Abstract
Sugarcane is one of the important crops for sustainable economy and global energy security with high phenotypic polymorphism. Precise identification and characterization of plant genetic resources is one of the most essential aspects for breeding programs. Sugarcane popular hybrid varieties and genetic stocks routinely being used as a parent in hybridization program were included in the study. Molecular cataloguing was accomplished using a set of 70 primer pairs, of which ~ 39% were found to be robust polymorphic and unique in all the genotypes used. The number of DNA bands amplified by expressed sequenced tags derived simple sequence repeat (EST-SSR) primers ranged from 1 to 10 with an average of 5 bands per primer pair and their fragment size ranged between 50 and 1690 bp. Polymorphism information content (PIC) values were ranged from 0.17 to 1.00 with an average of 0.58. Unique DNA fingerprints of thirteen elite sugarcane varieties were generated with 27 polymorphic sugarcane specific EST-SSR markers. Analyses of DNA profiles produced by microsatellite markers showed the unique bands specific for individual variety. Generated DNA profiles would facilitate accurate variety identification in the perspective of protection of the breeder’s intellectual property rights, farmer’s rights and their purity testing. In addition, developed reliable DNA fingerprinting system will be able to track a potent parent genotype for breeding programs. The identified genotype specific and unique EST-SSR markers will be valuable for conservation and management of sugarcane genetic resources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum sp. hybrid L.) is an important agro-industrial crop which contributes greatly to national gross domestic product (GDP) and provides employment opportunities to farmers, agriculture labourers, transporters and factory workers in India as well as in many other countries (Hoang et al. 2015). Sugarcane crop provides feedstock to industry for sugar and jaggery production and industrial cellulosic biomass reprocessed to electricity production and second-generation bioethanol production (De Souza et al. 2013). Ever-increasing number of sugarcane varieties being developed by the different public or/and private institutions in India and many other sub-tropical and tropical countries of the world. Variety diversification is the most important to improve farmers economic status and survival of the sugar industry. This is not only the desirableness of variety protection as well as the exigency of monitoring the genetic diversity in breeding programs. Therefore variety identification and characterization is extremely important for all agricultural practices (Anonymous 2012). Precise identification of the newly developed crop varieties is also essential for their registration, seed multiplication, trade and inspection, and germplasm conservation and management. Since genotype identification and characterization is the primary step in any crop improvement program, quick recognition of cultivars would hence provide value-aided information for crop breeding (Korir et al. 2012).
The traditional approach to germplasm identification is based on set of morpho-agronomical descriptors which involves observation and recording of gross morphological characters (Artschwager and Brandes 1958; LaBorde et al. 2008). In sugarcane there are the some major characteristics used to assess the distinctiveness, uniformity and stability (DUS) which includes; leaf sheath trichomes, shape of ligule, dewlap colour, leaf blade curvature, leaf blade width, internode colour, internode diameter, shape of internode, rind surface appearance, stalk waxiness, shape and size of bud, groove of bud, bud tip in relation to growth ring, number of millable canes (NMC) per stool, cane height, and others (Anonymous 2006). Phenotypic traits are often multigenic, not available at all growth stages and considerably influenced by prevailing environment (viz; light, ambience temperature, relative humidity, rainfall, atmospheric pressure, soil health and others) as well as geographical factors (latitude, altitude, tropical and sub-tropical) (Govindaraj et al. 2012; Singh et al. 2018). Thus, in present scenario a strong base is extremely required for the cultivar identification, protection of the breeder’s intellectual property rights, germplasm conservation and paternity testing of the released varieties (Hemaprabha et al. 2010) and early assessment of plantlets in sugarcane nursery (Silva et al. 2012).
With the devise of several molecular marker techniques, morpho-agronomical descriptors have supplemented with DNA markers in genotype identification and characterization (Singh et al. 2019a). Uniquely, DNA-based markers are not influenced by environmental fluctuations and detectable in all tissues regardless of plant age, growth and developmental stages, thus allowing precise germplasm identification, conservation and management across different environmental conditions (Singh et al. 2013, 2014a, 2015). Thus, molecular markers have emerged as powerful tools to identify the plant varieties. Several molecular marker techniques have been devised for DNA fingerprinting based genotype identification (Singh et al. 2011, 2017). Microsatellites or simple sequence repeat (SSR) marker technique preferred as a potent molecular tool owing to their desirable genetic attributes of reproducibility, relative abundance, multi-allelic nature, co-dominant inheritance and amenable to high throughput genotyping in sugarcane and allied grasses (Cordeiro et al. 2001; Varshney et al. 2002; Singh et al. 2014b, 2019b).
