Abstract
Introduction
Knowledge of the longitudinal and transverse relaxivities (r1 and r2) of a contrast agent (CA) is essential for its magnetic characterization. These parameters can be measured using Magnetic Resonance Imaging (MRI) clinical scanners with the advantage of characterizing the CA under the same experimental conditions where it will be employed. Nevertheless, when using MRI, there are several limitations to consider, and we provide ways to compensate for them to obtain accurate results.
Materials and Methods
We present a fast and robust methodology to determine the relaxivity of CA solutions using a 3 T MRI clinical scanner with a single-channel transmit-receive birdcage coil. We performed relaxivity measurements on a phantom consisting of five samples of copper sulfate at different concentrations.
Results
We optimized image acquisition for total scan time using three different pulse sequences. Post-processing steps following image acquisition were implemented in a semiautomatic MATLAB toolbox. Relaxation times were estimated using the three-parameter model with the Levenberg-Marquardt algorithm. Statistical comparisons demonstrate good reproducibility and robustness in the relaxivity estimation by each method.
Conclusions
This paper presented a methodology and a systematic discussion of experimental factors associated with relaxivity determination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Magnetic Resonance Imaging has become a powerful method in biomedical research because of its non-invasiveness, high spatial resolution and contrast, and the possibility to obtain quantitative information regarding dynamic processes. However, it has a relatively low sensitivity. One way to overcome this disadvantage is by using exogenous contrast agents (CA) (paramagnetic and superparamagnetic). The efficiency of the CA can be characterized by a parameter called relaxivity (longitudinal r1 and transverse r2) (Rohrer et al. 2005; Henoumont et al. 2009; Szomolanyi et al. 2019). Higher relaxivity allows a greater contrast enhancement in the image. Furthermore, higher relaxivity permits a lower dose for the same contrast enhancement, lowering CA toxicity (White et al. 2006; McDonald et al. 2015; McDonald et al. 2017).
The observed relaxation rate depends linearly on CA concentration. For a fast exchange regime between the inner sphere of the paramagnetic or superparamagnetic center (P/S-magnetic) and free water state, we get:
Here, the first term of the right member corresponds to the relaxation rate of the diamagnetic state. The second member corresponds to the P/S-magnetic contribution being [CA] the concentration and r1,2 the relaxivity. Relaxivities r1,2 depends on field strength, temperature and correlation times of the magnetic interactions. The linear relationship expressed by the Eq. (1) becomes more complex in vivo (Modo and Bulte 2007).
Although relaxivity is generally measured by NMR relaxometry (Rohrer et al. 2005; Henoumont et al. 2009; Haën et al. 2003; Jacques et al. 2010), there is an increasing trend to use MRI scanners for this purpose (Chen et al. 2020; Thangavel and Saritaş 2017; Knobloch et al. 2018). This approach has the advantage of characterizing the CA under the same experimental conditions (field strength, RF coil configuration, pulse sequences) where it is employed. However, we need to consider various sources of errors and inaccuracies like the proper selection of pulse sequence parameters to separate the different relaxation rates (\(1/{T}_{\mathrm{1,2}}\)) contribution to the MR signal, the influence of signal-to-noise ratio (SNR) on the precision of the relaxation time measurement; RF field inhomogeneity’s, susceptibility effects and temperature stability of the sample among others.
Given the increasing importance of relaxivity determination for CA characterization in MRI studies and the absence of a standardized procedure to this end, we present a systematic and brief evaluation of the factors that influence its determination using MRI clinical scanners. We suggest ways to prevent experimental errors and methods to compensate for them, developing a methodology and a semi-automated toolbox to determine the relaxivity of CA samples by MRI.
MRI experimental setup
Phantom characteristics
The methodology employs a phantom consisting of two sets of samples. The samples are contained in flat-bottomed polypropylene cylindrical vials with a volume of 2 mL and diameter of 10 mm, to facilitate the selection of homogeneous slices. The reference set consists of samples with known relaxation times while the second set contains CA samples whose relaxivities are to be determined. The CA sample concentration should not exceed the maximum dose allowed for the corresponding organ or tissue in which they will be employed. The sets will be prepared to obtain the same relaxation time range.
In all cases, sample volume should be enough to guarantee an adequate SNR and to permit the acquisition of at least two slices in a homogeneous region. Consequently, relaxation time estimation can be performed for each slice to compare and ensure that the sample is stable and homogeneous.
