Abstract
The molecular characterization of Anopheles gambiae complex mosquitoes in rice agroecosystems in Anambra State was studied from January to December 2020. The study was carried out in rice farms randomly selected from four local government areas in Anambra State. Mosquito larvae were collected with a WHO (350ml) standard dipper at the sampling point of each of the selected rice farms twice every month. The larval samples were taken to the insectary of the National Arbovirus and Vectors Research Centre (NAVRC) to be reared into adults. The molecular characterization of sibling species of Anopheles gambiae complex was accomplished with Polymerase Chain Reaction (PCR). A total of 289 An. gambiae complex mosquitoes were subjected to PCR and identified. The species were identified as An. gambiae s.s., 144 (49.8%) and An. coluzzii. 145 (50.1%). These observations indicated that malaria transmission in the Anambra state is mainly carried out by the Anopheles sibling species. This enhances, sustains, and extends the malaria disease transmission in this geographical area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa) is an important staple in Nigeria and is internationally consumed by all social and economic classes. Rice production is important in Nigerian agriculture and forms a vital element in the government's efforts to promote food security and curb food imports to feed the population (Amusa et al. 2020). Rice production in Nigeria has increased significantly. For instance, the quantity of milled rice in Nigeria increased from 2,818 million metric tonnes in 2010 to 5,000 million metric tonnes in 2021 (Sasu 2022). Land area under rice cultivation has also increased drastically. Despite the apparent increase in domestic rice production, local production in Nigeria has never met the growing demand and consumption of rice in the country. Kamai et al. (2020) noted that rice consumption in Nigeria is increasing rapidly due to some significant factors including changes in consumer preferences regarding rice, population growth, rising income levels, and rapid urban growth. Rice farming cuts across pre-harvest and post-harvest activities with a wide range of activities ranging from land selection, clearing, nursery, rice field preparation, and planting/transplanting, which require an aquatic environment (Amusa et al. 2020). Larvae of mosquitoes breed in rice farms, thereby putting people living around the farms and rice farmers potentially at risk of mosquito-borne diseases (Ukonze et al. 2017). Anopheline mosquitoes comprising of Anopheles gambiae complex are the major vector of human malaria and have been collected in rice farms in Omor, Anambra state breeding in large numbers (Ukonze et al. 2017). According to Emeka et al. (2021), the prevalence of malaria in Anambra state was as high as 74.2% which is quite alarming. Identification of mosquito species breeding in rice farms is based on morphological details. The sibling species of Anopheles are difficult to separate from their complexes using morphological features. There are at least eight related species that have a similar morphology that make up the Anopheles complex. The strong resemblance among the members of the An. gambiae complex mosquitoes make it very difficult for even an experienced eye to identify and differentiate them under the microscope. However, many mosquito species exist as complexes, such as Culex pipiens (s.l), Anopheles funestus (s.s), and Anopheles gambiae (s.l) (Cornel et al. 2012). Therefore, limiting their identification based on morphology alone may deter the knowledge of the genetic diversity of mosquito species.
Hence, the objective of this study is to characterize Anopheles gambiae complex mosquito’s species composition in rice agroecosystems in Anambra State, Nigeria. Therefore, molecular identification of An. gambiae complex species in the study area is necessary to provide baseline data on which members of the complex actively transmit malaria in the area and to develop effective management techniques.
Materials and methods
Anambra state lies in the south-eastern part of Nigeria at latitude 6.2758° N, and longitude 7.0068° E and covers 4,844 km2 (1,870 sq mi). The indigenous ethnic groups are the Igbo, which make up 98% of the population, and 2% of the Igala, who live mainly in the northwestern part of the state. Anambra has a tropical climate. It has two seasons, dry (November to March) and wet season (April to October). The average annual high temperature in Anambra is 32.88 °C (91.18 °F) and the average annual low temperature is 23.79 °C (74.82 °F). The warmest month of the year is February with an average temperature of 36.91 °C/98.44 °F. At 22.38 °C/72.28 °F on average, August is the coldest month of the year. The average annual rainfall is 212.36mm (8.36in). The driest month with 15.63mm/0.62in of rain is December. In September, the precipitation reaches its peak, with an average of 465.97mm/18.35in.
