Abstract
Angiotensin-converting enzyme (ACE) inhibitory peptides have lately attracted interest since functional foods that help maintain homeostatic regulations have been developed. Rarely discussed are the intrinsic ACE-peptide interactions and their positioning, both of which help illustrate the ACE inhibitory functionalities in food-derived peptides. In this study, 173 ACE inhibitory peptides were collated using the UWM-BIOPEP database. The sequences were grouped into short, medium, and long peptides. The hydrophobicity/hydrophilicity property of peptides was analyzed using Peptide2 and the peptide binding site on ACE was predicted using PepSite2. Peptide residues interacting with ACE were denoted as reactive amino acids. Molecular docking analysis was conducted to simulate ACE-peptide binding and delineate the roles of reactive amino acids at the ultimate, penultimate, and antepenultimate positions of N—(N1, N2, and N3) and C—(C1, C2, and C3) terminals. Peptide2 analysis suggested that hydrophobic property was prominent in the peptides. The C-terminals were prominent in ACE binding for long-chained peptides through interaction with ACE hotspots. Moreover, branched-chain amino acids (BCAA) such as leucine and isoleucine were crucial at the N-terminals. The bulky side chain of BCAA forms a hydrophobic shield that protects the Zn-peptide chelate complex from water attacks. The hydrophobic fence in turn stabilizes the disrupted tetrahedral Zn-coordinate complex of ACE. This finding provided a thorough exploration of how peptide structures are related and what function they play in ACE inhibitory action. The database analysis, therefore, gave a clearer insight and comprehensive understanding into the protein-peptide interactions and provided a mechanistic explanation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypertension is a widespread, severe chronic condition that affects roughly 25% of the world’s adult population with significant impact on public health and is a risk factor for heart disease, stroke, and renal failure, along with many other health complications (Jao et al. 2012; Wang et al. 2008). Notwithstanding the access to a variety of treatment options, only around one out of every five individuals has their blood pressure (BP) under check; as a result, hypertension control is becoming an essential healthcare objective (Ghatage et al. 2021). Angiotensin is a hormone that stimulates the constriction of blood vessels. The angiotensin-converting enzyme (ACE) inhibitors work by lowering blood pressure by inhibiting the synthesis of angiotensin (Ding et al. 2021). The precise ACE inhibitor mechanism is not precisely understood. The most common therapeutic therapy involves decreasing the activities of the ACE by suppressing angiotensin II, which decreases blood pressure (Ding et al. 2021). ACE is a zinc-metallopeptidase that catalyzes the release of angiotensin II from passive angiotensin I to a functional vasoconstrictive octapeptide, which plays an important role in blood pressure regulation (Parit and Jayavel 2021). It also deactivates bradykinin (a vasodilatory hormone) (Priyanto et al. 2015). Lisinopril, trandolapril, ramipril, moexipril, and quinapril hydrochloride are examples of chemically produced antihypertensive drugs (Ghatage et al. 2021). These medications are helpful, but they are usually recommended for a long-time use (Roy et al. 2010). Dry cough, elevated potassium levels in the blood, tiredness, dizziness, and other symptoms are all common adverse effects of ACE inhibitors. Furthermore, excessive potassium levels raise the risk of kidney failure, and altered sensory functions (Surma et al. 2021). However, ACE inhibitory peptides obtained from food-based dietary sources have been shown in studies to decrease high blood pressure without these adverse effects (Priyanto et al. 2015; Yamamoto 1997). Numerous antihypertensive peptides synthesized from food-based nutraceuticals functions primarily by blocking the ACE. Various studies have derived ACE inhibition peptides from many food-based products such as milk, egg white, cereals, chicken muscles, soybeans, pea, red algae, etc. (Meisel 1998; Villegas et al. 2014; Rawendra et al. 2014).
Mojica et al. (2017) characterized peptides of enhanced common bean varieties for possible bioactivities and contrasted the inhibitory effects of their digested isolated proteins and purified peptides on ACE inhibitory activities. In this study conducted by Mojica et al. (2017), the characteristics of the protein isolated digested and peptides tested validated the claim of the possible pharmacological activities of cowpea. The polypeptide in common bean amino acids may assist to suppress ACE, the increased activity linked to non-communicable conditions, such as high blood pressure and hypertension. Li et al. (2021) discovered four new bioactive peptides from bean paste with RGLSK exhibiting the highest IC50 (87 μmol/L) against ACE activity. ACE inhibitory activity and allergenicity of the crude extracts from peanuts of alcalde hydrolysates were tested by Yu et al. (2021). Also examined were the effects of alcalase hydrolysis on the solubility of protein and the extract inhibitory activity of the ACE. The results indicated that a smaller molecular fraction of hydrolysates shows greater ACE inhibitory activity (45–47%) at 1 mg/mL concentration, than the crude peanut flour hydrolysates at the same time and concentration. The anti-ACE hydrolysates of lightly roasted peanut flour were more than that of peanut flour hydrolysates from low concentrations of dark-roasted peanut flour.
A key potential risk in a variety of chronic illnesses among people is hypertension or high blood pressure. The gradual increase in blood pressure can progress into hemorrhagic stroke, heart attack, chronic renal disease, cognitive loss, and untimely mortality. Response to fluctuations in lifestyle and dietary patterns in the past few years has seen continuous growth of hypertensive individuals. Although synthetic medications are obtainable, there is unpredictability and severe toxic effects in some people as a result of medication. With long-term usage, medication side effects are likely to be present in individuals with metabolic diseases such as hypertension. Therefore, to precisely characterize the interplay of molecules (ligands and receptors) and to anticipate their binding patterns and affinities, researchers created a molecular docking technique based on sophisticated computers and software to investigate the connection between inhibitors and ACE (Xiang et al. 2021). Ligand-receptor binding must adhere to the reciprocity matching rules of shape, electrostatic attraction, analogous hydrogen bonding, and hydrophobic interactions, as well as energy compatibility. The location of the molecule and the dihedral angle of the intramolecular adaptable bonds may be adjusted using molecular docking capabilities in order to identify the optimum conformational structure of the bioactive peptide and the target (Xie et al. 2018).