Another factor to consider is the sample's relative position within the phantom. To prevent the mutual influence of the sample’s macroscopic susceptibility, which can produce artifacts, a distance of at least one diameter between any two samples is necessary, especially for superparamagnetic iron oxide nanoparticles at high magnetic fields. This distance was determined through a trial and error procedure considering previous studies on geometric distortions (Gonzalez 2006). The samples must be fixed with their vertical axes parallel to each other so that the excited slice plane is orthogonal to all samples.
In MRI, coil volume is usually larger than NMR relaxometers, which facilitates the characterization of several samples simultaneously, thus reducing the required time.
Pulse sequences
Relaxation times can be determined using spin-echo sequences (SE) for T1 and T2 and inversion-recovery spin-echo sequences (IR-SE) for T1.
For SE, the equation describing pixel intensity can be expressed as:
Since no diffusion sensitizing gradients are applied, the last exponential factor in Eq. (2) can be discarded. The parameter \(A(x,y)=1-\mathrm{cos}\theta (x,y)\) accounts for RF inhomogeneity, which is discussed in more detail below. Repetition time TR and echo time TE are adjusted to separate, as much as possible, the contribution from each relaxation time.
In SE T1 estimation, TR is varied while keeping TE minimum, to minimize T2 weighting. Equation (2) is simplified to:
The minimum TR attainable affects the T1 measurement error by the MRI equipment. The error is higher in cases of short T1 values.
For SE T2 measurement, TE is varied and TR is chosen to fulfill TR≈5T1, minimizing T1 weighting. Equation (2) now becomes:
The minimum TE attainable affects the T2 measurement error. The error is higher for short T2 values.
In the case of IR-SE sequence, the signal equation can be expressed as:
where TI is the inversion time. By choosing TR ≥ 5T1 and setting minimum TE, Eq. (5) is simplified to:
When using IR-SE, the T1 estimate standard deviation is proportional to Brown et al. (2014):
Here σ refers to image noise standard deviation and DR to signal dynamic range. Hence the IR-SE has a two-fold advantage over the SE because its dynamic range is twice that of the SE. However, the IR-SE method is slower than SE.
RF field inhomogeneity
The aforementioned spatial inhomogeneity of the B1 excitation field results in flip angle deviations depending on the spatial position according to:
For any RF coil configuration, the B1 inhomogeneities depends on several factors like RF pulse shape, RF penetration depth, standing-wave effects, among others. In general, all these factors must be considered but their contribution depends on the specific RF coil transmission and reception configuration. These inhomogeneities reduce the magnetization vector dynamic range, thus reducing the estimated T1.
This spatial distribution of flip angles due to B1 inhomogeneity can be mapped with the dual-angle method using a homogeneous phantom. This method acquires two images, S1 and S2, with flip angles related by θ2 = 2θ1, and the flip angle distribution is obtained by Cunningham et al. (2006):
Another point related to the B1 excitation to be taken into account in MRI is that the duration of the selective pulse is several times longer than the non-selective used in the relaxometry. Then, during excitation, flip angle is more affected for those spin systems having very short T1, which is equivalent to the B1 inhomogeneity.
In addition, to minimize the influence of B1 inhomogeneity, the phantom must be always placed in the more homogeneous region of the coils.
Signal to Noise Ratio
The image SNR is another factor influencing the relaxation time estimation. We estimated SNR for each sample by region of interest (ROI) measurements. Let Ssample be the ROI image intensity in a sample and Sbackground be the ROI image intensity on the image background (air surrounding the samples), then SNR can be calculated as (National Electrical Manufacturers Association 2021):
The factor 0.665 accounts for the Rayleigh distribution of the noise in the magnitude image. If a real image is evaluated no correction factor is needed.
Susceptibility effects
Susceptibilities effects result from variations in the local magnetic field occurring near the interfaces of substances with different magnetic susceptibilities, in this case, the sample-air interface. These variations produce signal loss from T2*-dephasing and spatial missmapping of the MR signal, which is more noticeable at high magnetic field strength. To prevent these effects, the region of interest (ROI) in the samples should be selected away from its boundaries.
Temperature
The behavior of relaxation rates with temperature is different for each paramagnetic species (Kraft et al. 1987), therefore, the temperature of the samples should be kept constant throughout the experiment, to avoid any influence on the determination of the relaxivity.
MRI acquisition
MRI acquisition was performed in a 3 T Siemens MAGNETOM Allegra clinical scanner with a single-channel transmit-receive birdcage coil. We optimized pulse sequence parameters to reduce total scan time while preserving an adequate SNR. Reducing scan time is essential for samples with poor solubility or aggregation problems. A fast way to reduce overall scan time and increase SNR simultaneously is to degrade base resolution. Additionally, slice thickness was also increased to further improve SNR.