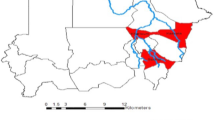
A longitudinal study was conducted at rice farm clusters in four of the local government areas of Anambra state (Fig. 1), which includes (Achalla) Awka North (6.1253417N, 6.771606E), (Anaku) Anyamelum (6.44914N, 6.926113E), (Umunze) Orumba South (5.943444N, 7.1913661E), and (Ufuma) Orumba North (6.083586N, 7.2064756E). These four local governments are known to be part of the rice-producing local government areas of Anambra State.
Study design
The larval survey was conducted over the course of two major seasons in Nigeria to assess Anopheles mosquito larvae. Sampling took place between January and December 2020 in the selected communities. A longitudinal study design was employed, with communities serving as study units. The four communities, each representing a local government region and their corresponding geographical coordinates were Anaku (6.44914N, 6.926113E), Achalla (6.1253417N, 6.771606E), Ufuma (6.083586N, 7.2064756E), and Umunze (5.943444N, 7.1913661E).
Mosquito larval sampling
Mosquito larvae were collected from the aquatic habitats of rice farms. The habitat was sampled for mosquito larvae twice every month (biweekly) and up to 20 dipper samples depending on the size of the aquatic habitat (ECDC Technical Report 2014). The mosquito larval population sampling method in the study area was determined using the sampling technique, according to Silver (2008) and ECDC Technical Report (2014). Samples were transported to the National Arbovirus and Vector Research Center Enugu (NAVRC) insectary. Culex larvae were separated from Anopheles larvae through careful observation of larval characteristics, including body shape, size, colouration, and the presence or absence of distinguishing structures such as respiratory siphons or palmate hairs, which allowed for initial differentiation. With the help of a magnifying glass or microscope to aid in the examination of finer details, the larvae were manually sorted and transferred into separate containers. Dead predators drowned in the larval water were removed from the bowls. The predators such as mosquitofish and chironomids were removed using micropipettes to pick them out. The Anopheles larvae were then placed in vials with water from the breeding sites, and labelled accordingly based on the sampling point and the farm from which it is collected. The labelled larval samples were reared into adults in an insectary prior to proper identification of the species.
Rearing of mosquito larvae to adults
The rearing of the mosquito larvae to adults was done according to the protocol by WHO (2013). The larvae were poured into bowls (500ml) and covered with a net to prevent oviposition by other mosquitoes and possible escaping of the emerging adult mosquitoes. The larvae were fed with ground pellets of fish feed daily. The water in the bowls was changed daily to prevent pollution. The larvae were monitored to know when they pupate so that the pupae can be introduced into pupae cups (5ml) and transferred into adult cages of 30 by 30cm3. Female mosquitoes were sorted, aspirated, and identified morphologically. They were placed in 1.5 ml Eppendorf tubes with labels and kept dry for later study on dried silica gel.
Morphological identification of Anopheles gambiae s.l
The identification of the adult mosquito raised was carried out morphologically using established identification keys by Gillies and Coetzee (1987) and Coetzee (2020) (Fig. 4). Adult morphology such as legs, antennae, wings, and position of the abdomen was observed under the stereomicroscope (Olympus SZ61). All Anopheles mosquitoes were stored in Eppendorf tubes filled with silica gel in their solid form. Each Eppendorf tube had 70% of the silica gel served as a preservative to prevent the mosquito from decaying during transportation. All the tubes containing the preserved mosquitoes were delivered to the Laboratory of Nigerian Institute of Medical Research, Yaba, Lagos State, for Polymerase Chain Reaction (PCR) studies.
Deoxyribonucleic acid extraction (DNA extraction)
The Wilkins et al. (2006) method of Anopheles gambiae complex discrimination was used for molecular identification. Genomic DNA was extracted from the wings and legs of each mosquito using a Blood-Animal-Plant DNA preparation Kit manufactured by Jena Bioscience, Germany, with slight modifications to the manufacturer’s protocol. The isolated genomic DNA (gDNA) was checked using NanoDrop 2000c to check for the quantity and purity of the samples. The genomic DNA extracted was also run on 1.5% agarose prepared with 1X TBE buffer at 80 V for 1 h to confirm the presence of DNA.
