Abstract
In this study, we aimed to characterize the distribution of Yersinia enterocolitica in a pork production chain in Brazil, as well as the virulence profile and antibiotic resistance of the obtained isolates. Samples from 10 pig lots obtained from finishing farms (water, feed, and barn floors, n = 30), slaughterhouse (lairage floors, carcasses at four processing steps, tonsils, and mesenteric lymph nodes, n = 610), and processing (end cuts, processing environment, n = 160) were obtained in Paraná state, Brazil, and subjected to Y. enterocolitica detection by ISO 10,273. The obtained isolates were identified based on biochemical and molecular features (16 s rRNA, inv, bioserotyping) and subjected to PCR assays to detect virulence (ail, ystA, ystB, virF, myfA, fepA, fepD, fes, tccC, ymoA, hreP, and sat) and multidrug resistance–related genes (emrD, yfhD, and marC). Also, isolates were subjected to disk diffusion test to characterize their resistance against 17 antibiotics from 11 classes and to pulsed field gel electrophoresis (PFGE) after XbaI macro-restriction. Y. enterocolitica was detected in a single sample (tonsil), and the obtained three isolates were characterized as serotype O:3, harboring ail, ystA, virF, myfA, tccC, ymoA, hreP, emrD, yfhD, and marC, and resistant to all tested antibiotics. The three isolates presented identical macro-restriction profiles by PFGE, also identical to isolates obtained from Minas Gerais, other Brazilian state; one selected isolate was identified as biotype 4. Despite the low occurrence of Y. enterocolitica in the studied pork production, the virulence potential and the antibiotic resistance profiles of the isolates demonstrated their pathogenic potential, and the macro-restriction profiles indicate strains descending from a common subtype in the pork production chain of two Brazilian States.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pork is the most produced and consumed animal protein in the world [1]. In Brazil, swine production has been showing relevant growth rates since the 1980s, mainly due to the intensive breeding of animals, associated with genetic, nutritional, and health improvement [2]. However, an intensive farming system can enhance the spread of important pathogens in the swine production, such as the use of collective drinking and feeders, high stool density, direct contact between animals, and number of animals per barn [3, 4].
Different foodborne pathogens can be associated to the swine chain. Yersinia enterocolitica is considered an emerging pathogen and pigs are described as reservoirs, once they can carry this pathogen asymptomatically into their lymph nodes, tonsils, and intestines [5, 6]. As consequence, poor hygienic conditions and inadequate procedures during pigs slaughtering and processing can facilitate the contamination and spread of Y. enterocolitica to carcasses, environment, utensils, and end products [7,8,9].
The pathogenic potential of Y. enterocolitica to humans is determined by virulence features encoded by genes located at the bacterial chromosome or in a plasmid: pYV [10, 11]. Virulence activities of chromosomal genes are mainly related to adhesion and invasion (ail, inv, mvf, hreP), toxin production (yst), enterotoxic activity (tccC), iron production (fes, fep), and virulence modulation (ymoA) [10, 12,13,14,15]. Among the pYV genes, virF is described as a transcriptional regulator of genes associated with important proteins, such as YadA (adhesin A) and Yops (other Yersinia proteins), related to adherence and activity against host immune cells [16, 17].
Besides the pathogenic potential of foodborne pathogens, there is a current worldwide concern related to antibiotic resistance [18]. Y. enterocolitica is usually susceptible to different antibiotics, such as aminoglycosides, tetracycline, chloramphenicol, extended spectrum cephalosporins, and trimethoprim-sulfamethoxazole [19]. However, as Y. enterocolitica can be hidden in lymphatic tissues since early ages of pigs, it is consequently subjected to all antibiotic treatments during the different stages of swine production and it leads to a natural development of resistance against these substances [20, 21]. In addition, the contact with other bacteria allows transference of genetic material, leading to modifications and acquisition of resistance-related genes and plasmids, resulting in increase of the antibiotic resistance characteristics by Y. enterocolitica strains [22, 23].
There are no official data or report of human yersiniosis associated to pork consumption in Brazil, but some studies have demonstrated the relevance of the pork production chain in harboring Y. enterocolitica [24,25,26,27]. Thus, studies that characterize the distribution and the virulence of Y. enterocolitica in Brazil are necessary to support the epidemiological characterization of this foodborne pathogen in the Brazilian pork production chain. Here, we characterized the distribution, virulence, and antibiotic resistance of Y. enterocolitica in Western Paraná, a Southern Brazilian region known by its relevance in the pork production.