However cost-effective DNA marker systems (SSRs) have been developed by several research groups over the world, but a huge number of varieties have been developed and used in breeding which makes fingerprinting job expensive in terms of laboratory resources, labor and time expenditure (Zhu et al. 2012; Singh et al. 2019a). In spite of that, it is inevitable to assemble the fingerprinting database of the elite commercial sugarcane varieties for frequent and unambiguous identification with the other developed similar varieties (Hemaprabha et al. 2010; Singh et al. 2017). In present study, EST-derived sugarcane microsatellites (EST-SSR) markers developed by our team (Singh et al. 2019a) in previous investigation were used to identify and characterize 13 popular Indian sugarcane varieties through DNA fingerprinting. Most of the selected varieties are under cultivation at medium to large scale in sub-tropical areas of India. Used varieties represent important genetic stock and frequently being used as parents to establish a cross for breeding high yielding variety. The variety specific molecular cataloging was carried out for each individual accession which maintained as a reference dataset at Sugarcane Research Institute (UPCSR), Shahjahanpur, (U.P.) India.
Materials and methods
Sugarcane varieties
A set of 13 promising sub-tropical CoS, (Coimbatore–Shahjahanpur) CoSe (Coimbatore–Seorahi) and UP (Uttar Pradesh) sugarcane varieties developed at Sugarcane Research Institute (UP Council of Sugarcane Research), Shahjahanpur, (UP), and its regional center Genda Singh Sugarcane Breeding & Research Institute (GSSBRI), Seorahi, (UP), India were selected. Although used all the 13 varieties are not frequently under cultivation at farmer’s fields but, they have ruled over the entire sugarcane growing areas of the northern India and are still being cultivated in many of the sugar factories of Uttar Pradesh. Since they are having some untapped useful alleles in their genome, owing to this reason these varieties are frequently used as proven parents in sugarcane genetic improvement programs to breed promising varieties for subtropical India. Genetically pure material of these genotypes was collected from experimental farms of the institute. Variety name, their parentage, maturity group, year of release, and originating center are detailed in Table 1.
Extraction of genomic DNA and PCR amplification
Genomic DNA was extracted from disease free immature fresh leaves of all the 13 sugarcane varieties using CTAB protocol described by Hoisington (1992) with minor modifications (Singh et al. 2011). To remove out the RNA content, raw DNA was treated with RNase enzyme for 1 h at 37 °C. The DNA was diluted to a final concentration of ~ 25 ng/µl as determined by agarose-gel electrophoresis using known concentration of uncut λ DNA as standard. Total 361 functionally informative microsatellite primers were designed from conserved nucleotide sequences SSR motifs of the non-redundant EST-database of sugarcane under our previous study (Singh et al. 2019a). To check the genetic uniformity of the SSR primers, ten individual DNA samples of each sugarcane variety were assayed with five sugarcane specific EST-SSR primers. Only monomorphic DNA profiles were observed between individuals within a genotype and thus establish confirmation to marker’s genetic uniformity. A set of seventy sugarcane EST-SSR markers were employed in genetic characterization assay, among them 27 primer pairs (Table 2) that generated unambiguous and reproducible profiles were used to construct DNA fingerprints of the 13 sugarcane varieties under study. Conserved genetic regions of the SSR motifs were PCR amplified with polymorphic EST-SSR primers in 10 µl reaction volume. PCR mixture were containing 25 ng of template DNA, 0.02 mM of both forward and reverse primers, 10 mM of each dNTPs (dATP, dTTP, dGTP, and dCTP), 0.5 U of Taq DNA polymerase (Bangalore Genei™, India) and 1.0 µl of 10× PCR buffer with 2.5 mM of MgCl2. The PCR reactions were performed in a thermo cycler (M J Research PTC 100) using the following thermal conditions as; initial denaturation at 94 °C for 5 min, followed by 30 cycles consisting of 94 °C for 1 min, primer annealing at optimised (50–60 °C) temperature for each primer pair for 1 min, and primer elongation at 72 °C for 2 min followed by a final primer extension step at 72 °C for 5 min. Three times PCR reactions were repeated for each primer pair to ensure the reproducibility of EST-SSRs.
Analysis of microsatellite (EST-SSR) data
The PCR products were resolved on a 7.5% native poly-acrylamide gel electrophoresis (PAGE) in vertical gel electrophoresis unit (Bangalore Genei™) using 0.5× TBE buffer. Visualization of bands was carried out by 0.5 μg/ml ethidium bromide (EtBr) and images were captured under UV light using gel documentation system (Alpha Innotech). Thirteen genotypes were compared with each other using their PCR amplified DNA profiles generated with specific markers. Only unambiguous, clear-cut, robust, and reproducible bands were scored and unclear and diffused bands ignored.