Molecular identification of sibling species of Anopheles gambiae complex using Polymerase Chain Reaction (PCR)
Deoxyribonucleic acid extract from the above described was used for the identification of mosquito siblings’ species. Mosquito DNA was amplified by PCR amplification of the ribosomal DNA (rDNA) (Wilkins et al. 2006). This method is based on species-specific single nucleotide polymorphisms (SNPs) in the intergenic spacer region (IGS). PCR amplification was carried out using 5X Hot FIREPol Blend Master Mix with 7.5 mM MgCl2 from Solis Bio-Dyne Estonia. A 12.5 μL reaction mixture was prepared using 2.5 μL of 5X Hot FIREPol, 0.5 μL each of primers (Table 1), 5.5μL of nuclease-free water and 1μL of template DNA. The PCR was run using the following conditions: initial denaturation at 95 °C for 5 min, 30 cycles for denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, elongation at 72 °C for 30 s, and 1 cycle for final elongation at 72°C for 5 min (Higa et al. 2010).
Gel electrophoresis
A total of 1.5% agarose gel was prepared and used to run electrophoresis for a period of 1 h and 30 min at 120 V. The gel was stained red by fluorescence dye ethidium bromide (EtBr) for 10 min. Next, 10 μl of the DNA ladder, PCR product, and the negative control was then added into each centrifuge well for electrophoresis and the gel was viewed using a gel documentation machine with the gel picture taken under UV light. The band was visualized with a dark reader trans-illuminator (Fig. 5). The expected band size for An. quadriannulatus is 637 bp, An. arabiensis 387 bp, An. gambiae s.s. 463 bp, An. melas 528 bp, An. merus 528 bp, An. coluzzii 333 bp, and An. gambiae 221 bp.
Data analysis
All the data obtained were analyzed using Statistical Package for Social Sciences (SPSS) software version 26.0. The means and standard deviations for the number of Anopheles mosquitoes collected for each species and measured study site were calculated. The differences in the numbers of Anopheles mosquitoes of each species collected from each location were analysed using one-way variance analysis (ANOVA). The Chi- square test (X2-test) was used to analyze and compare the abundance of the mosquito species in the four locations. P-Value less than 0.05, (P < 0.05) was considered not significant.
Results
Figure 2 shows the abundance of Anopheles mosquito larvae in relation to the locations of the rice farms in the study area. The data shows that the Anopheles mosquito larva was more abundant at Ufuma (15.6 ± 14.3), followed by Anaku (15.2 ± 18.2), Achalla (14.6 ± 18.8) and least abundant at Umunze (8.7 ± 9.5). There was no significant difference in the distribution of the Anopheles mosquito larva abundance in the selected rice farms with a p-value of 0.419 (P > 0.05).
Figure 3 show that the Anopheles mosquito larva was more abundant during the rainy season (April to October) and least abundant during the dry season (November to March). There was a significant difference in the distribution of the Anopheles mosquito larva in the seasonal abundance with a p-value of 0.00 (P < 0.05). For the interaction between the rice farms and the seasons, there was no significant interaction in the abundance of the mosquito larvae with regards to seasons and the rice farms in the different locations with a p-value of 0.56 (P > 0.050).
The morphological characteristics of female An. gambiae s. l. and Anopheles gambiae s.s. collected in this study was presented in Fig. 4. Other morphological identification keys used in the laboratory were those of Coetzee (2020).
A total of 289 (100%) An. gambiae s.l. complex mosquitoes collected at random from their habitats at different rice farms were subjected to PCR for molecular identification. Of this, 145 (50.1%) were An. coluzzii while 144 (49.8%) were An. gambiae s.s. An. coluzzii was more abundant in Anaku 36 (51.4%) and Achalla 59 (77.6%) while An. gambiae s.s. was more abundant in Ufuma 53 (67%) and Umunze 40 (62.5%). The Anopheles larvae were found in all four habitat types sampled at the different sampling locations. However, the abundance of the mosquito species was significantly different (P < 0.05) among the different locations (Table 2).
Figure 5 shows the electrophoresis gel image of the Anopheles gambiae complex speciation. The expected band size for An. coluzzii is 463 and 333 bp while that of An. gambiae is 463 and 221 bp. The first line is the DNA ladder showing the base pairs. Numbers 2 to 4 show the control sample identifying positive An. gambiae, a negative control, and a positive An. Coluzzii. The rest were the samples that were expressed. All the samples expressed were An. Coluzzii.