Although few investigations are closely related with this study, the results of their study have their own inherent limitations. For example, Ding et al. (2021) addressed the mechanism underlying the structure–activity correlation of certain chosen food-based ACE peptides. According to their findings, leucine showed greatest frequency (14.70%) at the N-terminal of peptide with ACE inhibition; proline had the greatest occurrence frequency at 23.21%, and the total number of occurrences for the remaining amino acids is less than 40%, suggesting that proline played a significant role in ACE inhibitory action. The limitation to this study is that the conclusion was arrived at using fewer numbers of peptides which might not be generally acceptable. Therefore, this study opts to analyze a larger number of food-derived ACE inhibitor peptides from the UWM-BIOPEP database, focusing on the structure activity relationship (SAR) between amino acid properties, side chains of peptides and ACE inhibitory activity. Deciphering the molecular structure of inhibitors and explaining their antihypertensive processes is critical for developing optimal antihypertensive medicines and advancing clinical expertise. In the biological system, and particularly in human existence, peptides perform critical functions. Peptides and proteins are widely employed in therapies, and more are being researched and developed as potential pharmacological targets. As a result, countless peptides and proteins are produced, synthesized, and tested for a range of therapeutic actions. Nonetheless, because there are so many different methods for arranging them, designing them and predicting their behavior continue to be among the biological sciences’ most difficult tasks. For an example, quantitative structure–activity relationship (QSAR) works which uses quantitative sequence-activity model (QSAM) to quantify the biosequence-activity or the function relationship for the peptides after validating the predictive power of resulting models that were built based on different descriptors such as size, charge distribution, hydrophobicity/hydrophilicity, polarity, as well as number of donor/acceptor atoms (Hemmateenejad et al. 2011; Bahadori et al. 2019; Mei et al. 2005; Barley et al. 2018). Apart from that, quantum topological molecular similarity (QTMS) system as a new source for amino acid indices was recently introduced. This system provides definite physical and chemical meanings. In addition, it has good capability in characterizing the peptide structure, thus, producing statistically significant QSAR models (Brandl et al. 2001). As a result, the topic of computational investigation of structure/activity connections in peptide design grows more appealing. Thus, the comprehensive database analysis performed in this study shed light on protein-peptide interactions and offered a mechanistic explanation. The results enable a systematic assessment of the relationship between peptide structures and the role they play in ACE inhibitory activity.
Synthesis of antihypertensive peptides from food sources
The isolation methods and sources have an effect on the structure and activity of different ACE inhibitory peptides (Xu et al. 2021). Enzymatic hydrolysis process may liberate these inactive peptides from the original protein's sequence. Moreover, an in vitro enzymatic or in vivo gastrointestinal digestion of the relevant precursor proteins can determine the structures of ACE inhibitory peptide sequences. The primary sources of antihypertensive peptides include dairy products, cheeses, nuts, etc. Increasingly, the synthesis of these important peptides has also been carried out using dietary sources other than milk. Examined in this section are the common methods previously used to synthesize ACE inhibitory peptides viz: enzyme hydrolysis, fermentation of food-based dietary sources, and genetic DNA recombination (Balgir et al. 2016).
Hydrolytic technique via digestive enzymes
Enzyme hydrolysis is still the most prevalent method of antihypertensive peptides production from dietary proteins, such as those produced using pepsin and trypsin as reported by Fitzgerald et al. (2004) and Gobbetti et al. (2002). Many dietary food sources have been synthesized with the use of enzymatic hydrolysis to yield antihypertensive peptides. Take, for instance, various enzymes were employed to degrade milk proteins to produce antihypertensive peptides. Samaei et al. (2021) synthesized hemp bran proteins, a component of the hemp seeds. These were isolated chemically and hydrolyzed to produce four fractions through the membrane ultrafiltration process. The ACE-inhibiting hydrolysate was significantly purified further via ultrafiltration. Thirty-five (35) strong ACE-inhibiting bioactive peptides were discovered out of the 239 peptides obtained (Samaei et al. 2021). The hypotensive efficacy and longevity of Moringa leaf peptides were studied by Ma et al. (2021). Leaves protein from Moringa was isolated and subsequently hydrolyzed to a protein hydrolysate using alcalase as a hydrolyzing enzyme. The protein hydrolysate was isolated by membrane ultra-filter to produce peptide fractions of different molecular weights. Two very potent ACE-inhibitory peptides were synthesized, namely Leu-Gly-Phe-Phe and Gly-Leu-Phe-Phe. In addition, using an in vivo model, the systolic blood pressure of induced diabetic rats were considerably lowered upon administration of the two peptides (Ma et al. 2021).
Gu et al. (2012) processed the silky black-bone muscle with alcalase and papain via a reversed-phase HPLC multi-stage separation. The test was later performed for ACE's inhibitory effects where a series of 29 peptides with a half maximum inhibitory activity of 253.07 and 6.66 μM were found to contain two unique ACE peptides of Leu-Glu-Arg and Gly-Ala-Gly-Pro, respectively. The anti-hypertensive effects of egg white protein were examined by Majumder et al. (2013). The results showed that Ile-Arg-Trp exhibited an anti-hypertensive property in in vivo hypertensive rate model, which was achieved through the inhibition of ACE and nitric oxide synthase. Inhibitory ACE peptides from fish origins were also isolated. The very first ACE-inhibitory peptide in sardine was discovered by Suetsuna and Osajima (1986) from more than three decades. From that time, numerous more peptides, including seafood, salmon, mackerel, sockeye, etc. have been identified from different species (Ryan et al. 2011). The isolate of pea protein, hydrolyzed by alcalase, showed inhibition effects to phosphodiesterase, ACE, renin and was related to calmodulin (Li et al. 2011). Four bioactive peptides (ITP, IIP, GQY, and STYQT) were isolated by the hydrolysis of protease enzyme from sweet potato protein in four sequence peptides. In vivo rat studies have revealed ITP peptide as the most powerful ACE inhibitor (Ishiguro et al. 2012).
Apart from that, proteinase K was used to hydrolyze camel milk casein in order to produce ACE inhibitory bioactive peptides (Rahimi et al. 2016). Fractionation and purification of the hydrolysates were performed, and strong ACE inhibitory activity (IC50 = 36 μg/mL) was obtained. Therefore, proteinase K was suggested as a proper enzyme for liberating peptides with ACE inhibitory activity. The commercial protease was also used to hydrolyze soy milk as investigated by Tomatsu et al. (2013). The RP-HPLC was used to isolate eight new peptides with improved ACE inhibition. Included among the new peptides are FFYY, WHP, FVP, LHPGDAQR, IAV, VNP, LEPP, and WNPR. The 50% inhibitory concentration of the ACE inhibitory peptides from grain, fowl, tuna, wines, etc. are similar as reported by Tomatsu et al. (2013). The characterization of ACE inhibitory peptides produced from wheat gluten protein hydrolyzed by trypsin was discussed by Asoodeh et al. (2014). Four peptides were synthesized based on computer simulations and docking characteristics and two peptides, i.e., IPALLKR and AQQLAAQLPAMCR gave IC50 values of 68 ± 2.8 and 43 ± 1.3 μM, respectively. The serine protease alcalase has been confirmed to become the most broadly utilized endoprotease for hydrolysis of different plant proteins such as canola, seed protein of sunflower, soybean, lentils, and mung and chickpeas with an increased inhibition potential (Pihlanto and Makine 2013). This has a vast volume and is the most commonly utilized endoprotease in hydrolysates of plant protectants (Pihlanto and Makine 2013).
