Abstract
Human Pegivirus Type 1 (HPgV-1), a ubiquitous commensal virus, has been recently suggested as a marker of immunologic function. There is scarce data for the presence, genotypes, and molecular characteristics of HPgV-1 among kidney transplant recipients. Therefore, the objective of this study was to examine the prevalence and the molecular characteristics (cycle threshold, genotypes) of this viral infection among kidney transplant recipients from the Brasília, Federal District of Brazil. HPgV-1 RNA detection in the plasma was assessed by RT-qPCR. Positive samples were submitted to sequencing and phylogenetic analysis of the 5´-UTR portion of the viral genome. The estimated HPgV-1 prevalence among renal-transplant recipients was 20%. The performed phylogenetic inference revealed that the most frequent genotype among these patients was HPgV-1 genotype 2 (78.9%) presented by its two subgenotypes (2 A and 2B), followed by genotypes 1 and 3 (10.5% each). This study presents new data about the HPgV-1 circulation and molecular characteristics among kidney transplant recipients from the Federal District of Brazil. Further work is fundamental to examine the effect of HPgV-1 among patients with immunological suppression, including kidney transplant recipients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human Pegivirus Type 1 (HPgV-1), formerly known as GBV-C or hepatitis G virus, is a single-stranded positive RNA virus belonging to the Pegivirus genus of the Flaviviridae family. The HPgV-1 genome contains approximately 9,400 nucleotides that encompass a single open reading frame (ORF) located between the untranslated regions (UTRs) at the 5′ and 3′ ends of the genome. The RNA translation results in the synthesis of a single polyprotein of approximately 3,000 amino acid residues which is cleaved into two structural proteins (envelope proteins E1 and E2) and six non-structural (NS) ones (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) [1]. Taxonomically, HPgV-1 is divided into seven genotypes that show specific geographic endemicity based on the phylogenetic analysis of the 5 ́UTR and E2 genomic regions of HPgV-1 [2].
Initially, the HPgV-1 was detected in patients suffering from acute hepatitis and was thought that is a cause of this disease named therefore Hepatitis type G. However, the association with hepatic disease or other acute clinical condition in humans has not been proven over the years, and currently the scientific community believes that HPgV-1 is a commensal virus due to its widespread in the human population [3]. Studies indicate that approximately 750 million people worldwide are infected with HPgV-1. HPgV-1 is efficiently transmitted parenterally, sexually and vertically [4]. Therefore, higher prevalence is observed in individuals who have been submitted to multiple transfusions including high levels of parenteral exposure, human immunodeficiency virus (HIV-1), HCV (hepatitis C virus), and Hepatitis B virus (HBV) and in transplant recipients [5,6,7,8].
Recently, several studies have evaluated the presence of the HPgV-1 RNA as a co-infection with HBV, Ebola, and especially HIV [8,9,10,11]. In HIV-infected individuals, HPgV-1 viremia appears to be associated with better outcomes, such as reduced HIV viral load, better response to antiretrovirals, lower rates of vertical transmission, and slower progression to AIDS [10, 11]. It is believed that the better outcome in HIV-infected patients harboring HPgV-1 is due to immunomodulatory effects of the latter [10]. Some studies were conducted to observe whether these immunomodulatory events influence the outcome of the HPgV-1 infected kidney recipients, since these patients are submitted to specific immunosuppression schemes for graft maintenance [12, 13]. Until now, there is no association between HPgV-1 and transplantation outcome.
Although there are various studies about the influence of HPgV-1 on renal transplantation, there is no information about the HPgV-1 genotypes that circulate among patients with kidney transplantation. In our previous survey that used viral metagenomics to characterize the virome of kidney transplant recipients from Central-West Brazil, we observed a high abundance of sequence reads that belonged to HPgV-1. This raised serious concerns about HPgV-1 frequency and genotypes that circulate among patients submitted to renal transplantation.
Materials and methods
Kidney transplant recipients and sample collection
Between June 2018 and August 2021, 95 kidney transplant recipients (54 men and 41 women) treated at the University Hospital of Brasília, Federal District, Brazil (Fig. 1) were included in this study. Briefly, 5 mL of total blood was collected in EDTA sterile vacutainers (Vacuette, Greiner Bio-One, Americana, São Paulo, Brazil) and after low-speed centrifugation (1931×g, 10 min) the plasma was separated and stored at -80 °C until use. The study was approved by the Research Ethics Committee of the Faculty of Ceilândia, University of Brasília, Distrito Federal, Brazil (CEP-FCE/UnB, CAAE 83811718.8.0000.8093).