Results and discussion
DNA fingerprints and molecular cataloging of identified varieties
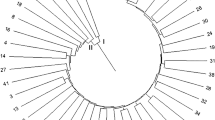
The elementary DNA fingerprints of the 13 popular sugarcane varieties were generated successfully with the 27 robust polymorphic EST-SSR markers. A set of 70 primer pairs were used to amplified, of which ~ 39% were found to be robust polymorphic. The number of DNA bands amplified by EST-SSR primers was ranged from 1 to 10 with an average of 5 bands per primer and their fragment size ranged between 50 and 1690 bp. Polymorphism information content (PIC) value was ranged from 0.17 to 1.00 with an average of 0.58 (Table 2). For instance, DNA profiles of the sugarcane variety CoS 767 was generated using a set of 22 EST-SSR markers (Fig. 1). The developed microsatellite markers were found to be potentially suitable for DNA fingerprinting based genotype identification and characterization. The molecular profiles and genotype specific unique DNA bands produced by different EST-SSR markers have maintained as a molecular catalogues of each identified variety (Table 3).
DNA profiling of sugarcane variety CoS 767 generated by 22 sugarcane microsatellites (SMS) markers. Note: M—50 bp ladder, lane 1—SMS1, 2—SMS3, 3—SMS4, 4—SMS9, 5—SMS10, 6—SMS11, 7—SMS14, 8—SMS15, 9—SMS17, 10—SMS18, 11—SMS23, 12—SMS24 13—SMS25, 14—SMS27, 15—SMS28, 16—SMS29, 17—SMS30, 18—SMS33, 19—MS37, 20—SMS41, 21—SMS42, 22—SMS43
The results of the DNA fingerprinting in promising sugarcane varieties have revealed the authenticity of EST-SSR marker system for the identification and characterization of genotypes (Jannoo et al. 2001; Piperidis 2003). DNA fingerprinting in sugarcane is relatively simple than the in seed propagated crops and found to be more uniform and stable fingerprints once generated (Singh et al. 2011). Thus, DNA profiles generated by these functional markers produced using genomic DNA create a unique identity of the analyzed genotype and hold promise the ultimate tool for biological individualization of sugarcane varieties (Singh et al. 2013).
Hence, DNA fingerprinting would provide accurate and stable varietal recognition in regards of plant variety protection, authenticity testing and germplasm conservation and utilization in addition to quality assurance for release of new varieties to the country (Singh et al. 2013, 2015). Molecular characterization to assign identity to the cultivars could also be used in DUS testing, variety registration, and in resolution of dispute (Hemaprabha et al. 2010). Moreover, to develop a DNA profile of plants has great importance in breeding, owing to facilitating for the identification of duplicates in the germplasm collection and the organization of crosses leads to an improved breeding outcome (Ruiz et al. 2011). The identification of varieties based on molecular markers is important to set up distinctness, uniformity and stability of protected cultivars (Hemaprabha et al. 2010; Swapna et al. 2011). Polymorphic microsatellites were used to generate unique molecular profiles for the identification of individual genotypes that can be used in breeding program to assist the protection of new sugarcane varieties (Silva et al. 2012). Accordingly, DNA fingerprinting is a valuable tool to serve in the protection of newly developing sugarcane varieties by the different breeding programs.