Fermentation process
In recent years, the utilization of plant and animal sources for active compounds with antihypertensive action has become more and more prominent. Fermented grain food items contain blood pressure-reduction peptides (Patel et al. 2021). Research has demonstrated the major source of bioactive peptides in common animal-plant diet. Grains, peas, and cruciferous vegetable plants are generally bioactive peptides of plant source food materials. The occurrence of bioactive peptides in grains and legumes has been reported to enhance the quality of the fermented functional dietary protein in everyday meals (Singh et al. 2021). Soy-based products are a fast-growing industry in the food industry, with a high demand for soy components with enhanced processing properties. Fermented soy items, which have long been utilized in various parts of the world, have been discovered to be a rich source of ACE inhibitory peptides. (Vallabha and Tiku 2014) described the identification of two significant ACE inhibitory peptide fractions from soy protein by fermentation using Lactobacillus casei spp. Premised on the N-terminal sequences of the polypeptide, an analog of Leu-Ile-Val-Thr-Gln was synthesized, and the impact of various residues on ACE enzymes was investigated. The researchers discovered the significance of glutamine and threonine residues in ACE inhibitory activities.
Furthermore, several commercially exploited dairy starter cultures are substantially proteolytic in nature and can be harnessed by fermenting the animal products for the synthesis of antihypertensive peptides. Based on this, a variety of products have been produced for drug trials to test their effectiveness with various hypertensive patients (Jakala and Vapaatalo 2010). Numerous dietary proteins, and in particular milk proteins, incorporate proteins pharmacologically coded in the bioactive peptides. Such peptides once synthesized and delivered, have distinct physiological roles via gastrointestinal digestion. Biologically active, antibacterial, immunological modulate, oxidative, anti-mineral-binding, and antioxidant characteristics are demonstrated to contain milk-derived peptides (Jakala and Vapaatalo 2010). Fermentation is therefore a viable process for the synthesis of ACE inhibitory peptides that are generated from milk proteins with the potential in reducing blood pressure. A few of these peptides also share the binding characteristics of the opioid receptor (Meisel 1998). Nakamura et al. (1995) reported a significant decrease in the blood pressure of spontaneously hypertensive rats (SHRs) using a fermented milk product comprising of physiologically bioactive peptides of Val-Pro-Pro and Ile-Pro-Pro. Short/medium peptides have been reported to be assimilated into the gastrointestinal system without any further breakdowns by the digestive enzymes. When administered to SHRs, the peptides Tyr-Pro and Lys-Val-Leu-Pro-Val-Pro-Gln isolated from fermented milk also demonstrated ACE inhibitory activity (Hernandez-Ledesma et al. 2007). Nurminen et al. (2000) similarly discovered the reduction in normal blood pressure and SHR in Tyr-Gly-Leu-Phe.
Recombinant DNA techniques
In addition to the methods described in previous sections, recombinant DNA techniques are another viable means of producing peptides with antihypertensive activities. This has been utilized to biosynthesize novel ACE inhibitory peptides with a strong hypertensive effect (Xue et al. 2021). In contrast to the enzymatic hydrolysis approach, the bacterial genetic recombinant method exhibits a better peptide output and reduced cost of production as reported by Lv et al. (2003). The initial investigation by Kim et al. (1999) involves the production and separation of recombinant α-1-casein in Escherichia coli. Rao et al. (2009) reported the cloning of an antihypertensive peptide in E. coli, with a subsequent expression of four peptide sequences of HHL, HVLPVP, FFVAPFPEVFGK, and GHIATFQER.
Losacco et al. (2007) described the design of innovative methods for highly pure fractions of three polypeptides synthesized from bovine β-casein potent as an in vitro ACE inhibitor, relying on recombinant DNA technology. The process comprises the production of peptides in E. coli as recombinant proteins, breakdown by a proteinase from the chosen Lactobacillus helveticus strain, and extraction of bioactive peptides by chromatographic techniques. The observed results further demonstrated the viability of pure peptides with greater inhibitory characteristics and novel pharmaceutical applications. Furthermore, commercial production of these ACE inhibitory peptides with anti-hypertensive properties is of future relevance using the genetic recombinant DNA techniques.
