionic stress due to the accumulation of toxic levels of Na + ions. It has been found that Saltol QTL contributes profoundly to salinity stress tolerance at the seedling stage by maintaining the Na +/K + homeostasis (Ganie et al. 2016). Due to this, oxidative stress is worsened by the accumulation of reactive oxygen species (ROS) that are mostly produced during electron transport in the mitochondria, chloroplast, and peroxisomes (del Río et al. 2006; Pospíšil 2012; Verma et al. 2019). Apart from the ROS group, a decline in photosynthesis rate and chlorophyll content sideways with a disruption of chloroplast structure is the most commonly observed phenomenon during salt stresses (Hu et al. 2020; Wang and Blumwald 2014). While cellular damage and cell death are frequently observed in salt-sensitive plants upon exposure to extreme salt conditions, the tolerant cultivars respond to these stresses with a multitude of mechanisms that enable them to withstand the deleterious effects of salinity. Several recent studies (Ma et al. 2020) have shed light on the cellular and molecular mechanisms by which rice responds to salt and develops tolerance to it.
On the other side, RNA-seq was utilized to detect genome-wide gene expression changes under salt stress and predicted potential molecular networks in rice (Kong et al. 2020; Mirdar Mansuri et al. 2020). Many differentially expressed genes (DEGs) have been identified among the contrasting samples through RNA-sequencing analysis (Kong et al. 2021a). This approach is effective in the detection of novel transcripts, which is one of the main findings of salinity stress-related studies (Kong et al. 2021b). A few salt transcriptome analyses dependent on RNA-Seq have been conducted with root samples. For instance, a meta-analysis of different rice genotypes at the vegetative stage based on 96 public microarray datasets, aims to identify the key salt-responsive genes and understand the molecular response mechanism of rice under salt stress. The recognized salt stress genes have been verified and a range of new salt-responsive genes are found (Ganie et al. 2021). The analysis reveals most notably that salinity translation regulations are an essential aspect of Pokali salt tolerance, but this control is less apparent in IR 29 which is salt-sensitive. In previous studies, 5559 genes were identified to be differentially expressed genes (DEGs) under salt stress, and 3210 DEGs were identified during the recovery process (Ibrahim et al. 2019). Similarly, Pokali is more effective in the maintenance of the integrity of the cell wall, detoxification of reactive oxygen, molecules translocation, and photosynthesis under salt stress.
The rice cultivar of the Gujarat region, Dandi has several other differentially expressed genes (s) during salinity stress which could be regulated under biotic stress conditions. More so, during exposure to abiotic stress, such regulatory genes are also upregulated. Furthermore, the Dandi cultivar is resistant to BLB and Blast, and moderately resistant resistance to sheath rot and WBPH. Due to multiple resistant mechanisms, Dandi can give the most significant impact on crop improvement through marker-assisted selection approaches.
Our gene expression studies focused on 24 cultivars comprising 14 genotypes in the initial stage, all these were initially morpho physiologically characterized and exposed to salt stress to identify the tolerance, further two varieties of rice belonging to the japonica group, demonstrated noteworthy differences in tolerance to salinity, were selected and analyzed to reveal the mechanisms responsible for salt tolerance. The comparison was performed at the vegetative stage, which is one of the two most salt-sensitive growth phases in rice. Analysis of tolerance mechanisms during the most sensitive stages of growth is considered essential to provide new information on how to generate salt-tolerant rice plants (Ismail and Horie 2017). Hence, molecular analysis was performed using RNA-seq to identify the metabolic and signalling pathways responsible for the physiological responses observed in the two varieties which respond to salinity differently. Keeping in view the research was carried out on rice for physiological characterization and transcriptome analysis involving qRT-PCR analysis in response to salinity stress. Furthermore, this piece of research may serve as a preliminary study for the elucidation of the molecular mechanism underlying the differences in the salt stress responses of the studied genotypes.
Materials and methods
Plant materials, growth conditions, and stress treatments
The Main Rice Research Station, Anand Agricultural University, Nawagam (14 genotypes), Central Soil Salinity Research Institute, Karnal (6 genotypes), and Kerala Agricultural University, Kerala (4 genotypes) supplied rice (Oryza sativa L.) seeds (Table 1). Rice seeds from different sources were surface sterilized as described by Ahmad et al. (2016). After imbibition, seeds were transplanted into Hoagland solution for 45 days, with the amount of iron doubled, in the hydroponic container. All the plants were grown at 28.8 °C/25.8 °C (50% humidity, 16-h-light/8 h—dark cycle; 700 mmol photons m22 sec21). Salt stress was initiated 120 h after the start of the light period by adding 150 mM NaCl in dilute (one-fourth) Hoagland solution, and after three days seedlings were transferred to 250 mM of NaCl (for 72 h) nutrient solution, and the latter one to 300 mM of NaCl for 48 h.
Two sets of the plantlets were grown; one was for the salinity treatment and the other one was for collecting control samples. After 45 days 150 mM of salt stress (NaCl) was applied for 24 h. Similarly at the same time interval 250 mM (48 h), and 300 mM (72 h.) treatments were given to individual plants. Samples (replicated pools of 3) were collected at three time points, at 24 h, 48 h, and 72 h after applying the targeted salt (NaCl) stress. Control samples or samples without salinity treatment were also collected at both time points. Shoot and root tissues were collected separately. We collected the whole shoot and root materials and extracted RNA for sequencing.
Furthermore, a diagrammatic depiction of the overall experimental design is presented (Fig. 1). Additionally, morphological parameters like fresh dry weight shoot length and root length (Mahmud et al. 2016), Photosynthetic rate per unit area (stomatal conductance and evaporation rate of the leaves were measured under different salinity treatments (Munawar et al. 2021). In addition to this, the shoot and root Na+/K+ concentration was also determined by a flame photometer (Asch et al. 2000). Standard evaluation system (SES) for salinity tolerance scoring was done when the salt-sensitive check was almost going to die or already dead. In this experiment, salt-sensitive check, GR-11 was dead after two weeks of salinization (Table 1). Scoring was done according to the modified standard evaluation system used in the rating of the visual symptoms of salt toxicity injury (IRRI 1967).