Material and methods
Sampling
A pork production chain from Western Paraná, a Southern Brazilian state, was selected for this study. Ten pig lots from different farms were selected and samples of water (n = 10, 25 mL), feed (n = 10, 25 g), and barn floors (n = 10, footprint, as described by Botteldoorn et al. [28]) were obtained. During the slaughtering of the selected pig lots, carcasses were surface sampled at different stages: after bleeding (n = 100), after singeing (n = 100), after evisceration (n = 100), and after final rinse (n = 100): four sterile molds of 100 cm2 were placed in different carcass sites and swabbed with pre-moistened sponges (3 M Microbiology, St. Paul, MN, USA), as described by ISO 17,604 [29]. Prior to slaughtering, the lairage floors of pig lots were sampled by footprint (n = 10). Mesenteric lymph nodes (n = 100, 12.5 g) and palatine tonsils (n = 100, 12.5 g) were also samples from the selected carcasses. During the processing of the selected lots, surface samples of end cuts (n = 40), knives (n = 40), steel gloves (n = 40), cutting boards (n = 20), and conveyor belts (n = 20) were also obtained, as described above. All samples were transferred to sterile bags and kept refrigerated until laboratory analysis.
Detection of Yersinia enterocolitica
The samples were subjected to Y. enterocolitica detection based on ISO 10,273 [30], with modifications. Aliquots of 25 mL of water and 25 g of feed were diluted in 225 mL peptone-sorbitol-bile (PSB, Sigma-Aldrich, St. Louis, MO, USA), homogenized in Stomacher Seward 400® for 1 min (230 rpm). Samples from surface, lymph nodes, and tonsil were diluted in 160 mL and 112.5 mL of 0.1% peptone saline, respectively, and homogenized. Forty milliliters of aliquots of these samples were transferred to falcon tubes and centrifuged at 2,000 × g for 15 min. The obtained pellet was suspended in 10 mL of PSB broth and incubated at 25° C for 72 h. After incubation, 0.5 mL aliquots of the PSB cultures were transferred to 4.5 mL 0.5% potassium hydroxide (KOH) solution for 20 s and then streaked on cefsulodin-irgasan-novobiocin agar (CIN, Oxoid, Basingstoke, England). The plates were incubated at 30 °C for 18 to 48 h, when typical colonies of Y. enterocolitica were observed (small colonies with “red bull’s eyes red” center). Up to three suspected colonies from each plate were selected, purified, and subjected to biochemical tests of urease, indole, citrate, glucose fermentation, glucose gas production, lactose fermentation, H2S production, mobility, and liquid disposal of lysine to confirm the results [30]. Y. enterocolitica subsp. enterocolitica ATCC 9610 was used as positive control.
Isolates that presented biochemical results coherent with Y. enterocolitica were subjected to DNA extraction by boiling [31] and PCR assays targeting inv and a specific region of 16 s rRNA, for Y. enterocolitica identification [32]. Also, PCR assays were performed targeting per, wbbU, wbcA, and wzt for characterization of serotypes O:9, O:3, O:8, and O:5,27, respectively [32]. Amplification reactions were conducted using Gotaq Green Master Mix (Promega Corp., Madison, WI, USA), 200 nMol of each primer, 40 ng of extracted DNA, and nuclease free water with 25 μL final volume. PCR products were visualized after 1.5% agarose gel electrophoresis in GelRed ™ Tris–borate-EDTA (TBE) buffer (Biotium, Inc., Fremont, CA, USA). Primer sequences, conditions used for PCR amplifications, and expected product sizes are specified in the Supplementary Table. Y. enterocolitica subsp. enterocolitica ATCC 9610 was considered as the positive control for PCR assays.
Isolates identified as Y. enterocolitica were subject to DNA macro-restriction with XbaI (Promega) and pulsed field gel electrophoresis (PFGE), as indicated by PulseNet (Centers for Disease Control and Prevention, Atlanta, GA, USA), following the protocol described by Ribot et al. [33]. The obtained band profiles were compared using the software Bionumerics 6.6 (Applied Maths, Ghent, Belgium), considering 5% optimization and 5% Dice coefficient. Band profiles from Y. enterocolitica isolates (n = 8) were included for a comparative analysis. These isolates were obtained from different steps of the pork production chain (tonsils, mesenteric lymph nodes, pork carcasses) in Minas Gerais state, Brazil, using the same isolation protocol adopted in this study and identified as bioserotype 4/O:3 [25].
Based on the obtained band profiles, one isolate was selected and biotyped in the Yersinia Research Reference Laboratory of the College of Pharmaceutical Sciences at the University of São Paulo (USP; Ribeirão Preto, SP, Brazil) using the protocol described by Petersen et al. [34].
Virulence-related genes
Isolates identified as Y. enterocolitica were subjected to PCR assays for detection of virulence-related genes, as described by Martins et al. [25]. virF, myfA, ystA, ystC, fepA, fepD, fes, tccC, ymoA, and hreP genes were screened by individual PCR assays using primers described by Bhagat and Virdi [35]. Primer sequences, PCR conditions, and expected product sizes are specified in the Supplementary Table.