Data collection and analysis methods
In this study, a total of 173 biologically active ACE inhibitory peptides originating from different food items were collected via the BIOPEP-UWM database with their respective amino acid sequence. The BIOPEP-UWM is a publicly available, and frequently updated bioinformatics database that is carefully arranged for the proteins sequence, allergic proteins along with their epitopes, sensory peptides, and amino acids (Minkiewicz et al. 2019). The bioinformatics database has a consistent one-letter code defining twenty amino acids and their d-enantiomers. In this present review, the UWM-BIOPEP database was used to collate large number (173) of ACE inhibitor peptides from different dietary food items with their matching amino acid sequences. Hydrophobic, acidic, basic, or neutral peptides were grouped according to their inhibitory characteristics. Following that, the Pepsite2 website was used to predict the binding of antihypertensive peptides to ACE. This resulted in a comprehensive understanding of the interaction between protein and peptide, as well as the mechanism of action of ACE inhibitor peptides generated from different natural-occurring food sources. Tables S1, S2, and S3 show the antihypertensive (ACE) related peptides collected from the database which were grouped into short (2–4 AAs), medium (5–10 AAs), and long peptides (> 10 AAs), respectively. There were 87 short, 44 medium and 42 long peptides in total. Following the three categories of the peptides sequence groupings, Pepsite2 webserver (http://pepsite2.russelllab.org/) was afterward deployed to predict the binding sites of antihypertensive peptides on ACE (PDB ID: 1O8A). One of the merits of these web servers is the rapid generation of results within a short time. Reactive amino acids are peptides residues involved in ACE binding whereas non-reactive amino acids did not, based on Pepsite2 prediction. The hydrophobic-aliphatic, hydrophobic-aromatic, hydrophilic-acidic, hydrophilic-basic and polar neutral characteristics of peptides were estimated using the Peptide2 webserver (https://www.peptide2.com/N_peptide_hydrophobicity_hydrophilicity.php). The proportion of reactive as well as non-reactive amino acids was also determined. The frequencies of reactive and non-reactive amino acids at both C- and N-terminals were calculated using Microsoft Excel. Molecular docking analysis was then conducted for clearer insight into the protein-peptide interactions and provides a mechanistic explanation of the workings of amino acid side chains. Briefly, the peptide structures were generated using the PepFold3 server (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) whereas the ACE structure was obtained from the RCSB Protein Data Bank (PDB ID: 1O8A). The structures of ACE and peptide were submitted to the HADDOCK 2.4 webserver (https://wenmr.science.uu.nl/haddock2.4/) for modelling of ACE-peptide binding interactions using the default protein-peptide docking parameters. The reactive residues were specified according to Pepsite2 results. Upon completion, protein–peptide docking solutions were clustered and ranked by HADDOCK score. The cluster with the lowest HADDOCK score, relatively large cluster size and more negative Z-score out of 200 predicted models was the most likely conformation. The enzyme-peptide binding energy was then computed by PRODIGY webserver (https://wenmr.science.uu.nl/prodigy/). Visualization of docking result was performed using the LigPlot + software version 2.2 (EMBL-EBI, Hinxton, Cambridgeshire, UK). Figure 1 illustrates the step-by-step schematic flow of how the ACE inhibitory data was analyzed.
Results and discussion
Overall preference of amino acid residue property on inhibitor peptides
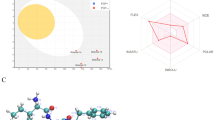
The overall classification based on inhibitory properties was derived by categorizing the amino acid of each peptide sequence. Take for instance the amino acids A, V, I, L, M, G, P fall under the hydrophobic-aliphatic; F, Y, W are hydrophobic-aromatic; N, C, Q, S, T are all polar neutral; D, E are hydrophilic-acidic while R, H, K are hydrophilic-basic. In Fig. 2, the inhibitory properties of novel peptides against ACE were classified as hydrophobic (aliphatic or aromatic), hydrophilic (acidic or basic), or polar neutral. Based on this classification, the overall categorization placed the hydrophobic-aliphatic and hydrophilic-acidic as the highest and least peptides properties, with 51.66% and 5.00%, respectively. The polar neutral (16.66%) and hydrophilic-basic (15.66%) properties were approximate of the same relative abundance, followed by the hydrophobic-aromatic properties (11.00%).
In terms of the length of the peptides, the hydrophobicity properties with aliphatic side chain were more prominent in 59% of the short peptides, 47% medium peptides, and 49% of long peptides, based on Fig. 3. The classification of the bioactive peptides based on their length shows the short peptides exhibiting the most dominant hydrophilic-aliphatic properties (59%) while showing the least dominance of just 1% hydrophilic-acidic properties. Moreover, in the long-peptides, only 49% of the peptides exhibited more hydrophobic-aliphatic properties when compared with 47% of the medium peptides. However, 16% of medium peptides show the highest hydrophobic-aromatic properties, followed by short (9%) and long (8%) peptides. In addition, 19% of the long peptides exhibited polar neutral amino acid properties, followed by short and medium peptides with 16% and 15%, respectively. As for the hydrophilic-acidic properties, 1%, 5% and 9% of short, medium, and long peptides respectively exhibited this amino acid property, while 15%, 17%, and 15% of short, medium, and long peptides exhibited the basic hydrophilic amino acid properties. Overall, hydrophobic is the most dominant property, with 59% of the short peptides reviewed conspicuously showing this property. Past studies have reported that a peptide’s amino acid structure has a measurable impact on its ACE inhibitory function, so the ACE tends to bind to a peptide chain comprising of a high proportion of hydrophobic amino acids (Sun et al. 2019). Ryan et al. (2011) indicated that the majority of ACE inhibitory peptides are small peptides with sequences that can work more effectively at the ACE binding site and hence exert higher inhibitory action (Li et al. 2018a).
Overall, the hydrophobicity properties with the aliphatic chain were more evident in short, medium, and long peptides. However, the hydrophilic properties with acidic chains only have 1% of the short peptides and 5% of medium peptides binding to amino acids based on the Peptide2 analysis. Meanwhile, in reactive amino acids, 52%, 11%, 17%, 3%, and 16% of the overall peptides sequence have hydrophobic-aliphatic, hydrophobic-aromatic, polar neutral, hydrophilic-acidic, and hydrophilic basic properties, respectively (Fig. 4). It is interesting to note that in non-reactive and reactive amino residue, the same level of hydrophobicity properties with the aliphatic chain was experienced among the short peptides but different with medium and long peptides. In total, 52%, 11%, 17%, and 16% of the amino acid were, respectively hydrophobic-aliphatic, hydrophobic-aromatic, polar neutral, and hydrophilic basic properties in nature, which is the same when compared with the reactive amino acid. The degree of hydrophobic properties with aliphatic chain in both reactive and non-reactive amino acids are the same which indicated that the short peptides contributed more to the higher degree of hydrophobic-aliphatic properties in both reactive and non-reactive amino acids.
Based on Fig. 5a, proline (16%), tyrosine (12.8%), lysine (9.9%) and leucine (9.1%) were the prominent amino acids with hydrophobic-aliphatic property for short peptides. Hence, proline, tyrosine, and leucine were majorly responsible for the aliphatic-hydrophobicity properties of the non-reactive amino acid residue in short peptide sequences. Meanwhile, in terms of the reactive amino acid, proline in both long and medium peptides exhibited the highest frequency of 63 and 45%, respectively, as shown in Fig. 5b. This was trailed by both glycine (37%) and glutamine (36%) in the representative long peptides. This indicated that the percentage of proline was more in medium and long peptides than among the majority of the short peptides. According to Brandl et al. (2001), proline is the top 5 donor in the Cα–H···Aro-π-interaction, Cα–H···Am-π-interaction and Cα–H···Arg-π-interaction. Jabs et al. (1999) reported that the interaction between aromatic residues could be stabilized by a Cα–H···aro-π-interaction by involving Pro-Cα–H and the aromatic ring, thus suggesting this Pro residue (as donor) in the peptide might highly interact with His353, His383, His387, His513, Tyr520 and Tyr523 (as acceptor) in ACE.