Diagrammatic depiction of the overall experimental design. In the first part, the group of tolerant and sensitive plants was categorized by using physiological factors for screening purposes. The experimental design has been shown sequentially from sample collection to RNA isolation and further analysis in separate sections. (Note This figure is inspired by the figure which was depicted in a article by Razzaque et al. 2019)
Stress susceptibility index and stress tolerance index
Stress susceptibility index (SSI) was calculated for the tested genotypes under 150, 250, and 300 mM salt stress and non-stress conditions by using the formula given by Fisher and Maurer (Askari et al. 2016) and stress tolerance index (STI) was calculated for the determination of stress tolerance potential using the formula given by Fernandez (1993).
RNA extraction, cDNA library preparation, and sequencing
Total RNA was extracted for leaf and root tissues collected from the vegetative parts for three biological replicates using the TRIzol kit (Invitrogen). Total RNA was then purified and checked using Bioanalyzer 2100 (Agilent). Each of the four libraries was quantified using the TBS 380 Fluorometer. The standard curve was generated, and library sample concentration was determined using Rapid Library Quantification Calculator while quality and quantity assessments were done by running 1 μl of single-stranded DNA library on an Agilent 2100 DNA High Sensitivity chip on Agilent 2100 Bioanalyzer as per the manufacturer’s protocol. Sequencing of clone amplified bead-immobilized DNA library was carried out by using GS FLX Titanium Sequencing Kit XLR70 on Genome Sequencer FLX Instrument using Pico Titer Plate Kit 70 × 75 mm.
Processing of RNA-sequencing data and differential expression analysis
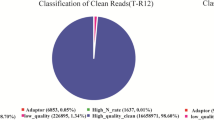
Adaptor sequences, empty reads, low-quality sequences with an 'N' percentage greater than 10%, and those containing more than 50% bases with a Q-value greater than 20% were eliminated using a Perl program built using the custom way of Programme editing. Filtered reads, were called clean reads, and used for downstream bioinformatics analysis using CLC genomics workbench 4.9. The retained high-quality reads were mapped to the reference mapped with available rice chromosomes (https://rapdb.dna.affrc.go.jp). Expression levels for each gene were calculated by quantifying the reads according to the RPKM (reads per kilobase per million reads) method. We used ‘FDR (false discovery rate) 0.001 and the absolute value of log2 RPKM ratio 1’ as the threshold to judge the significance of gene expression difference.
The quality of the fastq raw reads from sequencing data was checked using FastQC (Andrews et al. 2010). Further, the paired-end reads were filtered to eliminate the low-quality reads via a quality filter using Trimmomatic 0.36 (Bolger et al. 2014) with rigorous filtering criteria. The clean reads were mapped to the reference genome Oryza sativa IRGSP-1.0 (https://phytozome.jgi.doe.gov/pz/portal.html) genome using hisat2 (Kim et al. 2015). For evaluation of gene expression levels, Per Kilobase of exon per Million fragments mapped (FPKM) method was (Trapnell et al. 2010) using StringTie (Pertea et al. 2015). The differential expression analysis was done using edgeR (Anders and Huber 2010). The threshold for significant differential expression, log2 (Fold Change) of < − 2 or > 2 were set. The log2 (Fold change) values were used to generate the heatmap using the p-heatmap package in R. The annotation of differentially expressed genes (DEGs) was carried out with Oryza sativa IRGSP-1.0 (https://phytozome.jgi.doe.gov/pz/portal.html). PageMan (http://mapman.mpimp-golm.mpg.de/pageman/). The current analysis was performed to display the expression profiles of different clusters of differentially expressed genes (DEGs) (Kong et al. 2019).
Gene ontology (GO) and KEGG pathway enrichment analysis
To study the gene function, a GO study of differentially expressed genes was carried out and for the metabolic pathways of significant genes, the KEGG analysis was performed. Gene ontology (GO) enrichment analysis of differentially expressed genes was done by agriGo v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/) (Tian et al. 2017). A corrected p value of < 0.05 was significantly enriched by differentially expressed genes in the Go term. For KEGG enrichment analysis, a p value < 0.05 was used as the screening threshold.
Expression studies by real-time quantitative PCR (qRT-PCR) validation
The qRT-PCR reaction was performed based on SYBR Green chemistry using SYBR® GreenER™ qPCR SuperMix Universal Kit (Invitrogen), step one real-time PCR system (Applied Biosystem, California, USA.) and using the following program—one cycle of 95 °C for 10 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Out of three different endogenous specific primers Tubulin (Tub) was selected by screening against the cDNA pool by PCR. This endogenous gene was subjected to qRT-PCR and then the stability of their expressions was checked through geNorm and RefFinder (Reddy et al. 2016). The relative quantification methodology was used to obtain the Ct values of respective genes. The relative quantification of gene expression was calculated with kinetic PCR efficiency correction using the comparative Ct method (2(−ΔΔCt) to determine the relative expression of transcripts relative to endogenous control.
Results
Morphological parameters
Root and shoot lengths
The growth parameters root and shoot lengths were not significantly affected by NaCl treatment, except for Vytilla-1 and Narmada in which the shoot length was significantly increased at 300 mM compared to that in non-stress conditions. However, most of the genotypes had not shown considerable changes in the root (Supplementary S1-a) and shoot lengths (Supplementary S1- b). Based on STI, Vytila-3, GAR-13, CSR-30, and CSR-23 were found to be tolerant genotypes for root length under stress conditions. Cell activities are affected by salt stress, which leads to a diminution of root, and leaf development, and yield. The significant reduction in mean root length, mean root numbers per plant, and shoot length occurred under increased salt stress. Thus, root and shoot lengths are two indicators of rice plant response to salt stress. All the accessions were also cross-checked with SSI. In fewer than 300 mM salt stress conditions, CSR-10, GR-7, and Vytilla-1 were found to be susceptible to root length (Supplementary S2-a). Similarly, IR-64, CSR-30, Vyitlla lines, GAR-13, Pokkali, Dandi, and GR-11 showed more STI values for shoot length under stress conditions Supplementary S2-b).