Antibiotic resistance
Y. enterocolitica isolates were characterized according their antibiotic resistance based on disk diffusion assay, following the recommendations of the Clinical and Laboratory Standards Institute [36]. Seventeen antibiotics from eleven classes were considered: (1) aminoglycosides: gentamicin (10 μg) and amikacin (30 μg); (2) fluoroquinolones: ciprofloxacin (5 μg) and norfloxacin (10 μg); (3) tetracyclines: doxycycline (30 μg) and tetracycline (30 μg); (4) phenicols: chloramphenicol (30 μg); (5) third-generation cephalosporins: ceftriaxone (30 μg); (6) folate pathway inhibitors: trimethoprim (5 μg) and sulfonamide (300 μg); (7) carbapenem: meropenem (10 μg) and imipenem (10 μg); (8) quinolone: nalidixic acid (30 μg); (9) penicillins: ampicillin (10 μg) and amoxicillin (10 μg); (10) macrolides: azithromycin (15 μg); (11) lipopeptides: polymyxin B (300 IU). All antibiotics were purchased from Sigma-Aldrich. Escherichia coli ATCC 25,922 was considered as a pan-susceptible quality control. Results were interpreted according to enterobacterial susceptibility standards [36].
Also, the extracted DNA of the Y. enterocolitica isolates was subjected to PCR assays for detection of the antibiotic resistance–related genes yfhD, emrD, marC (multidrug resistance), and sat (streptogramins), as described previously [25, 35]. Primer sequences, PCR conditions, and expected product sizes are specified in the Supplementary Table.
Results
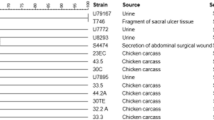
Considering all samples (n = 800), 108 (13.5%) presented characteristic colonies on CIN agar, allowing the selection of 307 typical isolates. After biochemical assays, 91 isolates from 48 samples presented results coherent with Y. enterocolitica, but only three, from a single sample (tonsil), were confirmed as such after PCR targeting 16 s rRNA and inv (Table 1). Based on these results, the frequency of Y. enterocolitica was 0.12% in the pork production chain, and 1.0% among tonsil samples. In addition, the three Y. enterocolitica isolates presented positive results only for wbbu, what characterize them as belonging to serotype O:3. The macrorestriction profiles of the three isolates are presented in Fig. 1. The isolates presented identical band profiles, also identical to 6 out of the 8 isolates obtained in Minas Gerais state [25]. These obtained isolates were grouped into two clusters with more than 86% genetic similarity between them. Once the obtained isolates presented identical macrorestriction profiles, one was selected and identified as belonging to biotype 4.
Schematic representation of macrorestriction profile (XbaI) by PFGE (Dice coefficient with 5% tolerance) and phenotypic profiles of antimicrobial resistance in disk diffusion test (CLSI, 2017) of Y. enterocolitica isolates obtained from state of Paraná and Minas Gerais. AMP, ampicillin; AMX, amoxicillin; TRI, trimethoprim; NAL, nalidixic acid; POL, polymyxin B; SUL, sulfonamides; DOX, doxycycline; NOR, norfloxacin
All isolates presented PCR amplification production for inv, ail, ystA, myfA, hrep, ymoA, tccC, and virF. However, fepA, fepD, and fes were not found in these isolates. Based on antibiotic resistance assays, the isolates presented PCR amplification products for yfhD, emrD, and marC. The three isolates were characterized as resistant to amoxicillin, ampicillin, and trimethoprim.
Discussion
The identification of the steps that have major influence on Y. enterocolitica contamination, from the farms to the animals’ slaughtering, is important for specific control measures and to monitor this pathogen along the production chain, resulting in safety of the end products [37]. Despite the low frequency of isolation, the identification of Y. enterocolitica in a palatine tonsil sample is consistent with the literature data, which indicates this site as the main location of this pathogen in pigs [8, 20, 25, 38, 39]. The presence of Y. enterocolitica in swine tonsils is considered as one of the main risk factors for carcass contamination in the slaughtering process and consequently in the end products, as these tissues are incised during inspection and slaughtering, increasing the chances of cross contamination [40, 41].
Low frequencies of Y. enterocolitica in pig slaughtering, especially in tonsils, have been reported in other studies conducted in Brazil. However, in São Paulo state, the occurrence of Y. enterocolitica positive samples was 8.0% in tonsils, tongues, submandibular, and mesenteric lymph nodes and knives [42]. Another study in São Paulo isolated 442 strains of Y. enterocolitica from a total of 792 samples collected from slaughtered pigs, slaughterhouse environment, and retail market [27]. From 400 samples of swine tonsils from Western Santa Catarina, 101 (25.25%) were positive for Y. enterocolitica [43]. In a similar study in Minas Gerais state, approximately 5% of swine tonsil samples were positive for Y. enterocolitica [25]. In other countries, especially in Europe, the presence of Y. enterocolitica in pig tonsils can be considered as high when compared to Brazilian studies, ranging from 13 to 90% [12, 38, 39, 44,45,46], while in the USA this prevalence was reported as 10% [47] and in China as 19.5% [48]. It is important to highlight that these differences can occur due to different isolation and detection approaches [17]. Conventional PCR and real-time PCR assays directly from samples and pre-enrichment broths have shown higher detection capacity of Y. enterocolitica than conventional procedures, and some studies already described that although the pathogen is present in samples, it is not always able to form colonies, which undermines the results obtained exclusively by culture plating [20, 21, 49]. Pork products usually contain a rich and diverse background microbiota, what can jeopardize the proper isolation of Y. enterocolitica by culture dependent methods [50]. Also, the low occurrence of Y. enterocolitica can be associated with the competing microbiota in the tested samples. It was already demonstrated that the serological prevalence of Y. enterocolitica is inversely proportional to the serological prevalence of Salmonella spp. in swine herds [51, 52]. In a parallel study with the same samples, Viana et al. [53] observed Salmonella occurrence of 45% in palatine tonsils, supporting this hypothesis.