All participants provided signed informed consent forms before sample collection. Prior to transplantation and annually post-transplantation, all patients underwent testing for HIV and HCV. None of the patients included in this study were infected with HCV and/or HIV. Furthermore, viral metagenomic analysis revealed the absence of HIV, HCV, or HBV reads, reinforcing the absence of these infections among the patients. The investigation was initiated due to the notable abundance of HPgV-1 sequence reads observed in the viral metagenomics analysis. Initially, samples were subjected to viral metagenomics, which revealed a significant prevalence of HPgV-1 sequence reads in the majority of sample pools. Out of the 19 tested sample pools, 14 exhibited the presence of HPgV-1 sequence reads. This prompted a more detailed investigation into this viral agent within the patient group, particularly considering the limited data available in the Federal District of Brazil and globally. Subsequently, all individual samples that composed the pools were tested for the presence of HPgV-1 RNA by RT-qPCR and sequenced to study the viral genotypes.
Viral RNA extraction and HPgV-1 detection by RT-qPCR
Viral nucleic acids were automatically extracted using EXTRACTA Kit FAST-DNA and RNA Viral in EXTRACTA 32 equipment (Loccus, Brazil) following the manufacturer’s instructions. HPgV-1 RNA was detected by RT-qPCR, applying the GoTaq® 1-Step RT-qPCR System kit (Promega, Madison, Wisconsin, USA) following the manufacturer’s protocol. The cycling consisted of 1 cycle of reverse transcription (RT) for 40 min at 40°C; inactivation of Reverse Transcription (RT)/start of denaturation (1 cycle) for 2 min at 95°C; amplification for 40 cycles of 95°C for 15 s and 60°C for 1 min. The following primers and probe corresponding to 5’ UTR of the viral genome were used: RTG1 5’-GTGGTGGATGGGTGATGACA-3’ (forward), RTG2 5’-GACCCACCTATAGTGGCTACCA-3’ (reverse) and NFQ 5’-FAM-CCGGGATTTACGACCTACC-3’ (hydrolytic probe) [14].
Amplification of HPgV-1 5’-UTR by RT-nested PCR
After the confirmation of HPgV-1 positive samples, the 5’-UTR region of the virus genome was amplified for further genotyping. Initially, the reverse transcription (RT) was performed using High-Capacity cDNA Reverse Transcription enzyme (Applied Biosystems, Waltham, Massachusetts, USA) according to the manufacturer’s recommendations using the primer GUTR.R01 (‘5- AATGCCACCCGCCCTCA-3’). The nested-PCR protocol was adapted from previous studies [15, 16]. In brief, both rounds of nested-PCR were performed in a 50 µL reaction containing 5 µL cDNA (the first reaction product was used for the second round), 1 × PCR buffer, 1.5 mMMgCl2, 400 nM of each primer, 200 µM of dNTPs and 1.25U/reaction of recombinant Taq DNA polymerase (Thermo Fisher Scientific, São Paulo, Brazil). For the first reaction, GUTR.F01, 5’−GGTTGGTAGGTCGTAAATCCCG − 3’ (forward primer) and GUTR.R01, 5’−ATGCCACCCGCCCTCA − 3’ (reverse primer) were used. For the second reaction, the GUTR.F02 5’−GTAGGTCGTAAATCCCGGTCA − 3’ (forward primer) and GUTR.R02 5’ −CGAAGGATTCTTGGGCTACC − 3’ (reverse primer) were used. Nested-PCR reactions were performed in Veriti 96-Well Thermal Cycler (Applied Biosystems, Waltham, Massachusetts, USA). For both rounds of nested-PCR the following protocol was used: denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s (40 cycles for second reaction), 55 °C for 30 s and 72 °C for 1 min. A final extension at 72 °C for 10 min was used. The Nested-PCR products (379-nucleotide band) were detected by agarose gel (2.0%) electrophoresis stained with ethidium bromide and visualized under ultraviolet (UV) illumination in an L-PIX TOUCH instrument (Loccus, São Paulo, Brazil).