Slice plane positioning should be perpendicular to the vertical axis of the vials so that all selected regions are at the same height to the bottom of the vial to minimize signal differences caused by B1 inhomogeneities.
The choice of TR will depend on the T1 relaxation times of the unknown samples. When characterizing for the first time, we set TR to the maximum available in the scanner to study the relaxation process of long-T1 samples. If the estimated T1 values are short compared to this maximum TR, we can progressively reduce TR until TR≈5T1 for every sample.
To perform a more accurate estimation, the number of time points should be higher at the beginning of the relaxation curves (for low TI, TR, and TE) when the magnetization vector, and so the signal, changes more rapidly.
The pulse sequence parameters for each method are detailed in Table 1
Software pipeline
We implemented the following semiautomatic post-processing steps as a MATLAB toolbox:
-
Automatic handling of DICOM files: The first post-processing step after image acquisition is to rename and organize the raw DICOM images in a specific folder (Slices) the toolbox automatically creates. Images are sorted in ascending order of TRs, TEs, or TIs according to the corresponding measurement
-
Automatic noise segmentation and ROI selection: Automatic noise segmentation is done based on the Rician distribution of pixel intensity in the MR image (Gudbjartsson and Patz 1995). We applied a thresholding method based on the image histogram to suppress pixels corresponding to background noise (Barbará Morales and Sánchez-Bao 2012). Upon completing this step, each sample ROI is identified, eroded, and low pass filtered to remove its boundaries. Alternatively, the user can select the ROI manually, avoiding susceptibility effects at the edges of the ROIs.
-
Calculation of mean and standard deviation from each ROI: We calculated the mean (Im) and standard deviation of pixel intensity from each previously selected ROI along the different time points (different TRs, TEs, or TIs) in the relaxation curve.
-
Estimation of relaxation times: We employed a nonlinear fit with Levenberg-Marquardt (L-M) algorithm (Gavin 2019) to estimate relaxation times from Eqs. (3), (4) and (6). The L-M is a robust and flexible algorithm that combines two numerical minimization algorithms: the gradient descent method and the Gauss-Newton method.
The objective equation to minimize is the sum of square errors for each signal equation:
$$\widehat{\beta }\in {\mathrm{argmin}}_{\beta }F\left(\beta \right)={\sum_{i=1}^{n}\left[{I}_{{m}_{i}}-S\left({T}_{i},\beta \right)\right]}^{2}.$$(11)where Imi are the mean intensities computed for each ROI in the images and Ti are the timing parameters TR, TE, and TI. The estimated parameter β is a vector consisting of the relaxation times T1 or T2, the parameter A and the equilibrium signal S0
-
Calculation of relaxivities: The relaxivities (expressed in mM/s−1) are calculated via a linear fit of Eq. (1) once the user inputs each sample concentration in mM.
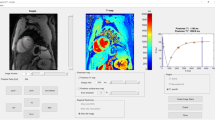
Figure 1 shows the graphical user interface (GUI) of the toolbox. The upper left part of the GUI comprises the buttons for DICOM file handling alongside image axes where the user can scroll through the different slices. The DICOM header info and the type of measurement are shown below these axes. At the bottom center, the user can find the buttons to extract information (mean and standard deviation of signal intensity) from each sample and visualize its signal evolution.
The signal evolution for each sample can be visualized in the upper right corner axes. Below this axis is a table filled with sample coordinates, relaxation times, and concentrations. There are also two buttons: one on the left to estimate the relaxation times and one on the right to calculate the relaxivity.
Figure 2 represents three fundamental steps in the determination of relaxivity. The first step is image acquisition, schematically represented in graphs (a) and (d). From each detected sample, we calculate the mean intensity of its corresponding ROI for each TE or TI. Using these intensities, we constructed the relaxation curves as shown in graphs (b) and (e). Subsequently, we estimated using the Levenberg-Marquardt algorithm the relaxation times for each sample. Finally, we obtained the relaxivity through a least squares fitting procedure using the known concentration as displayed in graphs (c) and (d).
Graphical scheme of three fundamental steps in the determination of relaxivity using MRI. Graphs (a) and (d) represent image acquisition for different TI (TE). Graphs (b) and (e) show the relaxation curves for each sample in a T1 (T2) calculation. Graphs (c) and (f) display the least square fit to determine the r1 (r2) relaxivity
To summarize the whole methodology Fig. 3, shows a general flowchart, comprising all the proposed steps.