Discussion
Anopheles gambiae s.s. is a particularly important vector for malaria transmission because it is highly efficient at transmitting the Plasmodium parasites to humans (Sinka et al. 2010). This species is found primarily in sub-Saharan Africa, where it is responsible for most malaria cases. Anopheles coluzzii is also a significant vector of malaria, although it is found primarily in West Africa. This species is less efficient at transmitting the plasmodium parasites than Anopheles gambiae s.s.(Caputo et al. 2021) however, it remains a significant contributor to the spread of malaria in the region. Also, neither species has distinguishing characteristics for rapid, accurate morphological identification, therefore, identification must rely on molecular methods (Coetzee et al. 2013).
In this study, the Anopheles mosquito larva was more abundant during the rainy season (April to October) and least abundant during the dry season (November to March). The highest peak abundance was experienced in July, which happens to be the beginning of the rice planting season and middle of the rainy season. Nothing was recorded in December, January, and February as that happens to be the peak of the dry season where little or no water body was identified. This finding coincides with Mwangangi et al. (2010), whose abundance and larval productivity were associated with rice cropping patterns and rainfall. Also, the abundance of Anopheles mosquito larvae in relation to the locations of the rice farms harbour similar amount of mosquito larvae. It indicates that the rice farms share the same ecological conditions, such as the type of crop planted, the climate and the kind of chemical pesticides used for pest control, among others.
In this study, 289 An. gambiae s.l. complex mosquitoes subjected to molecular identification using PCR. Anopheles gambiae s.s. and An. coluzzii was the only sibling species in the four rice farm communities identified in the study area. However, An. coluzzii was less dominant than the two sibling species identified. This is in accordance with work reported by Chukwuekezie et al. (2020), where only An. gambiae s.s. was reported in Anambra and Abia state and An. coluzzii in Ebonyi state. The factors responsible for the distribution of the sibling species in the study areas are unclear, yet few speculations such as habitat preference, breeding behaviour and climate and seasonal variation could be adduced to explain the pattern observed. However, ecological factors such as water availability, temperature and humidity, vegetation and landscape, human activities, and habitat modification, among others had been identified by different researchers as driving forces for the distribution of An. gambiae complex in West Africa (Oyewole and Awoola 2006; Noutcha and Anumudu 2009). In related studies, the presence of An. gambiae s.s. has been reported in Imo, Anambra, and Abia States (Irikannu et al. 2019; Chukwuekezie et al. 2020) while An. coluzzii was only reported in Ebonyi State (Chukwuekezie et al. 2020). In Ibadan Nigeria, An. gambiae s.s. and An. coluzzii were also reported with An. gambiae s.s. also being the dominant species (Okorie et al. 2015). In Osun state, An. gambiae s.s. was the only species found across all localities except for Osogbo and Ibokun where An. Arabiensis and An. gambiae s.s. were detected (Adeleke et al. 2018).
The observation from the study indicates that malaria transmission in the Anambra state is mainly carried out by the Anopheles gambiae sibling species, An. gambiae s. s. and An. coluzzii. Controlling the two mosquito species would be a bit difficult because although they are indoor biters, they exhibit different behaviour (Perugini et al. 2020) such as differences in the times of day that they are most active. Therefore, by understanding the epidemiology and biology of these two species, researchers can target their control interventions more strategically to achieve maximum effectiveness. For instance, they can be controlled majorly using indoor residual spraying (IRS) and impregnated treated nets (ITNs) inventions in which at the time of the study, the intervention has already taken place in 2014 and 2016 respectively “NMEP Strategic Plan 2014–2020” (2014) in Anambra state. Therefore, in this study area, rice agroecosystems should be included as target areas in the control interventions set up in the state.
The identification of An. gambiae s. l. is critical for the monitoring and control of mosquito-borne malaria. Correct identification is essential for maintaining accuracy in the findings, as well as applying the appropriate interventions. The polymerase chain reaction has been used successfully in identifying An. gambiae s. s. and An. coluzzii (Fredrick et al. 2016). This procedure has numerous advantages, such as being fast, reliable, and cost-effective. However, it also has a few disadvantages, such as potential contamination, the sensitivity of the reagents, and limitations in verifying results. The polymerase chain reaction is a powerful and useful tool for special identification data and provided invaluable information about the interactions between different species and the effects of human activity on these populations.