References
Anonymous (2006) Guidelines for the conduct of test for distinctiveness, uniformity and stability on sugarcane (Saccharum L.). Protection of Plant Varieties and Farmers’ Rights Authority (PPV & FRA), Government of India, New Delhi
Anonymous (2012) Louisiana sugarcane variety identification guide. Louisiana State University Agricultural Center, Baton Rouge, pp 1–22
Artschwager E, Brandes EW (1958) Sugarcane (Saccharum officinarum) origin, classification and descriptions of representative clones. Agriculture hand book no. 122. USDA, US Government Priority Office, Washington
Cordeiro GM, Casu RE, McIntyre CL (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
De Souza AC, Carvalho FP, Batista CF, Schwan RF, Dias DR (2013) Sugarcane bagasse hydrolysis using yeast cellulolytic enzymes. J Microbiol Biotechnol 23:1403–1412
Govindaraj P, Ramesh R, Appunu C, Swapna S, Priji PJ (2012) DNA fingerprinting of subtropical sugarcane (Saccharum spp) genotypes using sequence tagged microsatellites sites (STMS) markers. Plant Arch 12:347–352
Hemaprabha G, Krishna JA, Vincy P, Priji P, Swapna S, Govindaraj P (2010) DNA fingerprinting for identification and protection of elite sugarcane (Saccharum spp.) varieties. Electron J Plant Breed 4:420–425
Hoang NV, Furtado A, Botha FC, Simmons BA, Henry RJ (2015) Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front Bioeng Biotechnol 3:1–15
Hoisington D (1992) Laboratory protocol. CIMMYT Applied Molecular Genetics Laboratory, CIMMYT, Mexico
Jannoo N, Forget L, Dookun A (2001) Contribution of microsatellites to sugarcane breeding program in Mauritius. Proc Int Soc Sugarcane Technol 24:637–639
Korir NK, Han J, Shangguan L, Wang C, Kayesh E, Zhang Y, Fang J (2012) Plant variety and cultivar identification: advances and prospects. Crit Rev Biotechnol 33:111–125
LaBorde C, Legendre B, Bischoff K, Gravois K, Robert T (2008) Sugarcane variety identification Guide 2008. Louisiana State University Ag Center Publication, Baton Rouge
Piperidis G (2003) Progress towards evaluation of SSR as a tool for sugarcane variety identification. In: ISSCT IV. Molecular biology workshop, Montpellier, France
Ruiz M, Aguiriano E, Giraldo P, Carrillo JM (2011) Genetic redundancy among durum wheat accessions as assessed by SSRs and endosperm proteins. Span J Agric Res 9:156–165
Silva DC, Dos Santos JM, de Souza Barbosa GV, Almeida C (2012) DNA fingerprinting based on simple sequence repeat (SSR) markers in sugarcane clones from the breeding program RIDESA. Afr J Biotechnol 11:4722–4728
Singh RK, Singh RB, Singh SP, Sharma ML (2011) Identification of sugarcane microsatellites associated to sugar content in sugarcane and transferability to other cereal genomes. Euphytica 182:335–354
Singh RK, Singh RB, Singh SP, Mishra N, Rastogi J, Sharma ML, Kumar A (2013) Genetic diversity among Saccharum spontaneum clones and commercial hybrids through SSR markers. Sugar Tech 15:109–115
Singh RB, Srivastava S, Rastogi J, Gupta GN, Tiwari NN, Singh B, Singh RK (2014a) Molecular markers: exploited in crop improvement practices. Res Environ Life Sci 7:223–232
Singh RB, Srivastva S, Verma AK, Singh B, Singh RK (2014b) Importance and progress of microsatellite markers in sugarcane (Saccharum spp. hybrids). J Sugarcane Technol 29:1–12
Singh RB, Singh B, Singh RK (2015) Development of microsatellite (SSRs) markers and evaluation of genetic variability within sugarcane commercial varieties (Saccharum spp. hybrids). Int J Adv Res 3:700–708
Singh RB, Singh B, Singh RK (2017) Study of genetic diversity of sugarcane (Saccharum) species and commercial varieties through TRAP molecular markers. Indian J Plant Physiol 22:332–338
Singh RB, Singh B, Singh RK (2018) Evaluation of Genetic diversity in Saccharum species clones and commercial varieties employing molecular (SSRs) and physiological markers. Indian J Plant Genet Resour 31:17–26
Singh RB, Singh B, Singh RK (2019a) Development of potential dbEST-derived microsatellite markers for genetic evaluation of sugarcane and related cereal grasses. Ind Crops Prod 128:38–47
Singh RB, Singh B, Singh RK (2019b) Cross-taxon transferability of sugarcane expressed sequence tags derived microsatellite (EST-SSR) markers across the related cereal grasses. J Plant Biochem Biotechnol 28(2):176–188
Swapna M, Sivaraju K, Sharma RK, Singh NK, Mohapatra T (2011) Single-strand conformational polymorphism of EST-SSRs: a potential tool for diversity analysis and varietal identification in sugarcane. Plant Mol Biol Rep 29:505–513
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Zhu YF, Qin GC, Hu J, Wang Y, Wang JC, Zhu SJ (2012) Fingerprinting and variety identification of rice (Oryza sativa L.) based on simple sequence repeat markers. Plant Omics 5:421
Acknowledgements
Authors are thankful to Director, Uttar Pradesh Council of Sugarcane Research (UPCSR), Shahjahanpur, (UP) India for providing necessary facilities to perform this study.
Author information
Authors and Affiliations
Contributions
RBS conceived and designed the experiment, performed the experiments, and wrote the paper. BS was involved in acquisition and interpretation of data. RKS revised the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest directly or indirectly and informed consent to publish this study and that the manuscript complies with the ethical standards of the journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, R.B., Singh, B. & Singh, R.K. Identification of elite Indian sugarcane varieties through DNA fingerprinting using genic microsatellite markers. Vegetos 32, 547–555 (2019). https://doi.org/10.1007/s42535-019-00058-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42535-019-00058-9