Specificity of the inhibitor peptides towards the amino acid residues’ terminals
According to Fears et al. (2013), the location of specific amino acid residues at the C- or N-terminal is an important property of peptides. In this study, the angiotensin-converting inhibitory properties of bioactive peptides were centered on the ultimate, penultimate, and antepenultimate positions. The terminal indication of the ACE inhibitory peptide at the ultimate (C1 and N1), penultimate (C2 and N2), and antepenultimate (C3 and N3) positions were succinctly investigated for short, medium and long peptides, respectively, for both the reactive and non-reactive amino acids as presented as \(\mathrm{N}1-\mathrm{N}2-\mathrm{N}3-\mathrm{Xn}-\mathrm{C}3-\mathrm{C}2-\mathrm{C}1\) depicting the terminal indication of the inhibitor peptide, where N, C, and Xn represents the amino-terminal, carboxylic terminal other amino acid residues located in between the terminals, respectively.
N1 position
At the N1 ultimate terminal, 63% of the short peptides were hydrophobic-aliphatic followed by 50% and 39% of long and medium peptides, respectively, exhibiting high hydrophobicity characteristics (Fig. 6a). Other prominent properties were the hydrophobic-aromatic and hydrophilic-basic properties with 30% and 26% in medium and long peptides, respectively. Meanwhile, the hydrophobic-aliphatic properties in both the non-reactive and reactive amino acid residue are similar. Specifically, at the N1 position of the reactive amino acid in short peptides, the percentage of short, medium, and long peptides with hydrophobic–aliphatic properties were 63%, 36%, and 50%, respectively, showing a slight variation with the non-reactive peptides (Fig. 6b).
Furthermore, for individual amino acid based on the bioactive peptides length, isoleucine (15%), glycine (12%), alanine (9%), leucine (9%) and tyrosine (9%) were dominant in short peptides. For medium peptides, phenylalanine (14%), tyrosine (14%) and valine (11%) were dominant whereas for long peptides, lysine (12%) was dominant as presented in Fig. 7a. No long or medium peptides contained threonine and aspartic acid, while in terms of the histidine only in long peptides they can be found and not in short or medium peptides. This suggests that the shorter the peptide sequence the more likely it is to have threonine and aspartic acid, while the shorter the peptide sequence, the more likely it is not to contain histidine. Overall, 30% and 28% of the peptides contained leucine and tyrosine, while 24% contained an equal amount of isoleucine, valine, glycine, and lysine. Based on the peptide length of the reactive amino acid, 25% and 17% of long peptides contained arginine and leucine, respectively (Fig. 7b). It is interesting to know that no long or medium peptides contained isoleucine, glutamine, threonine, and aspartic acid. When compared with the non-reactive amino acid residue property, only aspartic acid was present in equal proportion (2.3%) in short peptides alone but absent in both long and medium peptides. This indicated that the longer the peptide sequence the less likely aspartic acid will be present. Overall, at the N1 ultimate position of the reactive amino acid residue property, leucine (38%) appears to be the most prominent amino acid in the N1 ultimate position, followed by tyrosine and arginine (29% each), alanine (25%) and proline (24%) (Fig. 7b).
ACE inhibitory peptide structure–activity relationship for reactive N1 amino acid
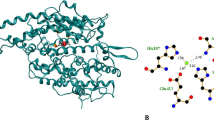
ACE is a zinc metallopeptidase comprising of two homologous domains, namely the N and C domains. Although both domains contain an independent functional active site characterised by the presence of a HEXXH zinc binding motif, the catalytic site action at the C domain was more efficient in catalysis hence play a more prominent role in blood pressure regulation (Fuchs et al. 2008; Acharya et al. 2003). At the C domain, Zn2+ is coordinated to H383, H387, E411 and a water molecule. Three additional subsites are found surrounding this catalytic site, namely the S1 subsite (A354, E384 and Y523), S2 subsite (contains N281, H353, K511, H513 and Y520) and S1’ subsite (E162). In addition, C domain is activated by two chloride (Cl−) ions. A Cl− binding site adjacent to the Zn2+ ion comprised of hotspots Tyr224 and Arg522 whereas another distal Cl− binding site comprised of hotspots Arg186, Trp485 and Arg489 (Fuchs et al. 2008; Acharya et al. 2003). Lisinopril was found to occupy these subsites and coordinate with the catalytically essential Zn2+. Another competitive inhibitor of ACE, captopril, was also found to occupy the S1 and S2 subsites of ACE which could prevent the putative binding of angiotensin I (Natesh et al. 2004). Based on these examples, peptides capable of binding to the Zn2+ site and the subsites S1, S2 and S1’ may potentially inhibit ACE in a similar competitive manner to confer anti-hypertensive effect. To determine the role of reactive N1 residue on ACE binding, molecular docking was performed using two example peptides from each reactive N1 amino acid i.e., leucine, tyrosine, arginine, alanine, and proline. Firstly, the N-terminal of peptide LHLPLPL protruded into the catalytic site and subsite S1 of ACE (− 10.9 kcal/mol) where the peptide N1 ultimate leucine residue hydrophobically interacted with Ala356, His387, Phe391, and His410 (Table 1; Fig. 8a). The interaction between the hydrophobic side chain of N1 leucine and the imidazole ring of His387 could weaken the catalytically essential tetrahedral Zn2+ coordinate complex in the ACE active site since His387 is a ligand of Zn2+ ion. Moreover, leucine is a branched-chain amino acid (BCAA). Its bulky side chain could form a hydrophobic shield to protect the Zn-peptide chelate complex from water attack. The neighboring histidine residue formed a salt bridge with hotspots Glu411 and Glu384 at the S1 subsite through its imidazole ring which could further hinder substrate access to the Zn2+ site to induce ACE inhibition.