Molecular methods such as PCR are essential for providing a reliable way to distinguish between An. gambiae s. l., but they should be supplemented with other methods, such as morphological assessments, Enzyme-linked Immunosorbent assay (ELISA), DNA sequencing, and Protein analysis. For example, TaqMan SNP genotyping (Walker et al. 2007) is a method of DNA analysis that uses PCR to amplify and detect specific DNA sequences in a sample. With an increasing understanding of the genetics and morphology of this species, additional diagnostic tests can be developed that hold the potential for more accurate identification.
Overall, Anopheles gambiae s. s. and Anopheles coluzzii are important species of mosquitos that are crucial to our understanding of malaria transmission, epidemiology, and control. And their presence in the rice agroecosystem enhances, sustains, and extends malaria disease transmission in this geographical area.
References
Adeleke MA, Adeyemi JA, Fassai KA, Oforka LC, Adeogun AO, Olatunde GO (2018) Molecular characterization of and insecticides susceptibility status of Anopheles gambiae complex (Giles 1902) in Osun State, Southwestern Nigeria. Nigeria J Entomol 34:69–76
Amusa TA, Igwe KC, Oti GO (2020) Environmental risks management practices among rice farmers in rice producing areas of Imo State, Nigeria. J Sustain Agric Environ 18(1):101–121. www.mouau.edu.ng. Accessed 10 Jun 2019
Caputo B, Pichler V, Bottà G, De Marco C, Hubbart C, Perugini E, della Torre A (2021) Novel genotyping approaches to easily detect genomic admixture between the major Afrotropical malaria vector species, Anopheles coluzzii and Anopheles gambiae. Mol Ecol Resour 21(5):1504–1516. https://doi.org/10.1111/1755-0998.13359
Chukwuekezie O, Nwosu E, Nwangwu U, Dogunro F, Onwude C, Agashi N, Gnanguenon V (2020) Resistance status of Anopheles gambiae (s.l.) to four commonly used insecticides for malaria vector control in South-East Nigeria. Parasit Vectors 13(1):1–10. https://doi.org/10.1186/S13071-020-04027-Z
Coetzee M (2020) Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar J 19:70. https://doi.org/10.1186/s12936-020-3144-9
Coetzee M, Hunt RH, Wilkerson R, Della Torre A, Coulibaly MB, Besansky NJ (2013) Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619:246–274 (PMID: 26131476)
Cornel A, Lee Y, Fryxell RT, Siefert S, Nieman C, Lanzaro G (2012) Culex pipiens sensu lato in California: a complex within a complex? J Am Mosq Control Assoc 28:113–121
ECDC Technical Report (2014) Field sampling methods for mosquitoes, sandflies, biting midges and ticks. https://doi.org/10.2900/416333
Emeka A, Catherine EC, Emenike OC, Emeka NK, Okemadu OC (2021) Prevalence and intensity of malaria infection and associated risk factors in Anambra State, Nigeria. World J Biol Pharm Health Sci 7(3):30–45. https://doi.org/10.30574/wjbphs.2021.7.3.0094
Fredrick GK, Anne MA, John BK, Enock M, Anne K, Anges Y, Ambrose WO (2016) Molecular identification of Anopheles gambiae sensu stricto Giles (formerly Anopheles gambiae Savannah Form) in Kamuli District, Uganda. Afr J Biotechnol 15(39):2124–2131. https://doi.org/10.5897/ajb2016.15444
Gillies MT, Coetzee M (1987) A supplement to the Anophelinae of Africa South of the Sahara. Publ S Afr Inst Med Res 55:1–143
Higa Y, Tomas T, Tsuda Y, Miyagi I (2010) A multiplex PCR-based molecular identification of five morphologically related, medically important subgenus Siegomyia mosquitoes from the Genus Aedes (Diptera: Cullicidae) found in the Ryukyu Archipelago, Japan. Jpn J Infect Dis 64:312–316
Irikannu KC, Onyido AE, Umeanaeto PU, Onwube O, Ogaraku JC, Egbuche CM et al (2019) Molecular characterization and malaria transmission potential of Anopheles gambiae complex in Awka, Anambra state, Nigeria. Int J Mosq Res 6(6):96–101
Kamai N, Omoigui LO, Kamara AY (2020) Guide to rice production in Northern Nigeria. www.iita.org. Accessed 22 Dec 2022
Mwangangi JM, Shililu J, Muturi EJ, Muriu S, Jacob B, Kabiru EW, Novak RJ (2010) Anopheles larval abundance and diversity in three rice agro-village complexes Mwea irrigation scheme, central Kenya. Malar J 9(1):228. https://doi.org/10.1186/1475-2875-9-228