Y. enterocolitica is described as an emerging pathogen in Europe due to the high number of reported yersiniosis cases in recent decades [5]. Fosse et al. [54] estimate that 77.3% of worldwide cases of yersiniosis may be associated with pork consumption. In Brazil, only a few studies describe the isolation and characterization of Y. enterocolitica from food, and the lack of epidemiological data from foodborne disease cases and outbreaks does not allow a proper characterization of the described yersiniosis, neither their potential link with contaminated pork products [26]. Y. enterocolitica isolates belonging to bioserotype 4/O:3 are described as the main pathogenic agents of yersiniosis in humans and animals worldwide, including in Brazil [10, 24, 32, 48, 55]. Martins et al. [25] identified Y. enterocolitica isolates from this bioserotype in a pork production chain in Minas Gerais, Brazil, and Paixão et al. [27] described that among 442 Y. enterocolitica isolates, all obtained from swine tonsils were characterized as serotype O:3.
The presence of the main virulence plasmid, pYV, is indicated by positive results for virF, responsible for coding one of the major pathogenicity factors of Y. enterocolitica, the type III secretion system [14, 17]. Despite presenting pYV, other chromosomal virulence genes are required for full pathogenicity capacity of Y. enterocolitica, such as inv, ail, ystA, myfA, hrep, ymoA, and tccC [14, 56]. These genes encode proteins that act in synergy for adhesion, internalization, and production of molecules necessary for enterotoxic activity to occur and to escape the host immune system [10, 12, 14, 24, 56, 57]. However, the absence of fepA, fepD, and fes indicates that the isolates obtained in this study have limited capacity for iron utilization, an essential factor for the development of most microorganisms [58].
Resistance to amoxicillin and ampicillin was already expected, once Y. enterocolitica is intrinsically resistant to these drugs [36]. All antibiotics usually recommended for yersiniosis treatment were effective against the obtained isolates: tetracycline, chloramphenicol, gentamicin, and third-generation cephalosporins [13]. Some studies have demonstrated increasing frequencies of multidrug-resistant Y. enterocolitica, as we observed based on phenotypical and molecular assays [25, 28, 31, 59]. Besides presenting amplification for yfhD, emrD, and marC, related to multidrug resistance, the three isolates also presented positive results for sat, related to resistance to streptogramins (virginiamycin), an antibiotic used as a feed additive [35, 60]. The use of antibiotics in animal production as growth promoters and prophylaxis is considered one of the main causes for the development of resistance, and the pork production is known by the wide use of drugs with these purposes when compared to other livestock systems [61]. Considering the strong epidemiological link between Y. enterocolitica and the pork production chain, monitoring the antibiotic resistance profiles of pork-related bacteria can be considered critical with regard to food safety and the performance of yersiniosis treatments in humans [19].
The results from the Minas Gerais’ strains are different probably because of the variances in pork production between these Brazilians states, and distinct drugs are probably being considered during production and resulting in these different profiles. Identification of Y. enterocolitica from Paraná and Minas Gerais states from a same serotype (O:3) and sharing identical band patterns suggests the low variability of this pathogen circulating in these two Brazilian states. Rusak et al. [26] reported a high similarity based on PFGE after XbaI macro-restriction among Y. enterocolitica from different bioserotypes and samples (swine, food, and clinical patients) in Brazil: all isolates identified as O:3 were grouped in a single cluster, with high similarity index.
Here we reported a low occurrence of Y. enterocolitica in a pork production chain, specifically in Western Paraná, a relevant pork production region in Southern Brazil. Despite the low occurrence, the Y. enterocolitica 4/O:3 isolates obtained presented high pathogenic potential and resistance to three antibiotics, but identical XbaI macro-restriction patterns with isolates obtained from other Brazilian state, Minas Gerais. These results suggest that strains descending from a common subtype may be circulating in the pork production chain of two states from different regions of Brazil, leading to further studies to elucidate their genomic profiles and potential clonality.
Data availability
Not applicable.
Code availability
Not applicable.