The toolbox code is available to download at (https://github.com/isra-RM/KRelax) as a public repository.
Comparison among methods
As the first step in data analysis, we tested for normality in the relaxivity values for each method using the Shapiro-Wilks test (p > 0.05). To demonstrate the robustness of the methodology, we analyzed the effect of the highest concentrated sample and the solvent in the relaxivity estimation. In addition, we compared the agreement in the equilibrium signal estimation across methods. If the relaxivity values comply with a normal distribution, a two-sample t-test (p > 0.05) will be used to analyze the abovementioned effects, otherwise its non-parametric equivalent Mann-Whitney U-test is employed.
The agreement and reproducibility of the estimated parameters are quantified using the percentage deviation to the mean (PDM) and the coefficient of variation (CV). The percentage deviation to the mean quantifying the agreement between any two variables S1 and S2 is defined as:
To assess the reproducibility of the estimates of a variable S, we calculated the coefficient of variation defined as:
Validation using copper sulfate solutions
For the validation of the methodology, several measurements of relaxation times, equilibrium signals, and relaxivities were performed on a phantom containing samples of copper sulfate solutions at five different concentrations (0.97 mM, 2.00 mM, 3.90 mM, 7.8 mM, and 15.7 mM). The first step in data analysis was to verify the normality of relaxivity values obtained for each method (Table 2).
The Shapiro Wilks normality test suggests that relaxivity measurements follow a normal distribution. In order to test the reproducibility of the methodology, the CV was calculated for the three methods (Table 3).
The CV values indicates a very low variability of all methods. As expected, the most stable method is the IR-SE.
To demonstrate the stability of the methodology, we considered the effect of removing the highest concentrated sample from the relaxivity estimation (Table 4). We compared the estimation using all samples with the estimation without the highest concentrated sample, using a two-sample t-test.
We obtained p-values of p = 0.123 for T1 IR-SE, p = 0.108 for T1 SE, and p = 0.506 for T2 SE, indicating that the highest concentrated sample does not significantly affect the relaxivity estimation. The reason to consider the effect of the highest concentrated sample is the high variance in its relaxation time estimation. Testing the effect of high-concentration samples allows finding an upper threshold in concentration, and hence a lower threshold in relaxation time, that can be estimated without affecting relaxivity.
Table 5 shows the effect of discarding the solvent (setting its relaxation rate to zero) from the relaxivity estimation.
We obtained, using a two-sample t-test, p-values of p = 0.143 for T1 IR-SE, p = 0.702 for T1 SE, and p = 0.077 for T2 SE, indicating that the presence of the solvent does not modify the relaxivity estimation.
Table 6 shows a comparison of the equilibrium signals estimated by each method with the experimental value. This value represents the highest intensity signal point in the relaxation curves.
As expected, the T1 IR-SE method shows the smallest PMD compared to the experimental value obtained from the T1 relaxation curve. As shown, PMD decreases as the concentration increases, reflecting that less concentrated samples (longer relaxation times) might not fully relax for the maximum TI available in the scanner. The PMD of the T2 SE method shows the opposite trend; the higher the concentration, the higher the PMD. The equilibrium signal in the T2 SE method is determined experimentally as the intensity of the first point in the decay curve, hence, the difference between this value and the L-M estimate becomes higher for samples with high concentrations (short T2 times).
Conclusions
In this paper, we presented a brief and systematic analysis of the relaxivity determination in clinical scanners identifying the sources of error and their respective compensation methods. We developed a fast and robust methodology, including a MATLAB toolbox, to determine the relaxivities of contrast agent samples using MRI clinical scanners. We optimized image acquisition using IR-SE and SE pulse sequences to minimize total scan time, which prevents problems derived from poor sample stability. Post-processing steps following image acquisition were implemented in a semiautomatic MATLAB toolbox to speed up relaxivity determination. Statistical comparisons of the estimated parameters demonstrate the reproducibility and accuracy of the toolbox.
References
Barbará Morales E, Sánchez-Bao R, González Dalmau E. Comparación de algoritmos de segmentación de ruido aplicados a imágenes de resonancia Magnética. Ing Electrónica, Automática y Comunicaciones. 2012;33(3):8–18–18.
Brown RW, Cheng YC, Haacke EM, Thompson MR, Venkatesan R. Magnetic resonance imaging: physical principles and sequence design. John Wiley & Sons; 2014.