NMEP (2014) National malaria strategic plan 2014–2020, pp 1–113. https://www.health.gov.ng/doc/NMEP-Strategic-Plan.pdf. Accessed 2 Jul 2023
Noutcha MAE, Anumudu CI (2009) Entomological indices of Anopheles gambiae sensu lacto at a rural community in Southwestern Nigeria. J Vector Borne Dis 46:43–51
Okorie PN, Ademowo OG, Irving H, Kelly-Hope LA, Wondji CS (2015) Insecticide susceptibility of Anopheles coluzzii and Anopheles gambiae mosquitoes in Ibadan, Southwest Nigeria. Med Vet Entomol 29:44–50
Oyewole IO, Awoola TS (2006) Impact of Urbanization on bionomics and distribution of malaria vectors in Lagos, Southwestern Nigeria. J Vector Borne Dis 43:172–178
Perugini E, Guelbeogo WM, Calzetta M, Manzi S, Virgillito C, Caputo B, Pichler V, Ranson H, Sagnon N, Torre AD, Pombi M (2020) Behavioral plasticity of Anopheles coluzzii and Anopheles arabiensis undermines LLIN community protective effect in a Sudanese-savannah village in Burkina Faso. Parasites Vectors 13:277. https://doi.org/10.1186/s13071-020-04142-x
Sasu D (2022, November 18) Nigeria: production of milled rice 2010–2022. Statista. Retrieved December 22, 2022, from https://www.statista.com/statistics/1134510/production-of-milled-rice-in-nigeria/. Accessed 22 Dec 2022
Silver JB (2008) Sampling the larval population. Mosquito Ecology: Field Sampling Methods 165(4193):137–338. https://doi.org/10.1038/165378a0
Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, Okara RM, Boeckel TV, Godfray HCJ, Harbach RE, Hay HI (2010) The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit Vector 3:117. https://doi.org/10.1186/1756-3305-3-117
Ukonze C, Umeh C, Ezihe CK, Ononye B, Duru V, Obiefule I (2017) Physico-chemical parameters associated with mosquito abundance in rice farms at Omor, Anambra state, Nigeria. Bioscientist 5(1):82–87. http://www.bioscientistjournal.com/pub_2017/vol5/publication_Ukonze_etal_2017.htm. Accessed 22 Dec 2022
Walker ED, Thibault AR, Thelen AP, Bullard BA, Huang J, Odiere MR, Vulule JM (2007) Identification of field caught Anopheles gambiae s.s. and Anopheles arabiensis by TaqMan single nucleotide polymorphism genotyping. Malar J 6(1):1–9. https://doi.org/10.1186/1475-2875-6-23/FIGURES/3
WHO (2013) Malaria entomology and vector control. Guide for participants. https://apps.who.int/iris/bitstream/handle/10665/85890/9789241505819_eng.pdf. Accessed 2 Jul 2023
Wilkins EE, Howell PI, Benedict MQ (2006) IMP PCR primers detect single nucleotide polymorphisms for Anopheles gambiae species identification, Mopti and Savanna rDNA types, and resistance to dieldrin in Anopheles arabiensis. Malar J 5(1):1–7. https://doi.org/10.1186/1475-2875-5-125
Acknowledgements
We would like to express our gratitude to the members of our research team, who worked tirelessly to collect the data for this study, and their valuable feedback and support throughout the writing process. We would also like to thank Dr. C. M. Egbuche at Nnamdi Azikiwe University Awka Anambra, Nigeria for analysing the data for this study. We would also like to thank the community heads and farm owners in Anambra State for the support and assistance they rendered to facilitate the successful completion of this study. Finally, we are grateful for the support and research facilities provided by the School of Biological Sciences, Universiti Sains Malaysia, Malaysia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Financial or proprietary interests
No funding was received to assist with the preparation of this manuscript. The authors have no financial or proprietary interests in any materials discussed in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ukonze, C., Ezihe, E., Nzeukwu, C. et al. Molecular characterization of Anopheles gambiae complex mosquitoes in rice agroecosystems in Anambra State, Nigeria. Int J Trop Insect Sci 43, 1911–1918 (2023). https://doi.org/10.1007/s42690-023-01089-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-023-01089-1