a Binding interactions between ACE and LHLPLPL. b Binding interactions between ACE and LVYPFPGPIPNSLPQNIPP. c Binding interactions between ACE and YLYEIAR. d Binding interactions between ACE and YINQMPQKSRE. e Binding interactions between ACE and RMLGNTPTK. f Binding interactions between ACE and RNEQMGAGRLGRLRK. g Binding interactions between ACE and AKYSY. h Binding interactions between ACE and ASQSIWLPGWL. i Binding interactions between ACE and PNSHP. j Binding interactions between ACE and PKHKEMPFPPKYPVEPFT
Peptide LVYPFPGPIPNSLPQNIPP was bound outside of the ACE active site (− 15.8 kcal/mol) and its N1 leucine formed hydrophobic interactions with Leu139, Glu143, Trp357, Pro515, Ser516, and Ser517 (Table 1; Fig. 8b). The large molecular weight of the peptide and its bulky side chains may cause large steric hindrances which hinder its passage through the narrow opening of the ACE active site (Natesh et al. 2003) However, the P4 residue of peptide hydrophobically interacted with Arg522, the ligand of the Cl− ion near the active site. Since Arg522 also indirectly contribute to the structural stability of Zn2+ binding site and subsite S1 (Yates et al. 2014), peptide interaction with this hotspot may disrupt substrate catalysis. On the other hand, peptides YLYEIAR and YINQMPQKSRE with N1 tyrosine residue showed a similar binding affinity (− 11.0 kcal/mol) towards the ACE active site. The N1 tyrosine of YLYEIAR hydrophobically interacted with the stabilizing residues His353, Ala354, His383, and Tyr523 in the catalytic site as well as subsites S1 and S2. The hydrophobic, aromatic side chain of tyrosine may favor the peptide interaction with these hydrophobic hotspots in the ACE active site. The N1 tyrosine amide nitrogen also contributed a hydrogen bond and coordinate bond with Glu384 and Zn2+ (Table 1; Fig. 8c). For YINQMPQKSRE, the reactive hydroxyl group attached to the phenyl ring of N1 tyrosine had formed a hydrogen bond with Zn2+ and Glu384 (Table 1; Fig. 8d). The aromatic ring also hydrophobically interacted with the stabilizing residues Ala354 and Tyr523 as well as the ligands of Zn2+ (His383, His387, and Glu411). These observations suggested that an N1 tyrosine residue could suppress ACE activity by distorting the catalytically essential tetrahedral Zn2+ coordinate complex. Next, RMLGNTPTK resided at the Zn2+, S1 and S2 sites of ACE C domain (− 12.0 kcal/mol) where its N1 ultimate arginine residue was hydrogen-bonded to Gln281, Thr282, and Glu376, hydrophobically interacted with Thr166, Asn277, Thr372, and Asn374 and formed a salt bridge with Glu376 and Asp377 (Table 1; Fig. 8e). For RNEQMGAGRLGRLRK (− 12.8 kcal/mol), the N-terminal was pointed away from the active site where its N1 arginine formed two salt bridges with Asn105 (Table 1; Fig. 8f). The peptide C-terminal, on the contrary, was bound to the catalytic and S1 site of ACE. Besides, AKYSY and ASQSIWLPGWL showed binding affinities of − 9.5 and − 13.2 kcal/mol towards ACE, respectively. The N1 alanine of AKYSY formed hydrogen bonds with Zn2+, Ala356, and Glu384 and hydrophobic interactions with Ala354, Ser355, His383, and His387 in the ACE active site (Table 1; Fig. 8g). Although the N1 alanine of ASQSIWLPGWL did not interact with Zn2+, it hydrophobically interacted with His387 and Ala356 at the S1 site (Table 1; Fig. 8h). Finally, PNSHP and PKHKEMPFPPKYPVEPFT were bound to ACE catalytic and S1 and S2 sites with binding affinities of − 8.5 and − 13.0 kcal/mol, respectively. The N1 proline residue of PNSHP was hydrogen-bonded to Asn66 and Asn70 and hydrophobically interacted with Tyr69, Leu81, and Glu143 (Table 1; Fig. 8i). For PKHKEMPFPPKYPVEPFT, the N1 residue formed hydrophobic interactions with the ligand of Zn2+, His383 and the subsite stabilizing residues His353, Ala354, Glu383, and Tyr523 (Table 1; Fig. 8j). Overall, majority of the ACE inhibitory peptides were bound to the Zn2+ catalytic site and the adjacent subsites S1 and S2 with none of them interacted with subsite S1’. Reactive amino acids leucine, tyrosine, arginine, alanine, and proline at the N1 ultimate position of anti-ACE peptides could interact with the enzyme hotspots. Low molecular weight peptides were postulated to occupy the active site groove of ACE more readily. This is because the entrance to the active site has an opening of approximately 3 Å which poses a major challenge for the passage of large molecular weight peptides with bulky side chains (Natesh et al. 2003). Still, the presence of an N1 ultimate proline residue may be beneficial for long-chained peptides (e.g. PKHKEMPFPPKYPVEPFT) to interact with the catalytic site hotspots. This is because proline features a unique cyclic structure which could introduce kinks in ACE structure to alter hotspot residue orientation in the active site to hinder substrate binding (Li et al. 2018b; Schmidt et al. 2016).
C1 position
In Fig. 9a, the C1 amino acid residue of the inhibitor peptides mostly exhibited hydrophobic properties with aliphatic chain (48% in short peptides, 45% in medium peptides, and 26% in long peptides). However, the hydrophobic-aliphatic properties at the C1 terminal of reactive peptides were more prominent with the long peptides (56%), followed by short and medium peptides with 48% frequencies each Fig. 9b. This suggests that the hydrophobicity aliphatic properties in the C1 terminal of the reactive peptides are more prominent when compared with the non-reactive peptides. The hydrophobic-aromatic is an essential property at the C1 terminal with short peptides having a frequency of 29%, followed by the medium peptides with 28% frequency. It is interesting to know that there are no short peptides that exhibit hydrophilic properties at the C1 terminal acidic chain for both non-reactive and reactive amino acids. Also, no polar neutral nor hydrophilic acidic properties were reported at the reactive C1 terminals of the medium peptides.
While hydrophobic aliphatic properties appeared to be more prevalent throughout most peptides, a range of C1 amino acid functionalities was observed. For instance, proline was the dominant amino acid for short and medium peptides with frequencies of 24% and 23%, respectively (Fig. 10a). This was followed by 17% and 16% of tyrosine for the short and medium peptides at the C1 terminal of the non-reactive peptides. Moreover, other amino acids that showed considerable prominence at the C1 terminal include lysine (14% each for medium and long peptides) and 14% of threonine for the long peptides. However, in the C1 terminal, proline is the most reactive amino acid for the short, medium, and long peptides with 24%, 34%, and 44% prominence, respectively (Fig. 10b). The correlations between ACE inhibitory peptide structure and activity level suggest that the substrate's C-terminal tripeptide sequence has a significant impact on ACE interaction. At the active site of ACE, the C-terminal tripeptide residues may bind with other sub-sites. At three locations from the C-terminus, ACE favors hydrophobic amino acid residues like proline, phenylalanine, and tyrosine as substrates or competitors (Xiang et al. 2021). Proline is found at the C-terminus of most naturally produced peptide inhibitors (Byun and Kim 2002).