Phylogenetic analysis of HPgV-1
The 5´-UTR sequencing was performed in the ABI 3500 XL DNA sequencer, using BigDye™ Terminator v 3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, São Paulo, Brazil) with the following protocol: 95ºC for 1 min for initial denaturation and 25 cycles of 96ºC for 10 s, 50ºC for 5 s, and 60ºC for 4 min.
The obtained electropherograms were initially analyzed using BioEdit (Ibis Therapeutics, Carlsbad, CA). For reference HPgV-1 sequences we obtained 71 complete genomes from the GenBank using the key words GBV-C, hepatitis G, Human pegivirus − 1 and complete genome. To align the sequences, we used MAFFT software [17] and the obtained alignment was manually edited using Aliview [18]. Maximum-likelihood trees were reconstructed using IQ-TREE software [19] with bootstrap support of 1,000 replicates. The most adequate substitution model was determined by using ModelFinder Plus (MFP). Finally, Figtree software was used for tree visualization and for phylogenetic signal analysis [20].
Statistical analysis
The online tool http://www.winepi.net was used to calculate the prevalence of HPgV-1 RNA found in our study and the estimated true prevalence considering the total number of kidney transplantations performed in the University Hospital of Brasília until August 2021. To do this, we considered the perfect sensitivity and specificity for the used diagnostic test for HPgV-1 RNA detection. Student’s t-test was used to compare age mean and Fisher’s exact test was used to compare HPgV-1 RNA prevalence between male and female.
Results
Blood samples were obtained from 95 kidney transplant recipients, of which 54 were male (56.8%) and 41 were female (43.2%) with a mean age of 45.3 years (± 14.1 years). The mean age was 47.5 (± 12.6 years) years for male and 43.2 (± 15.4 years) for female patients. No difference was observed between male vs. female mean age (p = 0.1708).
Initially, the samples were pooled (5 samples/pool) and evaluated by viral metagenomics. From 19 pools, 14 pools presented positivity for HPgV-1 RNA (unpublished data). Then, the positive pools (70 samples) were separately tested for HPgV-1 RNA by RT-qPCR. The estimated HPgV-1 frequency was 20% (n = 19/95, mean age 41.6, range ± 12.9). The positive samples presented a mean cycle threshold of 27.62 (± 3,86). Despite the absence of statistical significance (p = 0.0695), the prevalence of HPgV-1 RNA was higher in females compared to male kidney transplant recipients. The prevalence was 13% (n = 7/54, mean age 42.3, range ± 7.6) in males and 29.3% (n = 12/41, mean age 41.2, range ± 12.9) in females. The statistical evaluation demonstrated that the estimated true prevalence ranged between 13.1 and 26.9%, considering the total number of kidney transplantations performed at the University Hospital of Brasília until August 2021 (352 kidney transplantations). No statistical difference was observed between the mean ages of HPgV-1 RNA-positive male and female patients (Table 1).
The prevalence of HPgV-1 RNA by age group and time after kidney transplantation are shown in Table 2. To evaluate the distribution of HPgV-1 RNA among the different age groups of the kidney transplant recipients, they were divided into five age groups: 18–29 years, 30–39 years, 40–49 years, 50–59 years, and > 60 years. The observed prevalence was 18.8% among the group with 18–29 years, 26.6% in the group of 30–39 years, 28.5% in the group of 40–49 years, 17.2% in the group of 50–59 years, and 7.1% in the group of > 60 years. We also evaluated the prevalence HPgV-1 RNA among the recipients with different time after kidney transplantation was evaluated. The recipients were divided into five different groups: < 1 year, 1–3 years, 4–6 years, 7–9 years and > 10 years after transplantation. The observed prevalence was 7.1% among the group with < 1 years, 25% in the group of 1–3 years, 27.3% in the group of 4–6 years, 22.2% in the group of 7–10 years, and 0.0% in the group of > 10 years after transplantation (Table 2).