Chen A, et al. The effect of metal ions on endogenous melanin nanoparticles used as magnetic resonance imaging contrast agents. Biomater Sci. 2020;8(1):379–90. https://doi.org/10.1039/c9bm01580a.
Cunningham CH, Pauly JM, Nayak KS. Saturated double-angle method for rapid B1+ mapping. Magn Reson Med. 2006;55(6):1326–33. https://doi.org/10.1002/mrm.20896.
de Haën C, Cabrini M, Akhnana L, Ratti D, Calabi L, Gozinni L. Gadobenate dimeglumine 0.5 M solution for injection (MultiHance®): Pharmaceutical formulation and psysicochemical properties of a new magnetic resonance imaging contrast medium. Appl Radiol. 2003;32(4 SUPPL.):12–20.
Gavin HP. The Levenberg-Marquardt algorithm for nonlinear least squares curve-fitting problems. Duke Univ. 2019;1–19. http://people.duke.edu/~hpgavin/ce281/lm.pdf.
González E. Descriptores cuantitativos de calidad para Tomógrafos por Resonancia Magnética. Dissertation. Universidad de Oriente, Cuba. 2006.
Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med. 1995;34(6):910–4.
Henoumont C, Laurent S, Vander Elst L. How to perform accurate and reliable measurements of longitudinal and transverse relaxation times of MRI contrast media in aqueous solutions. Contrast Media Mol Imaging. 2009;4(6):312–21. https://doi.org/10.1002/cmmi.294.
Jacques V, Dumas S, Sun W-C, Troughton JS, Greenfield MT, Caravan P. High-relaxivity magnetic resonance imaging contrast agents part 2. Invest Radiol. 2010;45(10):613–24. https://doi.org/10.1097/rli.0b013e3181ee6a49.
Knobloch G, et al. Relaxivity of Ferumoxytol at 1.5 T and 3.0 T. Invest Radiol. 2018;53(5):257–63. https://doi.org/10.1097/RLI.0000000000000434.
Kraft KA, Fatouros PP, Clarke GD, Kishore PRS. An MRI phantom material for quantitative relaxometry. Magn Reson Med. 1987;5(6):555–62. https://doi.org/10.1002/mrm.1910050606.
McDonald RJ, et al. Gadolinium deposition in human brain tissues after contrast-enhanced MR imaging in adult patients without intracranial abnormalities. Radiology. 2017;285(2):546–54.
McDonald RJ, et al. Intracranial gadolinium deposition after contrast-enhanced MR imaging. Radiology. 2015;275(3):772–82. https://doi.org/10.1148/radiol.15150025.
Modo MM, Bulte JW. Molecular and cellular MR imaging. CRC Press; 2007.
National Electrical Manufacturers Association. NEMA Standards Publication MS 1-2008 (R2014, R2020). Determination Of Signal-To-Noise Ratio (SNR) In Diagnostic Magnetic Resonance Imaging. VA: NEMA; 2021.
Rohrer M, Bauer H, Mintorovitch J, Requardt M, Weinmann HJ. Comparison of magnetic properties of MRI contrast media solutions at different magnetic field strengths. Invest Radiol. 2005;40(11):715–24. https://doi.org/10.1097/01.rli.0000184756.66360.d3.
Szomolanyi P, et al. Comparison of the relaxivities of macrocyclic gadolinium-based contrast agents in human plasma at 1.5, 3, and 7 T, and blood at 3 T. Invest Radiol. 2019;54(9):559–64. https://doi.org/10.1097/RLI.0000000000000577.
Thangavel K, Saritaş EÜ. Aqueous paramagnetic solutions for MRI phantoms at 3 T: A detailed study on relaxivities. Turkish J Electr Eng Comput Sci. 2017;25(3):2108–21. https://doi.org/10.3906/elk-1602-123.
White GW, Gibby WA, Tweedle MF. Comparison of Gd(DTPA-BMA) (Omniscan) versus Gd(HP-DO3A) (ProHance) relative to gadolinium retention in human bone tissue by inductively coupled plasma mass spectroscopy. Invest Radiol. 2006;41(3):272–8. https://doi.org/10.1097/01.rli.0000186569.32408.95.
Acknowledgements
We would like to thank the Ministry of Science, Technology, and the Environment of the Republic of Cuba for the Financial Fund for Science and Innovation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Reyes Molina, I., Hernandez Rodriguez, A.J., Cabal Mirabal, C.A. et al. Semi-automated methodology for determination of contrast agent relaxivity using MRI. Res. Biomed. Eng. 39, 843–851 (2023). https://doi.org/10.1007/s42600-023-00309-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-023-00309-4