ACE inhibitory peptide structure–activity relationship for reactive C1 amino acid
According to Sitanggang et al. (2021), peptides with a C-terminal proline tend to bind strongly to ACE. For instance, LGPVRGPFP and VKKVLGNP were bound to the ACE active site with affinities of − 13.1 and − 10.3 kcal/mol, respectively. The C1 ultimate proline of LGPVRGPFP formed hydrogen bonds with Asn277 and Thr282 and hydrophobic interactions with Glu376 and Val380 which were adjacent to the ACE hotspots at the S1 site (Table 2; Fig. 11a). For VKKVLGNP, its C-terminal proline hydrophobically interacted with the hotspots Gln281, His513 and Tyr520 at the S2 subsite as well as Tyr523 at the S1 subsite (Table 2; Fig. 11b). The presence of cyclic side chain of proline might have altered the active site hotspot orientation which allowed peptide interactions with the catalytic essential Zn2+ deeply buried within the active site. A similar hypothesis by Wu et al. (2006) also suggested that a C-terminal proline residue was one of the structural features of strong ACE inhibitory di- and tripeptides. A typical example was the commercial peptide analog drug Lisinopril sequenced KP where its C-terminal proline could more readily interact with most hotspots in the ACE active site (Natesh et al. 2003).
N2 position
The amino acid residue at the N2 position was found to exhibit hydrophobic properties at the aliphatic chain as the prominent feature property for both medium (48%) and long (39%) peptides followed by hydrophilic-basic (29%) in short peptides, and medium (23%) peptides (Fig. 12a). Moreover, the long peptide has the most prominent polar neutral (27%) characteristic at the N2 position. Whereas there is a similar degree (29%) of hydrophobic-aliphatic, hydrophobic-aromatic, and hydrophilic basic properties by the short peptides. However, the properties of the reactive amino acid residue tend to be different with 40% and 30% of the medium peptides exhibiting hydrophobic properties at the aliphatic chain and hydrophilic-basic of the N2 terminal, respectively (Fig. 12b).
Moreover, 29% of the short peptides each exhibited hydrophobic-aliphatic, hydrophobic-aromatic, and hydrophilic-basic properties. It was observed that none of the amino acids of the short and long peptides exhibited hydrophilic properties at the acidic chain. Thirty-one percent (31%) of the long peptides exhibited hydrophobic-aliphatic properties, with an equal number exhibiting the polar neutral property. In the same way, similar properties were observed with 19% of long peptides equally exhibiting hydrophobic-aromatic and hydrophilic-basic properties. It is important to know that only 3% of the medium peptides exhibited hydrophilic-acidic property, whereas no short nor long peptides exhibited this property.
Short peptides appeared to have more isoleucine residue (29%) at the N2 position followed by lysine (16%) from medium peptides and glutamic acid (15%) from the long peptides. Despite that, N2 amino acid residues of short peptides tend to have a frequency of 14% each for phenylalanine, tyrosine, glutamine, arginine, lysine (Fig. 13a). Moreover, based on the length of the peptides no alanine, valine, leucine, methionine, glycine, proline, tryptophan, asparagine, cysteine, serine, threonine, aspartic acid, glutamic acid nor histidine was found among the short peptides reviewed. Also, no tyrosine was found among the long peptides. Tryptophan, aspartic acid, and histidine are predominantly found in medium peptides but not significant in both short and long peptides. However, isoleucine residue (29%) was the most observed reactive amino acid in short peptides followed by phenylalanine, tyrosine, glutamine, arginine, and lysine with 14% each interacting with the ACE active site (Fig. 13b). However, at the medium peptides lysine (20%), proline (13%) have the highest frequency, while at the long peptides arginine (19%), phenylalanine (19%), leucine (13%), and glutamine (13%) are the most dominant amino acids. Overall, isoleucine (29%) was found to have the highest number of amino acids found at the N2 position. A similar trend was observed in the reactive amino acid residues in which isoleucine (29%) are the most reactive amino acid residues. This indicated that irrespective of isoleucine interaction with the ACE active site, their frequency of occurrence remains approximately the same.
ACE inhibitory peptide structure–activity relationship for reactive N2 amino acid
To elucidate the role of N2 position residue in peptide binding to ACE, molecular docking was conducted for the example peptides with penultimate arginine, lysine, phenylalanine, isoleucine, glutamine, and asparagine at the N-terminal. Peptide ERKIKVYL with an N2 arginine residue stabilized peptide binding by forming hydrophobic interaction with Trp59, a salt bridge with Glu403, and a coordinate bond with Tyr360 (Table 3; Fig. 14a). The N2 arginine of RRQRRRRMRKDK also stabilized the peptide binding by forming hydrophobic interactions with Tyr62, Ile88, and Arg124 as well as a salt bridge with Tyr360 (Table 3; Fig. 14b). Next, the N2 lysine residues of KKIATYQER and VKKVLGNP could directly interact with the ACE hotspots in the active and Cl− binding site. The reactive N2 lysine of KKIATYQER occupied the Zn2+ binding site, S1 and S2 subsites where its side chain was hydrogen-bonded to Ala356 and Glu384, hydrophobically interacted with Zn2 + , His353, Ala354, His383, His387, His513, and Tyr523 (Table 3; Fig. 14c). The reactive N2 lysine of VKKVLGNP, on the other hand, formed a salt bridge with Glu411 at the catalytic site and hydrophobic interaction with Arg522 at the Cl− binding site (Table 3). For peptides with N2 phenylalanine residue (i.e. FFGRCVSP and KFHSGIQSEPKAIP), peptide FFGRCVSP did not bind to the active site of ACE (− 10.9 kcal/mol) but to the adjacent Cl− binding site. The N2 phenylalanine hydrophobically interacts with Trp59, Tyr62, Ile88, Tyr360, and Glu403 (Table 3; Fig. 14d).