References
OECD/FAO (2018) Organisation for economic co-operation and development-food and agriculture organization of the United Nations. OECD-FAO Agricultural Outlook 2018-2027, OECD Publishing, Paris/Food and Agriculture Organization of the United Nations, Rome. https://doi.org/10.1787/agr_outlook-2018-en
SEBRAE/ABCS (2016) Brazilian micro and small business support service / Brazilian association of swine breeders. Mapping of Brazilian Pork Chain. Brasília, DF, 2016, 376p. https://www.abcs.org.br/wpcontent/uploads/2020/06/01_Mapeamento_COMPLETO_bloq.pdf. Accessed 10 Aug 2021
Laukkanen R, Martínez PO, Siekkinen KM, Ranta J, Maijala R, Korkeala H (2009) Contamination of carcasses with human pathogenic Yersinia enterocolitica 4/O:3 originates from pigs infected on farms. Foodborne Pathog Dis 6:681–688. https://doi.org/10.1089/fpd.2009.0265
Virtanen S, Salonen L, Laukkanen-Ninios R, Fredriksson-Ahomaa M, Korkeala H (2012) Piglets are a source of pathogenic Yersinia enterocolitica on fattening-pig farms. Appl Environ Microbiol 78:3000–3003. https://doi.org/10.1128/AEM.07805-11
European Food Safety Authority, European Centre for Disease Prevention and Control, Food Safety Authority E, Boelaert F, Stoicescu A, Amore G, Messens W, Hempen M, Rizzi V, Antoniou S-E, Baldinelli F, Dorbek-Kolin E, Van der Stede Y, ECDC staff members Taina Niskanen the, Haussig J, Kaczmarek M, Gomes Dias J, Barco L, Mancin M, Mantovani C, Sardella A, Antonelli P, Leati M, Anna Lettini A, Losasso C, Istituto Superiore di Sanita the, and staff members I, Morabito S, Scavia G, Knijn A, Tozzoli R, Iacoponi F, Moro O, Luca MD, Gattuso A, Suffredini E, Di Bartolo I, Delibato E, Anniballi F, Ianiro G, Altieri I (2021) The European Union One Health 2019 Zoonoses Report. EFSA journal Eur Food Saf Auth 19:e06406. https://doi.org/10.2903/j.efsa.2021.6406
Bancerz-Kisiel A, Szweda W (2015) Yersiniosis – a zoonotic foodborne disease of relevance to public health. Ann Agric Environ Med 22:397–402. https://doi.org/10.5604/12321966.1167700
Laukkanen R, Martínez PO, Siekkinen KM, Ranta J, Maijala R, Korkeala H (2008) Transmission of Yersinia pseudotuberculosis in the pork production chain from farm to slaughterhouse. Appl Environ Microbiol 74:5444–5450. https://doi.org/10.1128/AEM.02664-07
Laukkanen-Ninios R, Fredriksson-Ahomaa M, Korkeala H (2014) Enteropathogenic Yersinia in the pork production chain: challenges for control. Compr Rev Food Sci Food Saf 13:1165–1191. https://doi.org/10.1111/1541-4337.12108
Poljak Z, Dewey CE, Martin SW, Rosendal T, Christensen J, Ciebin B, Friendship RM (2010) Prevalence of Yersinia enterocolitica shedding and bioserotype distribution in Ontario finisher pig herds in 2001, 2002, and 2004. Prev Vet Med 93:110–120. https://doi.org/10.1016/j.prevetmed.2009.10.003
Bottone EJ (2015) Yersinia enterocolitica: revisitation of an enduring human pathogen. Clin Microbiol Newsl 37:1–8. https://doi.org/10.1016/j.clinmicnews.2014.12.003
Gnanasekaran G, Na EJ, Chung HY, Kim S, Kim YT, Kwak W, Kim H, Ryu S, Choi SH, Lee JH (2017) Genomic insights and its comparative analysis with Yersinia enterocolitica reveals the potential virulence determinants and further pathogenicity for foodborne outbreaks. J Microbiol Biotechnol 27:262–270. https://doi.org/10.4014/jmb.1611.11048
Drummond N, Murphy BP, Ringwood T, Prentice MB, Buckley JF, Fanning S (2012) Yersinia enterocolitica: a brief review of the issues relating to the zoonotic pathogen, public health challenges, and the pork production chain. Foodborne Pathog Dis 9:179–189. https://doi.org/10.1089/fpd.2011.0938
Fàbrega A, Vila J (2012) Yersinia enterocolitica: pathogenesis, virulence and antimicrobial resistance. Enferm Infecc Microbiol Clin 30:24–32. https://doi.org/10.1016/j.eimc.2011.07.017
Sabina Y, Rahman A, Ray RC, Montet D (2011) Yersinia enterocolitica: mode of transmission, molecular insights of virulence, and pathogenesis of infection. J Pathog 2011:1–10. https://doi.org/10.4061/2011/429069
Tennant SM, Skinner NA, Joe A, Robins-Browne RM (2005) Homologues of insecticidal toxin complex genes in Yersinia enterocolitica biotype 1A and their contribution to virulence. Infect Immun 73:6860–6867. https://doi.org/10.1128/IAI.73.10.6860-6867.2005