Plant fresh and dry weights
In rice plants, NaCl treatment significantly reduced plant biomass in five genotypes CSR-10, Dandi, CSR-30, Jaya, and GR-12, and a similar trend was also noted in other genotypes. However, GR-12, CSR-10, and CSR-30 had the least difference in fresh weight between non-NaCl and NaCl-treated plants (Supplementary S1-c). Plant fresh weight was significantly reduced in GR-7. Similarly, based on SSI and STI, all were found to be susceptible to fresh weight and GR-9, GR-7, Vytilla-2, Vytilla-1, IR-64, and Dandi for dry weight under stress conditions (Supplementary S2-a and Supplementary S2-b). The fresh and dry weights were least affected by salt stress in GR-7 and Vytilla lines, except for GR-11, which showed susceptibility under 300 mM salt stress for the dry weight (Supplementary S1-d). Although salt stress had a significant effect on plant biomass, Vytilla lines, Dandi, and CSR series improved significantly by maintaining their dry weights compared to that under control. Salinity generally reduces the shoot growth of crops more than root growth. The results indicated the effect of salinity on the plant height of different cultivars which might be due to the genetic potentiality of the varieties. Tree diagrams based on different morphological, physiological parameters, and nutrient concentrations for different rice genotypes under control and salt stress conditions are depicted in supplementary data (S3–14).
Physiological parameters
Stomatal conductance and photosynthetic rate
Salt stress significantly decreased stomatal conductance in GR-11, GR-7, Narmada, and IR-64 (Supplementary S1- e). However, stomatal conductance was maintained in Dandi, Pokkali, CSR, and Vytilla lines under stress and non-stress conditions. The photosynthetic rate was maintained in all the genotypes except for GR-6, GAR-13, CSR-10, Gurjari, GR-12, and CSR-10 (Supplementary S1- f). Photosynthetic pigments play a key function in photosynthesis by harvesting light energy into chemical energy, but salinity stress causes critical pigment damage. Also, it has been well established that photosynthetic activity, one of the most important processes for plant production, is affected by many environmental factors. In this study, we found that salinity stress decreased the photosynthetic CO2 fixation rate in rice cultivars. Stomatal conductance and transpiration rate were also highly decreased. Based on SSI, GR-11, GR-7, and IR-64 were found to be more sensitive to fewer than 300 mM salt stress conditions (Supplementary S2-a).
Transpiration rate and chlorophyll content
Mostly in genotypes, salt stress decreased the transpiration rate. However, the least differences in salt stress for transpiration rate were observed in 10 genotypes. Nevertheless, in IR-64, the transpiration rate was observed as increased under 300 mM salt stress (Supplementary S1- g). Thus, based on SSI and STI, vytila, GR-12, and CSR lines were found to be slightly tolerant at less than 150 mM but sensitive to severe 300 mM dS/m salt stress (Supplementary S2-a and Supplementary S2-b). Similarly, the transpiration rate was significantly decreased in GR-12 under 150 mM salt stress (Supplementary S1- h). However, SLR, GR-104, Gurjari, GR-7, GAR-1, and Vytilla-2 maintained transpiration rates, which showed that they were the least affected by salt stress. The value of chlorophyll parameters was widely distributed in each subgroup, which could reflect the extensive genetic variation in photosynthetic efficiency in rice. Chlorophyll content did not differ significantly in all the genotypes between salt-treated and non-treated plants before and after salt stress was applied (chlorophyll content data before salt stress were not provided). This indicated that NaCl does not affect the efficacy of light during photosynthesis.
Na+ and K+ influx in rice shoot and root under salinity stress
Ionic homeostasis (Na +/K + Ratio)
Ionic stress is one of the most important components of salinity and is brought about by excess Na + accumulation, especially in the aerial parts of plants. Since Na + interferes with K + homeostasis, and especially given its involvement in numerous metabolic processes, maintaining a balanced cytosolic Na+/K+ ratio has become a key salinity tolerance mechanism (Assaha et al. 2017). NaCl accumulation negatively affects ion homeostasis, i.e., Na +, K +, and Na +/K + ratio. Na + content was found to be increased in NaCl-treated plants, especially under the increased salt concentration, i.e., 300 mM. Significant differences in Na +, K +, and Na +/K + ratios were observed among the genotypes. In GR series and CSR lines, ions accumulated more in leaves as compared to K + ions, and an increase in Na +/K + ratio was observed in root and shoot (Supplementary S-3). The maximum value was recorded for K + ion in Pokkali under the 300 mM salt stress condition of the shooting part. Moreover, K + content in the root of rice was maintained in GR-7, Narmada, GR-9, GAR-13, and CSR-13. Salt stress considerably increased Na + content, but decreased K +, in shoots and roots of all rice cultivars compared to the non-salt control. Genotypic differences in Na + and K + contents under salt stress were more obvious in shoots than in roots, indicating a significant difference in shoot Na + translocation among these cultivars. The most explored salt tolerance trait is the ability to maintain ion homeostasis, in terms of a high K +/Na + ratio in the cytosol and low Na + content in leaves, by activating K + and Na + transport systems, both at the local level (i.e., root cells directly affected by the salt) or systemically (reduction of sodium in the xylematic flow).
Correlation analysis among Morpho-physiological traits
Correlation (Pearson test) for morpho-physiological traits under control, and salt stress conditions was performed (Supplementary S-4) by using XLSTAT 2012.1.02 with a 95% confidence interval details are explained in Supplementary data files (Supplementary File S 5 to S12). Moreover, ANOVA for all morphological and physiological parameters explained (Table 2) indicated that shoot length, fresh and dry weights, and physiological parameters were highly significant except root length, which was non-significant under salt stress and non-stress conditions.
Cluster analysis and principal component analysis
All details for PCA analysis are mentioned in Supplementary data files S13–S 25.
Sequencing analysis and reference genome alignment of two rice varieties Dandi and GR-11
A total of 71,022 and 166,774 reads from control Dandi and GR-11 in shoot and 95,373 and 60,312 high-quality reads from treated Dandi and GR-11 from shoot sample. Moreover, a total of 173,096 and 108,350 high-quality filtered reads were generated from control Dandi and GR-11 in root while 125,886 and 63,107 reads were generated from treated Dandi and GR-11 in root samples from illumine sequencing. The average filtered rate was 98.05% and the percentage of GC content ranged from 52 to 53%. Overall alignment was 84.37% and 91.30% in Control Dandi and GR-11 condition while 87.45% and 83.87% in the treated Dandi and GR-11 condition in the shoot sample (Table 3). In the root sample, Overall alignments were 68.87% and 66.35% in Control Dandi and GR-11 condition while 72.46% and 70.37% in the treated Dandi and GR-11 condition (Table 3).