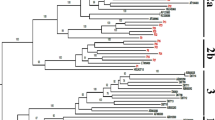
The performed phylogenetic analysis demonstrated that the obtained sequences from 19 kidney transplant recipients from Central-West Brazil: were characterized as HPgV-1 genotypes 1, 2 and 3. As expected, the most prevalent genotype was 2, and it was represented by 15 positive samples (78.9%). Genotype 2 was presented with both subgenotypes 2 A and 2B. One sequence belonged to sub-genotype 2 A (1/19, 5.3%) and 14 sequences belonged to sub-genotype 2B (14/19, 73.6%). Interestingly, HPgV-1 presented at lower frequency in Brazil were also identified, once 2 samples belonged to genotype 1 (2/19, 10.5%) and 2 samples belonged to genotype 3 (2/19, 10.5%) (Fig. 2). Finally, the phylogenetic signal analysis demonstrated that only 13.5% of the taxa did not have their topologies grouped, indicating a strong phylogenetic signal (Fig. S1).
Maximum-likelihood phylogenetic tree of the 5´UTR region of the Human Pegivirus-1 (HPgV-1) obtained from kidney transplant recipients from Central-West Brazil. HPgV-1 isolated from kidney transplant recipients were characterized as genotypes 1 (2/19), 2 (1/19 sub-genotype 2 A and 14/19 sub-genotype 2B) and 3 (2/19). The HPgV-1 isolates detected in kidney transplant recipients are marked by red color and identified by the respective accession number in NCBI (OR449336 to OR449354)
Discussion
In our study, where we estimated the frequency of HPgV-1 RNA among kidney transplant recipients from University Hospital of Brasília, Central-West Brazil, we detected an overall prevalence of 20% HPgV-1 RNA (estimated true prevalence between 13.1 and 26.9%). The prevalence in our cohort was in line with other studies that examined the HPgV-1 infection among Brazilian kidney transplant recipients, that is in Central-West Brazil (city of Goiania), where the detected prevalence was 16.7% [21] and in Rio de Janeiro (southeastern Brazil) reporting a very high HPgV-1 RNA prevalence of 36.1% [13]. Additionally, few studies evaluated the prevalence of HPgV-1 RNA among kidney transplant recipients worldwide. Higher prevalences were observed in Turkey (42%) [22] and India (52.9%) [23], and similar prevalences were observed in Italy (24%) [24] and Spain (26.1%) [12] compared to the present study. We believe that the differences in the prevalence of HPgV-1 may be explained by epidemiological variations (size and the clinical features of the patients), transmission routes, level of immunosuppression, living habits and sensitivity of the detection methods [21, 25].
On the contrary, studies that examine the HPgV-1 prevalence in other study groups showed lower values. For example, Brazilian studies that investigated the HPgV-1 prevalence among healthy blood donors showed lower prevalence which varied between 4.2% and 10% [15, 16, 26,27,28,29]. In general, the higher prevalence rates among kidney transplant recipients were expected due to two main reasons (i) high levels of parenteral exposure, such as previous hemodialysis and (ii) the levels of drug immunosuppression in these patients that maintain the functional characteristics of the transplanted kidney along life. In this regard, studies performed in South America showed HPgV-1 RNA prevalence rates varying between 12.8% and 17.6% in patients submitted to hemodialysis [21, 30,31,32].
We also observed a gradual increase of the HPgV-1 RNA prevalence which started with 21.4% in the age groups of 18–29 years and peaked in the group of 40–49 years (28.5%) with a gradual decrease in the group of 50–59 years (17.2%) and > 60 years (7.1%). These results are similar to the Brazilian studies, especially those investigating blood donors [15, 31]. Although the authors indicate that sexual activity influences this age distribution of prevalence for healthy blood donors, this conclusion cannot apply clearly for kidney transplant recipients due the enhanced possibility of parenteral transmission. This is strengthened by the evidence that HPgV-1 RNA-positive recipients have higher intraoperative transfusion requirements and increased number of pre-transplant transfusions compared to HPgV-1 RNA-negative recipients [12].