Peruzy MF, Murru N, Perugini AG, Capuano F, Delibato E, Mercogliano R, Korkeala H, Proroga YTR (2017) Evaluation of virulence genes in Yersinia enterocolitica strains using SYBR Green real-time PCR. Food Microbiol 65:231–235. https://doi.org/10.1016/j.fm.2017.03.004
Zadernowska A, Chajecka-Wierzchowska W, Łaniewska-Trokenheim Ł (2014) Yersinia enterocolitica: a dangerous, but often ignored, foodborne pathogen. Food Rev Int 30:53–70. https://doi.org/10.1080/87559129.2013.853775
Hashempour-Baltork F, Hosseini H, Shojaee-Aliabadi S, Torbati M, Alizadeh AM, Alizadeh M (2019) Drug resistance and the prevention strategies in food borne bacteria: an update review. Adv Pharm Bull 9:335–347. https://doi.org/10.15171/apb.2019.041
Frazão MR, Andrade LN, Darini ALC, Falcão JP (2017) Antimicrobial resistance and plasmid replicons in Yersinia enterocolitica strains isolated in Brazil in 30 years. Brazilian J Infect Dis 21:477–480. https://doi.org/10.1016/j.bjid.2017.04.006
Fredriksson-Ahomaa M, Stolle A, Stephan R (2007) Prevalence of pathogenic Yersinia enterocolitica in pigs slaughtered at a Swiss abattoir. Int J Food Microbiol 119:207–212. https://doi.org/10.1016/j.ijfoodmicro.2007.07.050
Bari ML, Hossain MA, Isshiki K, Ukuku D (2011) Behavior of Yersinia enterocolitica in foods. J Pathog 2011:1–13. https://doi.org/10.4061/2011/420732
Baym M, Stone LK, Kishony R (2016) Multidrug evolutionary strategies to reverse antibiotic resistance. Sci 80(351):aad3292–aad3292. https://doi.org/10.1126/science.aad3292
Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI (2017) Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat Rev Microbiol 15:689–696
Falcão JP, Falcão DP, Pitondo-Silva A, Malaspina AC, Brocchi M (2006) Molecular typing and virulence markers of Yersinia enterocolitica strains from human, animal and food origins isolated between 1968 and 2000 in Brazil. J Med Microbiol 55:1539–1548. https://doi.org/10.1099/jmm.0.46733-0
Martins BTF, Botelho CV, Silva DAL, Lanna FGPA, Grossi JL, Campos-Galvão MEM, Yamatogi RS, Falcão JP, Bersot LS, Nero LA (2018) Yersinia enterocolitica in a Brazilian pork production chain: tracking of contamination routes, virulence and antimicrobial resistance. Int J Food Microbiol 276:5–9. https://doi.org/10.1016/j.ijfoodmicro.2018.03.028
Rusak LA, Reis CMF, Barbosa AV, Santos AFM, Paixão R, Hofer E, Vallim DC, Asensi MD (2014) Phenotypic and genotypic analysis of bio-serotypes of Yersinia enterocolitica from various sources in Brazil. J Infect Dev Ctries 8:1533–1540. https://doi.org/10.3855/jidc.4533
Paixão R, Moreno LZ, Sena de Gobbi DD, Raimundo DC, Hofer E, Matté MH, Ferreira TSP, Gomes VTDM, Costa BLP, Moreno AM (2013) Characterization of Yersinia enterocolitica biotype 1A strains isolated from swine slaughterhouses and markets. Sci World J 2013:769097. https://doi.org/10.1155/2013/769097
Botteldoorn N, Heyndrickx M, Rijpens N, Grijspeerdt K, Herman L (2003) Salmonella on pig carcasses: positive pigs and cross contamination in the slaughterhouse. J Appl Microbiol 95:891–903. https://doi.org/10.1046/j.1365-2672.2003.02042.x
ISO (2015) International organization for standardization. Microbiology of the food chain — Carcass sampling for microbiological analysis (ISO 17604:2015). https://www.iso.org/standard/62769.html. Accessed 10 Aug 2021
ISO (2003) International organization for standardization. Microbiology of food and animal feeding stuffs — Horizontal method for the detection of presumptive pathogenic Yersinia enterocolitica (ISO 10273:2003). https://www.iso.org/standard/34564.html. Accessed 10 August 2021
De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L (2003) Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype Enteritidis in poultry. Appl Environ Microbiol 69:3456–3461. https://doi.org/10.1128/AEM.69.6.3456-3461.2003
Garzetti D, Susen R, Fruth A, Tietze E, Heesemann J, Rakin A (2014) A molecular scheme for Yersinia enterocolitica patho-serotyping derived from genome-wide analysis. Int J Med Microbiol 304:275–283. https://doi.org/10.1016/j.ijmm.2013.10.007
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ (2006) Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. https://doi.org/10.1089/fpd.2006.3.59