Analysis of differential expression genes (DEGs)
A total of 617 and 676 significant DEGs with log2-fold change in the range of ≥ + 2.0 to ≤ − 2.0 were identified from the comparative analysis of the two data sets DS (Dandi Shoot) and GS (GR-11Shoot) (Dandi vs. Control, GR-11 vs. Control) in the shoot sample. Furthermore, a total of 510 and 508 significant DEGs with log2-fold change in the range of ≥ + 2.0 to ≤ − 2.0 were identified from the comparative analysis of the two data sets DR (Dandi Root) and GR (GR-11 Root) (Dandi vs. Control, GR-11 vs. Control) in the root sample. All genes from the Dandi and GR-11in shoot and root along with expression patterns in the range of FPKM > 1 and FPKM < 300 were evenly mapped onto 12 Oryza sativa chromosomes using a circos plot. Out of 617 and 676 DEGs in shoots, 469 and 560 were up-regulated (≥ + 2.0-fold) and 148 and 116 were down-regulated (≤ − 2.0-fold) in DS and GS compared to control (Fig. 2a and b). Moreover, out of 510 and 508 DEGs in roots, 365 and 421 were up-regulated (≥ + 2.0-fold) and 145 and 87 were down-regulated in DR and GR as compared to the control (Fig. 2c and 2d). There were 443 and 534 genes that were uniquely up-regulated in DS and treatment as compared to control (Fig. 2a) and 143 and 111 were uniquely down-regulated in DS and GS treatment as compared to control (Fig. 2b). Moreover, 339 and 395 genes were uniquely up-regulated in DR and GR treatment as compared to control (Fig. 2c and d) and 112 and 81 genes were uniquely down-regulated in DR and GR treatment as compared to control (Fig. 3d). The Circos depicting the histogram of FPKM of the differentially expressed shoot and root genes mapped onto 12 Oryza sativa chromosomes is illustrated in Fig. 3a and b, respectively. Salt-responsive transcriptional regulation is a frequent adaptive strategy of plants that mitigates the undesirable effects of high salt concentration by restoration of cellular ion homeostasis and osmotic balance. In the experiment, assessing the transcript abundance in response to salt stress could also give a reliable estimate of specific gene activation or downregulation pertaining to the osmotic, ionic, and oxidative modules of salt stress tolerance, respectively. The list of all DEGs along with their gene description and fold change values analyzed under two conditions is mentioned in Supplementary data-Excel sheet-SA.
Gene ontology (GO) enrichment analysis of DEGs
Go functional groups are categorized into three main groups, i.e., biological processes (BP), cellular component (CC), and molecular function (MF). An analysis of GO terms suggested that the carboxylic acid metabolic process (5% in DS and 3.48% in GS), cellular nitrogen compound metabolic process (4.31% in DS and 3.29% in GS), and generation of precursor metabolites and energy (4.09% in DS and 2.71% in GS) were the most dominant terms in the biological process while ligase activity (2.5% in DS and 2.3% in GS), oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor (1.59% in DS and 1.16% in GS) and cofactor binding (4.77% in DS and 3.68% in GS) were leading in molecular function and cellular component, cell part (77.72% in DS and 79.45% in GS), intracellular organelle (66.36% in DS and 65.11% in GS) and ribonucleoprotein complex (4.09% in DS and 3.10% in GS) in shoot up-regulated DEGs (Supplementary 27a) whereas, molecular function and cellular function, cell part (80.14% in DS and 73.07% in GS) and membrane-bounded vesicle (10.63% in DS and 14.42% in GS) in shootdown-regulated DEGs (Supplementary 27b).
In root samples, regulation of RNA metabolic process (7.63% in DR and 7.31% in GR), response to chemical stimulus (4.58% in DR and 0% in GR), and lipid biosynthetic process (3.81% in DR and 0% in GR) were most dominant in the biological process while pyrophosphatase activity (0% in DR and 7.31% in GR) and pyrophosphatase activity (0% in DR and 7.31% in GR) were leading in molecular function and cytoplasmic membrane-bounded vesicle (16.03% in DR and 13.41% in GR) and membrane (16.03% in DR and 19.51%) in root down-regulated DEGs (Supplementary 27b).
The biological process in root samples was dominated by the amine metabolic process (4.17% in DR and 5.02% in GR), small molecule metabolic process (8.95% in DR and 9.29% in GR), and generation of precursor metabolites and energy (3.58% in DR and 4.02% in GR), while structural molecule activity (3.58% in DR and 4.27% in GR) and lyase ac (Supplementary S27b).
Kyoto encyclopedia of genes and genomes (KEGG) pathway mapping
To identify metabolic pathways in which DEGs were involved and enriched, pathway-based analysis was performed using the KEGG pathway database (Kumar et al. 2008). The DEGs were examined against the KEGG database to explore the biological pathways of Dandi and GR-11 in the shoot as well as root samples. Results demonstrated that 226 and 397 metabolic pathways in DS and GS, respectively (Supplementary data-SB), and 218, and 285 metabolic pathways were mapped to DR and GR. The top 30 DEGs enriched pathways DS and GS are shown in Supplementary S28a and Supplementary S28b showed the DEGs enriched pathways in DR and GR genes. Biosynthesis of secondary metabolites was highly abundant in GR-11 in comparison to Dandi in the shoot as well as in the root (Supplementary data-SB). Further PageMan analysis showed that there was a significantly higher number of DEGs (p < 0.05, ≥ ± 2.0-fold) involved in signalling pathways including secondary metabolism, hormone metabolism, abiotic stresses, and development (Supplementary 29a and Supplementary 29b), which have significant regulatory functions in shoot and root of Dandi and GR-11 variety of rice. These annotations provide a valuable resource for investigating specific processes, functions, and pathways of salt-stress responses in different tissues.
qRT-PCR for validation
To confirm the differential gene expression of unigene candidates underlying salinity tolerance in rice (Oryza Sativa L.), qPCR was performed based on the gene expression information obtained from the RNA seq Data (Supplementary data-S-26). The most promising genes that were either up-regulated or down-regulated during salinity stress were chosen based on fold change and pathway regulation under salinity stress for qPCR validation based on functional characterization. RNA expression of salt-stressed-induced genes was normalized against a Tibuine housekeeping gene (Fig. 4). The qRT-PCR results confirmed RNA-seq data for up-regulated and down-regulated salt-induced genes. These results demonstrate the reliability of the differential gene expression analysis.