The evaluation of the HPgV-1 infection kinetics demonstrated that the rate of HPgV-1 infection increased from 14.7% at pre-transplantation to 19.1% at 1-year post-transplantation [12]. Here, we observed that the prevalence rate of HPgV-1 RNA increased from 7.1% in patients with > 1-year post-transplantation to 25% in patients with 1–3 years post-transplantation. The prevalence rate remains higher in patients with 4–6 years (27.3%) and 7–9 years post-transplantation (22.2%), decreasing after 10 years. We are unaware if HPgV-1 RNA was already present in patients before transplantation and, therefore, it is impossible to guess if the respective patients had a previous HPgV-1 infection. However, the increasing prevalence within the post-transplantation period might be a result of infection obtained by the transplanted organ, received blood components or the presence of underlying low-level HPgV-1 infections due to the maintenance of immunosuppressive therapy. Current detection methods may not be effective in detecting an HPgV-1 infection with low-levels of replication [33] and the prevalence in the first-year post-transplantation may be higher than previously assumed.
The performed phylogenetic analysis showed the presence of three genotypes of HPgV-1 (genotypes 1, 2, and 3) in kidney transplant recipients from Central-West Brazil. The genotype 2 was the most frequently detected (15/19, 78%), with 1 specimen belonging to sub-genotype 2 A (5.3%) and 14 sequences belonging to sub-genotype 2B (73.6%). These results were expected, once the genotype 2 is the most frequently detected HPgV-1 genotype in Brazil in different patient groups like HIV-1 infected individuals [21, 29, 34], healthy blood donors [15, 16, 35,36,37], kidney transplant recipients [21, 38] and patients with fulminant hepatitis [38]. Additionally, HPgV-1 genotype 2 is the most prevalent one in European countries and the USA [39, 40].
At lower percentages, we also detected genotypes 1 and 3, with 2 specimens belonging to genotype 1 and 2 specimens belonging to genotype 3. Genotype 1 is identified predominantly in Africa [41, 42], while genotype 3 is endemic for Asian and Amerindian populations [43, 44]. Some Brazilian studies also detected, at lower prevalence, HPgV-1 genotypes 1 and 3 among blood donors [15], HIV-1 infected individuals [29] and kidney transplant recipients [21, 38]. We believe that the presence of HPgV-1 genotypes like 1 and 3 (rare for Brazil) might be related with the miscegenation of the country, where descendants of African and Amerindian origin are present.
Like any study, ours also exhibits several limitations, which are important to acknowledge as they may potentially influence the outcomes. These limitations encompass the restricted number of patients, who were sourced from a single center. Moreover, although we employed a genomic region traditionally utilized for HPgV-1 analysis, its size was relatively small. Consequently, acquiring complete HPgV-1 genomes is imperative for comprehending the molecular evolution of this viral agent among renal transplantation patients.
Conclusion
In our study, it was possible to observe a prevalence rate relatively close to other studies carried out in kidney transplanted recipients. Genotype 2 was the most frequent genotype (78.9%), followed by genotypes 1 and 3 (10.5% each), which corroborates other studies that point to genotype 2 as the most common genotype in Brazil. Many aspects of the HPgV-1 infection remain unclear, and we believe that other studies are necessary to examine the possible immunomodulatory functions of HPgV-1 among kidney transplant recipients and if this virus can influence the outcome of the kidney transplantation.
References