Petersen JM, Gladney LM, Schriefer ME (2015) Yersinia. In: Jorgensen JH, Carroll KC, Funke G, Pfaller MA, Landry ML, Richter SS, Warnock DW, Carroll KC, Funke G, Bernard KA, Dumler JS, Miller MB, Petti CA, Vandamme PAR (eds) Manual of clinical microbiology, 11th ed. John Wiley & Sons, Ltd, pp 738–751. https://doi.org/10.1128/9781555817381.ch39
Bhagat N, Virdi JS (2007) Distribution of virulence-associated genes in Yersinia enterocolitica biovar 1A correlates with clonal groups and not the source of isolation. FEMS Microbiol Lett 266:177–183. https://doi.org/10.1111/j.1574-6968.2006.00524.x
CLSI (Clinical and Laboratory Standards Institute) (2021) Performance standards for antimicrobial susceptibility testing. (M100-ed31), 31st edition. 352p. Wayne, Pennsylvania
Van Damme I, Berkvens D, Vanantwerpen G, Baré J, Houf K, Wauters G, De Zutter L (2015) Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: distribution, quantification and identification of risk factors. Int J Food Microbiol 204:33–40. https://doi.org/10.1016/j.ijfoodmicro.2015.03.016
Fondrevez M, Minvielle B, Labbé A, Houdayer C, Rose N, Esnault E, Denis M (2014) Prevalence of pathogenic Yersinia enterocolitica in slaughter-aged pigs during a one-year survey, 2010–2011, France. Int J Food Microbiol 174:56–62. https://doi.org/10.1016/j.ijfoodmicro.2013.12.027
Van Damme I, Habib I, De Zutter L (2010) Yersinia enterocolitica in slaughter pig tonsils: Enumeration and detection by enrichment versus direct plating culture. Food Microbiol 27:158–161. https://doi.org/10.1016/j.fm.2009.09.011
Biasino W, De Zutter L, Mattheus W, Bertrand S, Uyttendaele M, Van Damme I (2018) Correlation between slaughter practices and the distribution of Salmonella and hygiene indicator bacteria on pig carcasses during slaughter. Food Microbiol 70:192–199. https://doi.org/10.1016/j.fm.2017.10.003
Van Damme I, Mattheus W, Bertrand S, De Zutter L (2018) Quantification of hygiene indicators and Salmonella in the tonsils, oral cavity and rectal content samples of pigs during slaughter. Food Microbiol 71:120–128. https://doi.org/10.1016/j.fm.2017.03.012
Saba RZ, Rossi OD Jr, Kamimura BA, Lavezzo LF, Bürger KP, Vidal-Martins AMC (2013) Isolamneto de Yersinia enterocolitica em suínos de abate. Ars Vet 29:92. https://doi.org/10.15361/2175-0106.2013v29n4p92
Wildemann P, Gava D, Sfaciotte RAP, Melo FD, Ferraz SM, da Costa UM, Vaz EK (2018) Low occurrence of pathogenic Yersinia enterocolitica in pig tonsils at slaughter in Southern Brazil. Trop Anim Health Prod 50:671–675. https://doi.org/10.1007/s11250-017-1437-y
Bucher M, Meyer C, Grötzbach B, Wacheck S, Stolle A, Fredriksson-Ahomaa M (2008) Epidemiological data on pathogenic Yersinia enterocolitica in Southern Germany during 2000–2006. Foodborne Pathog Dis 5:273–280. https://doi.org/10.1089/fpd.2007.0076
Martínez PO, Fredriksson-Ahomaa M, Pallotti A, Rosmini R, Houf K, Korkeala H (2011) Variation in the prevalence of enteropathogenic Yersinia in slaughter pigs from Belgium, Italy, and Spain. Foodborne Pathog Dis 8:445–450. https://doi.org/10.1089/fpd.2009.0461
Martínez PO, Fredrlksson-Ahomaa M, Sokolova Y, Roasto M, Berzins A, Korkeala H (2009) Prevalence of enteropathogenic Yersinia in Estonian, Latvian, and Russian (Leningrad Region) pigs. Foodborne Pathog Dis 6:719–724. https://doi.org/10.1089/fpd.2008.0251
Wesley IV, Bhaduri S, Bush E (2008) Prevalence of Yersinia enterocolitica in market weight hogs in the United States. J Food Prot 71:1162–1168. https://doi.org/10.4315/0362-028X-71.6.1162
Liang J, Wang X, Xiao Y, Cui Z, Xia S, Hao Q, Yang J, Luo L, Wang S, Li K, Yang H, Gu W, Xu J, Kan B, Jing H (2012) Prevalence of Yersinia enterocolitica in pigs slaughtered in Chinese abattoirs. Appl Environ Microbiol 78:2949–2956. https://doi.org/10.1128/AEM.07893-11
Teodoro VAM, Pinto PSA, Vanetti MCD, Bevilacqua PD, Moraes MP, Pinto MS (2006) PCR technique application in Yersinia enterocolitica detection in non-inspected swine. Arq Bras Med Vet e Zootec 58:9–14. https://doi.org/10.1590/s0102-09352006000100002