Discussion
Rice is the most salt-sensitive cereal: as soon as the electrical conductivity in the soil reaches 6 dS/m, the yield is impacted (Hanin et al. 2016). Since the rice vegetative stage is very delicate and sensitive to abiotic stresses as apparent from several previous studies (Pires et al. 2015; Roy et al. 2014) this stage was chosen for our salt stress experiments. We have initially characterized the morphological and physiological responses of all the 24 genotypes involved in the study (Supplementary S1a-h).
In our experiment, initial screening is done for salt tolerance by exposing them to saline conditions (0, 150, 200, 250 mM, and 300 mM NaCl). Twenty-four genotypes were characterized for their morphological (Supplementary S1a-d) and physiological responses (Supplementary S1 e–g). Among the 24 rice varieties tested, we have selected the two demonstrating the greatest difference in response to salt exposure: the tolerant variety, Dandi, and the sensitive variety, GR-11 however both varieties have great economic importance for local markets in India. In morphological observation, shoot and root lengths of salt-exposed plants were analyzed, and the findings revealed that treatment with NaCl had no impact on growth parameters root and shoot lengths, and similar observations were recorded by Munawar 2021. Except for Vytilla-1 and Narmada, whose shoot length was significantly enhanced at 300 mM compared to non-stress circumstances, other genotypes did not demonstrate significant changes in root and shoot lengths. In different investigations carried out using rice plants, we found a considerable reduction in both fresh and dried weights. A loss in plant biomass was found to be a reliable measure for determining salt stress tolerance in different crops, as sensitive genotypes exhibited a significant reduction than tolerant genotypes (Munawar et al. 2021) and similar findings were recorded in the current study. The rice genotypes CSR-10, Dandi, CSR-30, Jaya, and GR-12, reduced plant biomass and other genotypes, also followed a similar pattern when exposed to salt treatment (Supplementary S1a-b). On the same line, several researchers reported that salt stress affects biomass accumulation and dry weight (Higbie et al. 2010). Even though salt stress had a major impact on plant biomass, contrasting results were also observed in the same study, where the Vytilla lines, Dandi, and CSR series benefited greatly by retaining their dry weights in comparison to the control group (Supplementary S1 a-g). Salt stress had the least effect on fresh and dry weights in the GR-7 and Vytilla lines, excluding GR-11, which demonstrated susceptibility to dry weight under 300 mM salt stress. This might be because of the ability of different cultivars to combat salt stress. Moreover, salt stress diminished stomatal conductance in the GR-11, GR-7, Narmada, and IR-64 plants under stress and non-stress conditions, however, stomatal conductance was maintained in the Dandi, Pokkali, CSR, and Vytilla lines.
The photosynthetic rate was also focused to analyze the response of different varieties where, except for GR-6, GAR-13, CSR-10, Gurjari, GR-12, and CSR-10, all genotypes maintained their photosynthetic rate. However, as per the SSI results, under 300 mM salt stress, GR-11, GR-7, and IR-64 were found to be more sensitive based on SSI (Supplementary S2b). In contrast to this, in previous studies, researchers used the PAM imaging approach and demonstrated a significant decrease in PSII maximum quantum yield (8PSII) in all leaves of the sensitive plants after the 6th day of salt exposure. However, tolerant plants were only affected by salt in the first and second leaves, and the leaves which emerged in the presence of salt showed the same photosynthetic efficiency as the control samples (Formentin et al. 2018). Similarly, it reduced the rate of transpiration in most genotypes; however, ten genotypes had no significant change in transpiration rate for salt stress. The Vytila, GR-12, and CSR lines were found to be marginally tolerant under 150 mM but sensitive to severe 300 mM dS/m salt stress based on SSI and STI, while under the 150 mM salt stress, GR-12 transpiration rate was also drastically reduced (Supplementary S2b). In earlier experiments (Munawar et al. 2021) between salt-stressed and non-stressed plants, chlorophyll concentration was found to be non-significant in all genotypes. (Higbie et al. 2010) in their experiments on salt tolerance observed that the fluorescence of chlorophyll does not alter when exposed to NaCl. Excess Na + accumulation, particularly in the aerial portions of plants, causes ionic stress, which is one of the most essential components of salinity. Maintaining a balanced cytosolic Na +/K + ratio becomes a crucial salt tolerance mechanism since Na + interacts with K + homeostasis, especially given its involvement in multiple metabolic processes (Assaha et al. 2017). The effects of NaCl on ionic homeostasis in the plant, such as Na +, K +, and the Na +/K + ratio, were substantial. The quantity of Na + was shown to be higher in NaCl-treated plants, especially at higher salt concentrations, such as 300 mM. The genotypes showed significant variations in Na +, K +, and the Na +/K + ratio. Ions accumulated more in leaves in the GR series and CSR lines than K + ions, and the Na +/K + ratio increased in the root and shoot. In Pokkali, the greatest value for K + ion was recorded under 300 mM salt stress in the shoot section. Furthermore, in GR-7, Narmada, GR-9, GAR-13, and CSR-13, the K + content in the rice root was sustained.
In an investigation (Yu et al. 2020), under salinity stress, a salt-sensitive rice genotype exhibited differential regulation of a greater number of transcripts than tolerant genotypes, whereas, under desiccation stress, the authors also determined that drought susceptible cultivar (IR64) had a larger number of differentially regulated genes than drought-tolerant. Under salt stress, however, a smaller number of transcripts were differentially expressed in susceptible cultivar (GR-11) compared with tolerant cultivar (Dandi) This might be related to genotype-specific adaptations, stress adaptive strategies, or the transcriptome analysis approach used in previous studies (Le Gall et al. 2015; Lenka et al. 2011; Walia et al. 2005).