Yu Y, Wan Z, Wang JH, Yang X, Zhang C (2022) Review of human pegivirus: prevalence, transmission, pathogenesis, and clinical implication. Virulence 13(1):324–341. https://doi.org/10.1080/21505594.2022.2029328
Miao Z, Gao L, Song Y, Yang M, Zhang M, Lou J, Zhao Y, Wang X, Feng Y, Dong X, Xia X (2017) Prevalence and clinical impact of human Pegivirus-1 infection in HIV-1-Infected individuals in Yunnan, China. Viruses 9(2):28. https://doi.org/10.3390/v9020028
Santos Bezerra R, Santos EV, Maraninchi Silveira R, Silva Pinto AC, Covas DT, Kashima S, Slavov SN (2020) Molecular prevalence and genotypes of human pegivirus-1 (HPgV-1) and SENV-like viruses among multiply transfused patients with beta-thalassemia. Transfus Apher Sci 59(2):10269. https://doi.org/10.1016/j.transci.2019.102697
Samadi M, Salimi V, Haghshenas MR, Miri SM, Mohebbi SR, Ghaemi A (2022) Clinical and molecular aspects of human pegiviruses in the interaction host and infectious agent. Virol J 19(1):41. https://doi.org/10.1186/s12985-022-01769-3
Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK (1995) Isolation of novel virus-like sequences associated with human hepatitis. Nat Med 1:564–569. https://doi.org/10.1038/nm0695-564
Marano G, Franchini M, Farina B, Piccinini V, Pupella S, Vaglio S, Grazzini G, Liumbruno GM (2017) The human pegivirus: a new name for an ancient virus. Can transfusion medicine come up with something new? Acta Virol 61:401–412. https://doi.org/10.4149/av_2017_402
Chivero ET, Stapleton JT (2017) Tropism of human pegivirus (formerly known as GB virus C/hepatitis G virus) and host immunomodulation: insights into a highly successful viral infection. J Gen Virol 96:1521–1532. https://doi.org/10.1099/vir.0.000086
Bhattarai N, Stapleton JT (2017) GB virus C: the good boy virus? Trends Microbiol 20:124–130. https://doi.org/10.1016/j.tim.2012.01.004
Lauck M, Bailey AL, Andersen KG, Goldberg TL, Sabeti PC, O’Connor DH (2015) GB virus C coinfections in west African Ebola patients. J Virol 89:2425–2429. https://doi.org/10.1128/JVI.02752-14
Horemheb-Rubio G, Ramos-Cervantes P, Arroyo-Figueroa H, Ávila-Ríos S, García-Morales C, Reyes-Terán G, Escobedo G, Estrada G, García-Iglesias T, Muñoz-Saucedo N, Kershenobich D, Ostrosky-Wegman P, Ruiz-Palacios GM (2017) High HPgV replication is associated with improved surrogate markers of HIV progression. PLoS ONE 12(9):e0184494. https://doi.org/10.1371/journal.pone.0184494
Blackard JT, Ma G, Welge JA, Taylor LE, Mayer KH, Klein RS, Celentano DD, Sobel JD, Jamieson DJ, King CC (2017) Cytokine/chemokine expression associated with human pegivirus (HPgV) infection in women with HIV. J Med Virol 89(11):1904–1911. https://doi.org/10.1002/jmv.24836
Fernández-Ruiz M, Forque L, Albert E, Redondo N, Giménez E, López-Medrano F, González E, Polanco N, Ruiz-Merlo T, Parra P, San Juan R, Andrés A, Aguado JM, Navarro D (2022) Human pegivirus type 1 infection in kidney transplant recipients: replication kinetics and clinical correlates. Transpl Infect Dis 24(1):e13771. https://doi.org/10.1111/tid.13771
Savassi-Ribas F, Pereira JG, Horta MAP, Wagner TCS, Matuck TA, Monteiro de Carvalho DB, Mello FCA, Varella RB, Soares CC (2020) Human pegivirus-1 infection in kidney transplant recipients: a single-center experience. J Med Virol 92(12):2961–2968. https://doi.org/10.1002/jmv.25764
Campos AF, Tengan FM, Silva SAA, Levi JE (2011) Influence of hepatitis G virus (GB virus C) on the prognosis of HIV-infected women. Int J STD AIDS 22:209–213. https://doi.org/10.1258/ijsa.2011.010283
Slavov SN, Maraninchi Silveira R, Hespanhol MR, Sauvage V, Rodrigues ES, Fontanari Krause L, Bittencourt HT, Caro V, Laperche S, Covas DT, Kashima S (2019) Human pegivirus-1 (HPgV-1) RNA prevalence and genotypes in volunteer blood donors from the Brazilian Amazon. Transfus Clin Biol 26(4):234–239. https://doi.org/10.1016/j.tracli.2019.06.005