Peruzy MF, Aponte M, Proroga YTR, Capuano F, Cristiano D, Delibato E, Houf K, Murru N (2020) Yersinia enterocolitica detection in pork products: evaluation of isolation protocols. Food Microbiol 92:103593. https://doi.org/10.1016/j.fm.2020.103593
Nathues C, Grüning P, Fruth A, Verspohl J, Blaha T, Kreienbrock L, Merle R (2013) Campylobacter spp., Yersinia enterocolitica, and Salmonella enterica and their simultaneous occurrence in German fattening pig herds and their environment. J Food Prot 76:1704–1711. https://doi.org/10.4315/0362-028X.JFP-13-076
Von Altrock A, Roesler U, Waldmann KH (2011) Herd factors associated with the serological Yersinia prevalence in fattening pig herds. Foodborne Pathog Dis 8:1249–1255. https://doi.org/10.1089/fpd.2011.0883
Viana C, Sereno MJ, Pegoraro K, Yamatogi RS, Call DR, Bersot LS, Nero LA (2019) Distribution, diversity, virulence genotypes and antibiotic resistance for Salmonella isolated from a Brazilian pork production chain. Int J Food Microbiol 310. https://doi.org/10.1016/j.ijfoodmicro.2019.108310
Fosse J, Seegers H, Magras C (2008) Foodborne zoonoses due to meat: a quantitative approach for a comparative risk assessment applied to pig slaughtering in Europe. Vet Res 39:01. https://doi.org/10.1051/vetres:2007039
Wang X, Qiu H, Jin D, Cui Z, Kan B, Xiao Y, Xu Y, Xia S, Wang H, Yang J, Wang X, Hu W, Xu J, Jing H (2008) O:8 serotype Yersinia enterocolitica strains in China. Int J Food Microbiol 125:259–266. https://doi.org/10.1016/j.ijfoodmicro.2008.04.016
Revell PA, Miller VL (2006) Yersinia virulence: more than a plasmid. FEMS Microbiol Lett 205:159–164. https://doi.org/10.1111/j.1574-6968.2001.tb10941.x
Heusipp G, Young GM, Miller VL (2001) HreP, an in vivo-expressed protease of Yersinia enterocolitica, is a new member of the family of subtilisin/kexin-like proteases. J Bacteriol 183:3556–3563. https://doi.org/10.1128/JB.183.12.3556-3563.2001
Schubert S, Fischer D, Heesemann J (1999) Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J Bacteriol 181:6387–6395. https://doi.org/10.1128/jb.181.20.6387-6395.1999
Ye Q, Wu Q, Hu H, Zhang J, Huang H (2016) Prevalence and characterization of Yersinia enterocolitica isolated from retail foods in China. Food Control 61:20–27. https://doi.org/10.1016/j.foodcont.2015.09.016
Seoane A, García Lobo JM (2000) Identification of a streptogramin a acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother 44:905–909. https://doi.org/10.1128/AAC.44.4.905-909.2000
Van Boeckel TP, Glennon EE, Chen D, Gilbert M, Robinson TP, Grenfell BT, Levin SA, Bonhoeffer S, Laxminarayan R (2017) Reducing antimicrobial use in food animals. Sci 80(357):1350–1352. https://doi.org/10.1126/science.aao1495
Acknowledgements
The authors are thankful to CAPES (Financial code 001), CNPq, Fundação Araucária, and FAPEMIG and to Juliana Pfrimer Falcão and Fábio Campioni for the biotype identification (Yersinia Research Reference Laboratory of the College of Pharmaceutical Sciences at the University of São Paulo, Ribeirão Preto, SP, Brazil).
Funding
CAPES (financial code 001), CNPq, and FAPEMIG.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by all authors. The first draft of the manuscript was written by Kadigia Pegoraro and Luciano dos Santos Bersot, and all authors commented on previous versions of the manuscript. Final review was conducted by Luís Augusto Nero and Luciano dos Santos Bersot. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
On behalf of all authors, the corresponding author states that all authors agreed in participating in this manuscript.
Consent for publication
On behalf of all authors, the corresponding author states that all authors agreed in publishing this manuscript in BJM after analysis, review, and acceptance.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Mariza Landgraf.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Mariza Landgraf
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pegoraro, K., Sereno, M.J., Viana, C. et al. Pathogenic potential and antibiotic resistance of Yersinia enterocolitica, a foodborne pathogen limited to swine tonsils in a pork production chain from Southern Brazil. Braz J Microbiol 52, 2335–2342 (2021). https://doi.org/10.1007/s42770-021-00591-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00591-3