Plant cell wall undergoes substantial cell wall remodeling to elicit an adaptive strategy for survival under stress (Tenhaken 2015). Many proteins have been discovered to assist in the maintenance of the osmotic potential inside the cell during situations of stress (Brutus et al. 2010). In the rice cultivars, we discovered an enrichment of genes involved in cell wall maintenance that may contribute to salt-tolerant phenotypes. Particularly, in the current study up-regulation enrichment of transcripts involved in the different cell-related processes were recorded Dandi, (salt-tolerant cultivar), such as cellular nitrogen compound metabolic process, cellular ketone metabolic process cellular catabolic process, cellular amino and metabolic process, cellular amine metabolic process. Interestingly, the enrichment of transcripts associated with cellular response to stimulus and cellular carbohydrate metabolic process were recorded only in the Dandi, tolerant cultivar, and not in its susceptible counterpart. In the cellular component section (CC), the following order was followed by the different processes when DS was compared Macro-molecular complex > Protein Complex > Ribonucleo-protein complex > Ribosome > chloroplast > Organelle membrane > Protein-DNA complex > Plasma membrane (Supplementary S 27a). Whereas for GS the order is, Macro-molecular complex > Membrane part > Intrinsic to membrane > Integral to membrane > Ribonucleo-protein complex > > Plasma membrane > chloroplast > Organelle membrane > protein-DNA complex (Supplementary S 27a). Surprisingly, no transcripts related to Ribosome, Protein-DNA-complex, Protein Complex, and Organelle Membrane were recorded in the GS sample. However, the Membrane part, Intrinsic to the membrane, and Integral to membrane-associated transcripts were absent in DS samples and this contrast is observed at several parameters in comparison of Ds and GS samples.
In the case of molecular functions, hydrolase activity acting on acid anhydrides, pyrophosphates activity, structural molecular activity, co-factor, coenzyme factor, and molecular transducer activity transcript enrichment was observed in the DS sample. To gain further insight into differentially regulated transcripts' involvement in diverse biological processes under stress conditions in rice, GO enrichment analysis was performed for differentially regulated transcripts under control and stress conditions. The most dominant terms in the biological process, according to an analysis of GO terms, were carboxylic acid metabolic process, cellular nitrogen compound metabolic process, and generation of precursor metabolites and energy while all of these were found highly overexpressed transcripts in DS versus GS. In molecular -function and cellular components, ligase activity, oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor, and cofactor binding were leading (Fig. 2a). The most dominant terms in the biological process where regulation of RNA metabolic process was observed in DS as well as GS (7.80 percent in DS and 8.65% in GS) indicated that these pathways might have a modulating impact on the regulation of salt-inducible gene expression.
Interestingly, transcripts related to response to chemical stimulus and ligase activity were absent in GS whereas, no expression of oxoacid metabolic process was observed in DS when compared with GS. Lipid biosynthetic process-related transcripts were the most dominant in DR, whereas absent in GR However, the pyrophosphatase activity was absent in DR but present in GR samples (Supplementary S 27 b and Supplementary data-SC). Functional categorization of the differentially expressed transcripts depicted that cell wall precursor synthesis and cell wall precursor synthesis sugar kinases were found to be enriched in Dandi shoots. Lipid metabolism was up-regulated and secondary metabolites, Iosprenoids non-mevalonate pathway (HDS) were also greatly up-regulated in Dandi shoot when compared with Gr-11 shoot. Along with these Hormone metabolism, cytokinin signal transduction, gibberellin signal transduction, Stress-related, biotic and abiotic stress, stress, abiotic drought, and salt-significant upregulation were also recorded in the Dandi shoot (Supplementary S 27 a–b). Whereas in roots, hormone metabolism, cytokinins, ethylene signal transduction Stress biotic and stress biotic regulation of transcription were observed to be common and significantly upregulated Dandi Roots as well shoots as compared to GR-11. Furthermore, the secondary metabolism of flavonoids and flavonols were found to be also greatly enriched in Dandi roots as compared to the GR-11 roots. This result suggested that there could be considerable differences in physiological processes between the leaves and roots of vegetative during salt stress. These annotations provide a valuable resource for investigating specific processes, functions, and pathways of salt-stress responses in different tissues.
The PageMan analysis showed that there was a significantly higher number of DEGs (p < 0.05, ≥ ± 2.0-fold) involved in signalling pathways including secondary metabolism, hormone metabolism, abiotic stresses, and development (Supplementary S29a and Supplementary S29b), which have significant regulatory functions in shoot and root of Dandi and GR-11 variety of rice. The role of plant hormones in abiotic stress responses is also well documented (Grant and Jones 2009). The genes participating in several hormone signalling pathways, like ABA, SA, ethylene, and jasmonic acid (JA) were found to be enriched in N22 under desiccation stress (Shankar et al. 2016). In an earlier study, a salt-sensitive rice genotype exhibited differential regulation of a larger number of transcripts as compared to the tolerant genotype under salinity stress Biosynthesis of secondary metabolites was highly abundant in GR-11 in comparison to Dandi in the shoot as well as in roots. In the present study, in roots analysis, hormone metabolism, cytokinin and cytokinin synthesis, degradation, and hormone metabolism, ethylene signal transduction, and stress biotic and stress abiotic regulation of transcription were significantly up-regulated in Dandi Root as compared to GR-11 root. The secondary metabolism of flavonoid flavonols was found to be significantly enriched in Dandi roots as compared to the GR-11 roots. The production of secondary metabolites was also recorded to be crucial in stress-adaptive mechanisms in plants by various researchers (Garg et al. 2014, 2015; Nakabayashi et al. 2014).
Phenylpropanoid biosynthesis genes have also been found to be induced by a variety of stresses (Shankar et al. 2016). In previous studies, authors found that genes involved in flavonoid derivatives and phenylpropanoid metabolism to be specifically up-regulated in a drought-tolerant cultivar, N22 under desiccation stress, whereas genes involved in carotenoid and terpenoid metabolism were found to be up-regulated in Pokkali, which is a tolerant cultivar under salinity stress. The above shreds of evidence suggest that stress-adaptive responses in salt and drought rice cultivars are governed by both common and specific genes involved in various secondary metabolic pathways (Shankar et al. 2016). Shoot analysis demonstrated that cell wall precursor synthesis sugar kinases were found to be enriched in Dandi shoots (Supplementary S29-a and Supplementary S29-b). Also, our results revealed that enrichment of genes involved in cell wall maintenance was significantly upregulated in DS as compared to GS and similar results were observed (Shankar et al. 2016) in drought and salt-tolerant rice cultivars. These might be the genes that possibly contribute to the drought and salinity-tolerant phenotype. For instance, wax being the major component of the cell wall outer cuticle layer inhibits the excess water evaporation from leaves, which acts as a possible adaptive mechanism in plants to evade the detrimental effects of salinity stress. Recently, the over-expression of genes involved in wax biosynthesis has led to the development of salinity tolerance in crop plants (Shepherd and Wynne Griffiths 2006; Zhou et al. 2013).