Slavov SN, Silveira RM, Rodrigues ES, Diefenbach CF, Zimmermann AM, Covas DT, Kashima S (2019) Human pegivirus-1 (HPgV-1, GBV-C) RNA prevalence and genotype diversity among volunteer blood donors from an intra-hospital hemotherapy service in Southern Brazil. Transfus Apher Sci 58(2):174–178. https://doi.org/10.1016/j.transci.2019.01.002
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. https://doi.org/10.1093/bioinformatics/btu531
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Rambaut A (2018) FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree
Filho RR, Carneiro MADS, Teles SA, Dias MJ, Cardoso DDP, Lampe E, Yoshida CFT, Martins RMB (2004) GB virus C/hepatitis G virus infection in dialysis patients and kidney transplant recipients in central Brazil. Memórias do Instituto Oswaldo Cruz 99(6):639–643. https://doi.org/10.1590/S0074-02762004000600019
Erensoy S, Zeytinoglu A, Göksel S, Ozacar T, Ozkahya M, Ok E, Tuumlrkoglu S, Bilgiç A (2002) GB virus C/hepatitis G virus infection among renal transplant recipients in Izmir, Turkey: molecular analysis of phylogenetic groups. Int J Infect Dis 6:242–243. https://doi.org/10.1016/s1201-9712(02)90121-9
Abraham P, John GT, Raghuraman S, Radhakrishnan S, Thomas PP, Jacob CK, Sridharan G (2003) GB virus C/hepatitis G virus and TT virus infections among high risk renal transplant recipients in India. J Clin Virol 28:59–69. https://doi.org/10.1016/s1201-9712(02)90121-9
De Filippi F, Lampertico P, Soffredini R, Rumi MG, Lunghi G, Aroldi A, Tarantino A, Ponticelli C, Colombo M (2001) High prevalence, low pathogenicity of hepatitis G virus in kidney transplant recipients. Dig Liver Dis 33:477–479. https://doi.org/10.1016/s1590-8658(01)80025-6
Odeh RA, Yasin S, Nasrallah G, Babi Y (2010) Rates of infection and phylogenetic analysis of GB virus-C among Kuwaiti and Jordanian blood donors. Intervirology 53(6):402–407. https://doi.org/10.1159/000317290
Goubau P, Andrade FB, Liu H-F, Basilio FP, Croonen L, Barreto-Gomes VAF (1999) Prevalence of GB virus C/hepatitis G virus among blood donors in north-eastern Brazil. Tropical Med Int Health 4:365–367. https://doi.org/10.1046/j.1365-3156.1999.00407.x
Pinho JRR, Zanotto PMA, Ferreira JLP, Sumita LM, Carrilho FJ, Silva LC, Capacci ML, Silva AO, Guz B, Gonçales FL Jr, Gonçales NSL, Buck GA, Meyers GA, Bernardini AP (1999) High prevalence of GB virus C in Brazil and molecular evidence for intrafamiliar transmission. J Clin Microbiol 37:1634–1637. https://doi.org/10.1128/JCM.37.5.1634-1637.1999
Levi JE, Contri DG, Lima LP, Takaoka DT, Garrini RH, Santos W, Fachini R, Wendel S (2003) High prevalence of GB vírus C/hepatitis G vírus RNA among Brazilian blood donors. Rev Inst Med Trop S Paulo 45:75–78. https://doi.org/10.1590/S0036-46652003000200004
Mota LDD, Finger-Jardim F, Silva CM, Germano FN, Nader MM, Gonçalves CV, Luquez KYS, Chies JAB, Groll AV, Hora VPD, Silveira J, Basso RP, Soares MA, Martínez AMB (2019) Molecular and clinical profiles of human pegivirus type 1 infection in individuals living with HIV-1 in the Extreme South of Brazil. Biomed Res Int 2019:8048670. https://doi.org/10.1155/2019/8048670
Watanabe MA, Milanezi CM, Silva WA Jr, de Lucena Angulo I, Santis G, Kashima S, da Costa JA, Neto MM, Covas DT (2003) Molecular investigation of GB virus C RNA in hemodialysis and thalassemics patients from Brazil. Ren Fail 25:67–75. https://doi.org/10.1081/jdi-120017469
Lampe E, Saback FL, Yoshida CFT, Niel C (1997) Infection with GB virus C/hepatitis G virus in Brazilian hemodialysis and hepatitis patients and asymptomatic individuals. J Med Virol 52:61–67.