In the current study, analysis of shoot has shown the enrichment of sugar kinases-related transcripts. These results can be supported and justified by the findings of previous researchers (Le Gall et al. 2015; Tenhaken 2015) who stated that plant cell wall undergoes substantial cell wall remodeling to elicit an adaptive strategy for survival under stress. Many proteins have also been discovered to assist in the maintenance of the osmotic potential inside the cell during situations of stress (Brutus et al. 2010). Glutathione metabolism-related genes were enriched in the case of Dandi shoot, (DS) samples (Supplementary S 28a and S28b). Our results are following the findings of (Shankar et al. 2016), where, they have demonstrated that genes involved in the salicylic acid (SA) hormone signaling pathway, transcription regulation, and glutathione-S-transferase were found to be enriched in Pokkali (salinity tolerant) rice cultivar under salinity stress. Lipid metabolism-related genes were also found to be upregulated and along with secondary metabolites Iosprenoid's non-mevalonate pathway HDS were also highly upregulated in Dandi shoot when the results were compared with GR-11shoot. Hormone metabolism cytokinin signal transduction, gibberellin signal transduction. Stress, biotic and abiotic stress, stress abiotic drought, and salt were upregulated in DS versus GS. Signaling in Sugar and nutrient physiology were greatly upregulated in DS versus GS samples. To explore the biological pathways of Dandi and GR-11 in the shoot and root, the DEGs were analyzed in the KEGG database (Supplementary S 28a–b). Supplementary S28a represents the 30 DEGs enriched pathways in the DS and GS genes, whereas Supplementary S28b shows the DEGs enriched pathways in the DR and GR genes. In comparison to Dandi, GR-11 demonstrated the most extensive biosynthesis of secondary metabolites in the shoot and root (Supplementary S28a-28b). Further PageMan analysis revealed a significantly higher number of DEGs (p 0.05, 2.0-fold) involved in signaling pathways such as secondary metabolism, hormone metabolism, abiotic stresses, and development all of which have significant regulatory functions in the Dandi, and GR-11 rice varieties' shoot and root.
The qRT-PCR was performed using at least three technical replicates of each biological replicate for each cDNA sample. Rice Tebuline was used as a suitable internal control gene as reported in various previous Expression patterns of transcripts obtained from both qRT-PCR and RNA-Seq analysis were plotted in Microsoft Excel 2007. The correlation coefficient was calculated using the R programming package. All the primer sequences used for qRT-PCR are provided in Supplementary data S26.
Conclusions
With all the results it can be concluded that screening at the vegetative stage of shoot and root would provide a clue about the potential morphological markers for salinity screening of different genotypes. To investigate the productivity in saline fields of Gujarat, a highly salt-tolerant rice line, Dandi can be utilized as a genetic resource to discover the functions of salt-tolerant genes. It can be further used for differential gene expression and identification of major salt-tolerant genes. Based on transcriptome analysis, the genes specifically activated and differentially over-expressed during salinity stress belongs to different functional categories mostly located on chromosome 1, 2, 3, 4, 8, and 11 in both Dandi and GR-11. Hence, the results of this study could be combined with QTL analysis to identify salinity tolerant genes useful in rice breeding programs, and several shoots and root-specific genes with similar transcript profiles observed across the varieties presented a set of candidate genes whose functions concerning salinity tolerance should be further investigated.
Data availability
All the supplementary data is available in the supplementary data section. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
References
Ahmad M, Nangyal H, Imran M, Ullah F (2016) Optimization of protocol for surface sterilization and callus induction for three rice varieties. J Am-Eurasian J Agric Environ Sci 16(2):357–361
Anders S, Huber WJNP (2010) Differential expression analysis for sequence count data. In: J Nature Precedings. pp 1–1
Andrews S, Krueger F, Segonds-Pichon A, Biggins L, Krueger C, Wingett S (2010) FastQC
Asch F, Dingkuhn M, Dörffling K, Miezan K (2000) Leaf K/Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica 113(2):109–118
Askari H, Kazemitabar SK, Zarrini HN, Saberi MH (2016) Salt tolerance assessment of barley (Hordeum vulgare L.) genotypes at germination stage by tolerance indices. Open Agric 1(1):37–44
Assaha DV, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8:509
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci 107(20):9452–9457
Challabathula D, Analin B, Mohanan A, Bakka K (2022) Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and-tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J Plant Physiol 268:153583
del Río LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB (2006) Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol 141(2):330–335
Fernández-Varea J, Mayol R, Liljequist D, Salvat F (1993) Inelastic scattering of electrons in solids from a generalized oscillator strength model using optical and photoelectric data. J Phys Condens Matter 5(22):3593
Formentin E, Sudiro C, Perin G, Riccadonna S, Barizza E, Baldoni E, Lavezzo E, Stevanato P, Sacchi GA, Fontana P (2018) Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front Plant Sci 9:204
Ganie SA, Borgohain MJ, Kritika K, Talukdar A, Pani DR, Mondal TK (2016) Assessment of genetic diversity of Saltol QTL among the rice (Oryza sativa L.) genotypes. Physiol Mol Biol Plants 22(1):107–114
Ganie SA, Wani SH, Henry R, Hensel G (2021) Improving rice salt tolerance by precision breeding in a new era. Curr Opin Plant Biol 60:101996
Garg R, Verma M, Agrawal S, Shankar R, Majee M, Jain M (2014) Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res 21(1):69–84
Garg R, Bhattacharjee A, Jain M (2015) Genome-scale transcriptomic insights into molecular aspects of abiotic stress responses in chickpea. Plant Mol Biol Rep 33(3):388–400
Grant MR, Jones JD (2009) Hormone (dis) harmony molds plant health and disease. Science 324(5928):750–752
Grattan S, Zeng L, Shannon M, Roberts S (2002) Rice is more sensitive to salinity than previously thought. J Calif Agric 56(6):189–198
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci 7:1787
Higbie SM, Wang F, Stewart JM, Sterling TM, Lindemann WC, Hughs E, Zhang JJIJoA (2010) Physiological response to salt (NaCl) stress in selected cultivated tetraploid cotton
Hu S, Ding Y, Zhu C (2020) Sensitivity and responses of chloroplasts to heat stress in plants. Front Plant Sci 11:375
Ibrahim W, Qiu CW, Zhang C, Cao F, Shuijin Z, Wu F (2019) Comparative physiological analysis in the tolerance to salinity and drought individual and combination in two cotton genotypes with contrasting salt tolerance. Physiol Plant 165(2):155–168
International Rice Research Institute (1967) Screening rice for salinity tolerance An Annual report for 1967, Manila (Philippines), pp 308
Ismail AM, Horie T (2017) Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol 68:405–434
Kazemi K, Eskandari H (2011) Effects of salt stress on germination and early seedling growth of rice (Oryza sativa) cultivars in Iran. Afr J Biotech 10(77):17789–17792
Khan MIR, Palakolanu SR, Chopra P, Rajurkar AB, Gupta R, Iqbal N, Maheshwari C (2021) Improving drought tolerance in rice: ensuring food security through multi-dimensional approaches. Physiol Plant 172(2):645–668
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Kong W, Zhong H, Gong Z, Fang X, Sun T, Deng X, Li Y (2019) Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants 8(3):64
Kong W, Zhang C, Qiang Y, Zhong H, Zhao G, Li Y (2020) Integrated RNA-Seq analysis and Meta-QTLs mapping provide insights into cold stress response in rice seedling roots. Int J Mol Sci 21(13):4615
Kong W, Zhang C, Zhang S, Qiang Y, Zhang Y, Zhong H, Li Y (2021b) Uncovering the novel QTLs and candidate genes of salt tolerance in rice with linkage mapping, RTM-GWAS, and RNA-seq. Rice 14(1):1–12
Kong W, Sun T, Zhang C, Deng X, Li Y (2021a) Comparative transcriptome analysis reveals the mechanisms underlying differences in salt tolerance between indica and japonica rice at seedling stage. Front Plant Sci 12
Kumar A, Bernier J, Verulkar S, Lafitte H, Atlin G (2008) Breeding for drought tolerance: direct selection for yield, response to selection and use of drought-tolerant donors in upland and lowland-adapted populations. Field Crop Res 107(3):221–231
Le Gall H, Philippe F, Domon J-M, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants 4(1):112–166
Lenka SK, Katiyar A, Chinnusamy V, Bansal KC (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9(3):315–327
Ma Y, Dias MC, Freitas H (2020) Drought, and salinity stress responses and microbe-induced tolerance in plants. Front Plant Sci 11:1750
Mahmud S, Sharmin S, Chowdhury BLD, Hossain MA, Bhuiyan MJH (2016) Mitigation of salt stress in rice plant at germination stage by using methyl jasmonate. Asian J Med Biol Res 2(1):74–81
MirdarMansuri R, Shobbar Z-S, BabaeianJelodar N, Ghaffari M, Mohammadi SM, Daryani P (2020) Salt tolerance involved candidate genes in rice: an integrative meta-analysis approach. BMC Plant Biol 20(1):1–14
Munawar W, Hameed A, Khan MKR (2021) Differential morphophysiological and biochemical responses of cotton genotypes under various salinity stress levels during early growth stage. J Front Plant Sci 12:335
Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77(3):367–379
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295
Pires IS, Negrão S, Oliveira MM, Purugganan MD (2015) Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiol Plant 155(1):43–54
Razzaque S, Sabrina M. E, Taslima H, Sudip B, Nurnabi A., Sazzadur R., Xiaoyu W., Abdelbagi M. l, Harkamal W., Thomas E. J. and Zeba I. S (2019). Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. Sci Rep 8249(9):10–24. https://doi.org/10.1038/s41598-019-44757-4
Reddy DS, Bhatnagar-Mathur P, Reddy PS, Sri Cindhuri K, Sivaji Ganesh A, Sharma KK (2016) Identification and validation of reference genes and their impact on normalized gene expression studies across cultivated and wild cicer species. PLoS ONE 11(2):e0148451
Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Curr Opin Biotechnol 26:115–124
Shankar R, Bhattacharjee A, Jain M (2016) Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci Rep 6(1):1–15
Shepherd T, Griffiths W (2006) The effects of stress on plant cuticular waxes. New Phytol 171(3):469–499
Shereen A, Mumtaz S, Raza S, Khan M, Solangi S (2005) Salinity effects on seedling growth and yield components of different inbred rice lines. J Pak J Bot 37(1):131–139
Tenhaken R (2015) Cell wall remodeling under abiotic stress. Front Plant Sci 5:771
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1):W122–W129
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Verma G, Srivastava D, Tiwari P, Chakrabarty (2019) ROS modulation in crop plants under drought stress. Genetics 311–336
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Close TJ (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139(2):822–835
Wang S, Blumwald E (2014) Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26(12):4875–4888
Yu X, Liu H, Hamel KA, Morvan MG, Yu S, Leff J, Guan Z, Braz JM, Basbaum AI (2020) Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. J Nat Commun 11(1):1–12
Zeng L, Shannon MC (2000) Salinity effects on seedling growth and yield components of rice. J Crop Sci 40(4):996–1003
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. J Plant Physiol 161(3):1375–1391
Acknowledgements
Authors are thankful to The Department of Agri. Biotechnology, Anand Agriculture University, Anand, Gujarat (India) for providing the experimental facility.
Funding
No potential funding is involved to carry out this study.
Author information
Authors and Affiliations
Contributions
A.V. did the overall execution of the experiment, analytical work, and collection of data after morpho-physiological and biochemical analysis of leaves, organization of resulting data, and writing up and revision of the manuscript. Y.D. and N.B. contributed to the planning, designing, and finalization of the basic idea of the experiment and overall supervision during analytical work carried out the statistical analysis of data using XL-STAT software did the presentation of the resulting data in the form of graphs, and revised and finalized the manuscript. R.T., C.J., and S.N. the arrangement and provision of rice seeds contributed to the study's basic idea and planning of the glasshouse experiment and revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Tommaso Martinelli.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vala, A.K.G., Bano, N., Deshmukh, Y. et al. Transcriptome analysis identifies novel gene(s) and pathways for salt stress responses in Dandi cultivar. CEREAL RESEARCH COMMUNICATIONS 51, 351–365 (2023). https://doi.org/10.1007/s42976-022-00319-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-022-00319-5