Fernandez JL, Valtuille R, Hidalgo A, Del Pino N, Lef L, Rendo P (2000) Hepatitis G virus infection in hemodialysis patients and its relationship with hepatitis C virus infection. Am J Nephrol 20:380–384. https://doi.org/10.1159/000013620
Graninger M, Aberle S, Görzer I, Jaksch P, Puchhammer-Stöckl E (2021) Human pegivirus 1 infection in lung transplant recipients: prevalence, clinical relevance and kinetics of viral replication under immunosuppressive therapy. J Clin Virol 143:104937. https://doi.org/10.1016/j.jcv.2021.104937
Alcalde R, Nishiya AS, Casseb J, Inocencio L, Fonseca LA, Duarte AJ (2010) Prevalence and distribution of the GBV-C/ HGV among HIV-1 infected patients under antirretroviral terapy. Virus Res 151:148–152. https://doi.org/10.1016/j.virusres.2010.04.008
De Oliveira LA, Martins RMB, Carneiro MADS, Teles SA, Silva SA, Cardoso DDP, Lampe E, Yoshida CFT (2002) Prevalence and genotypes of GB Virus C/Hepatitis G virus among blood donors in Central Brazil. Memórias Do Instituto Oswaldo Cruz 97(7):953–957. https://doi.org/10.1590/S0074-02762002000700005
Lampe E, Saback FL, Viazov S, Roggendorf M, Niel C (1998) Age-specific prevalence and genetic diversity of GBV-C/ Hepatitis G virus in Brazil. J Med Virol 56:39–43.
Silva ASN, Silva CP, Barata RR, da Silva PVR, Monteiro PDJ, Lamarão L, Burbano RMR, Nunes MRT, de Lima PDL (2020) Human pegivirus (HPgV, GBV-C) RNA in volunteer blood donors from a public hemotherapy service in Northern Brazil. Virol J 17(1):153. https://doi.org/10.1186/s12985-020-01427-6
da Silva AS, de Campos GM, Villanova MG, Bezerra RDS, Santiago LMM, Haddad R, Covas DT, Giovanetti M, Alcantara LCJ, Elias MC, Sampaio SC, Kashima S, Slavov SN (2023) Human Pegivirus-1 detection and genotyping in Brazilian patients with fulminant Hepatitis. Pathogens 12:1122. https://doi.org/10.3390/pathogens12091122
Kennedy N, Tong CY, Beeching NJ, Lamden K, Williams H, Mutton KJ, Hart CA (1998) Hepatitis G virus infection in drug users in Liverpool. J Infect 37(2):140–147. https://doi.org/10.1016/s0163-4453(98)80168-0
Neibecker M, Schwarze-Zander C, Rockstroh JK, Spengler U, Blackard JT (2011) Evidence for extensive genotypic diversity and recombination of GB virus C (GBV-C) in Germany. J Med Virol 83(4):685–694. https://doi.org/10.1002/jmv.22029
N’Guessan KF, Anderson M, Phinius B, Moyo S, Malick A, Mbangiwa T, Choga WT, Makhema J, Marlink R, Essex M, Musonda R, Gaseitsiwe S, Blackard JT (2017) The impact of human pegivirus on CD4 cell count in HIV-Positive persons in Botswana. Open Forum Infect Dis 4(4):ofx222. https://doi.org/10.1093/ofid/ofx222
Xiang J, Sathar MA, McLinden JH, Klinzman D, Chang Q, Stapleton JT (2005) South African GB virus C isolates: interactions between genotypes 1 and 5 isolates and HIV. J Infect Dis 192(12):2147–2151. https://doi.org/10.1086/498170
Yu ML, Chuang WL, Dai CY, Lu SN, Wang JH, Huang JF, Chen SC, Lin ZY, Hsieh MY, Tsai JF, Wang LY, Chang WY (2001) The serological and molecular epidemiology of GB virus C/hepatitis G virus infection in a hepatitis C and B endemic area. J Infect 42(1):61–66. https://doi.org/10.1053/jinf.2000.0785
Lee CK, Tang JW, Chiu L, Loh TP, Olszyna D, Chew N, Archuleta S, Koay ES (2014) Epidemiology of GB virus type C among patients infected with HIV in Singapore. J Med Virol 86(5):737–744. https://doi.org/10.1002/jmv.23893
Funding
This research was funded by Fundação de Amparo à Pesquisa do Distrito Federal, project numbers 00193–00000932/2019-14 and 00193–00000751/2021-11.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Nucleotide sequence accession number
The nucleotide sequences of HPgV-1 have been deposited in the NCBI database under accession number OR449336 to OR449354.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Responsible Editor: Luis Augusto Nero.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
da Silva, F.G., Moura, D.R., de Oliveira, P.M. et al. Molecular characterization and frequency of human pegivirus type 1 (HPgV-1) in kidney transplant recipients from Central-West Brazil. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01490-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01490-z