a Binding interactions between ACE and ERKIKVYL. b Binding interactions between ACE and RRQRRRRMRKDK. c Binding interactions between ACE and KKIATYQER. d Binding interactions between ACE and FFGRCVSP. e Binding interactions between ACE and KFHSGIQSEPKAIP. f Binding interactions between ACE and YIPIQY. g Binding interactions between ACE and YQEPVLQPVR. h Binding interactions between ACE and GQLGEHGGAGMG. i Binding interactions between ACE and GNPVGGVGHGTTGT
Peptide KFHSGIQSEPKAIP, on the other hand, was found to span across the active site and chloride binding site of ACE (Table 3; Fig. 14e). While the peptide N1 lysine formed a hydrogen bond with Zn2+ and Tyr523, salt bridges with Glu411, and hydrophobic interaction with His387 and Phe391 in the active site, the N2 phenylalanine hydrophobically interacted with Met223, Glu403, and Arg522 in the chloride binding site. The chloride binding site is closely located to Zn2+ ion in the active site (Natesh et al. 2003) and the binding of a Cl− ion to its major ligand Arg522 is necessary for the activation of ACE activity (Liu et al. 2001; Masuyer et al. 2014; Tzakos et al. 2003). Therefore, peptides with N2 phenylalanine could suppress enzyme actions by interfering with the Cl- activation of ACE. Next, peptide YIPIQY was bound to the active site of ACE (− 10.6 kcal/mol) where its N-terminal tyrosine formed hydrogen bonds with His383 and Glu411. The penultimate isoleucine did not interact with any of the hotspots but hydrophobically interacted with Ser355 and Ala356 in the S1 site (Table 3; Fig. 14f). A similar outcome was also observed from YINQMPQKSRE (− 11.0 kcal/mol) where the N2 isoleucine formed hydrophobic interactions with Asp358, Tyr360, and Phe391 (Table 3).
Since isoleucine is a BCAA, its side chain may form a hydrophobic fence to shield water attack which in turn stabilize the N1 tyrosine and the disrupted tetrahedral Zn-coordinate complex of ACE (Kathuria et al. 2016). The N2 isoleucine was thus suggested as a supportive residue in the overall binding interaction between peptide and ACE. Besides, the peptides YQEPVLQPVR and GQLGEHGGAGMG were bound near the chloride binding site of ACE with binding affinities of − 12.8 and − 13.2 kcal/mol, respectively. The N2 glutamine residue of YQEPVLQPVR only interacted with Ser355 via hydrophobic interaction (Table 3; Fig. 14g) whereas that of GQLGEHGGAGMG interacted with Trp59, Tyr62, Asn85, Ile88, and Tyr360 (Table 3; Fig. 14h). For peptides PNSHP and GNPVGGVGHGTTGT, their N2 asparagine residues seemed to stabilize peptide binding to the Zn2+ ion by interacting with the surrounding active site residues. The N2 asparagine of PNSHP formed hydrogen bonds with Ser355 and Lys368 and hydrophobic interactions with Asn70, Glu143, Val351, His353, and Phe512 (Table 3) whereas the N2 asparagine of GNPVGGVGHGTTGT formed a hydrogen bond with Asn66 and hydrophobic interaction with Trp357 (Table 3; Fig. 14i). Overall, it seemed that the reactive amino acids at the N2 position played supportive roles in the peptide binding and inhibition of ACE. The N2 residues could either stabilize its adjacent peptide residue involved in ACE inhibition or hinder ACE activation by interfering with the Cl− ligand at the Cl− binding site. The presence of N2 lysine could directly inhibit the catalytic actions of ACE by interrupting the stabilizing residues and the catalytically essential Zn-coordinate complex.
C2 position
The amino acid residue at the C2 position was found to exhibit hydrophobic-aliphatic and hydrophilic-basic as the dominant property for the short peptides in an equal proportion of 50% (Fig. 15a). Moreover, the hydrophobicity at the aliphatic chain of the long peptides exhibits a sample percentage of occurrences which cumulates to 50% of the total long peptides reviewed. Overall, the hydrophobic-aliphatic and hydrophilic-basic are the dominant properties of the amino acid residue at the C2 terminal. A close look at the other properties revealed that short peptides do not exhibit any of the hydrophobic-aromatic, polar neutral, and hydrophilic-acidic properties as presented in Fig. 15a. However, the reactive amino acid residue of the short peptides reviewed shows hydrophobic-aromatic as the most significant property with 89% dominance (Fig. 15b). This is followed by the hydrophobic-aliphatic of the long and medium peptides with an observed frequency of 46% and 43%, respectively. From the short peptides reviewed none of the reactive amino acids was found to be hydrophilic-acidic, hydrophilic-basic nor polar neutral.
C2 terminal of short peptides feature equal proportion of amino acids, i.e., 50% leucine and lysine. This is followed by 23% of proline and 17% of phenylalanine from medium and long peptides at the C2 terminal. Overall, only leucine and lysine were found in the short peptides, while other amino acids were absent at the C2 position of the ACE inhibitor peptides (Fig. 16a). However, the overall reactive amino acid residue in the C2 terminal of short inhibitor peptides is proline (33%), tyrosine (33%), valine (17%), and leucine (17%) (Fig. 16b). The long peptides consisted of phenylalanine (17%), glutamine (13%), and arginine (13%) at the reactive site of the C2 terminal of the amino acid residue (Fig. 16b). Overall, while leucine and lysine were the most predominant amino acid at the non-reactive site of the C2 terminal, proline and tyrosine are the dominant reactive amino acid at the C2 position. Moreover, in all the short peptides reviewed at the reactive site of the C2 terminal the following amino acid were not present viz: methionine, glycine, phenylalanine, tryptophan, asparagine cysteine, glutamine, serine, threonine, aspartic acid, glutamic acid, arginine, histidine, and lysine.
ACE inhibitory peptide structure–activity relationship for reactive C2 amino acid
For C2 reactive amino acids, peptides SKVYPFPGPI and GEHGGAGMGGGQFQPV with penultimate proline whereas SKVYP and ERKIKVYL with tyrosine were chosen for molecular docking study. The C2 proline residues of SKVYPFPGPI and GEHGGAGMGGGQFQPV were found outside of the ACE active site where the former hydrophobically interacted with Asn70, Glu143, Ser355, and Phe512 (Table 4; Fig. 17a) and the latter Thr166, Asn277, Glu376, and Asp377 (Table 4; Fig. 17b). The C-terminal penultimate tyrosine of SKVYP was predicted to form hydrogen bonds with His383, Glu384, and His387, as well as a coordinate bond with Zn2+ through its phenyl side chain (Table 4; Fig. 17c). As a result, the Zn2+ binding site may be disrupted to exhibit reduced ACE catalyzing activity. This aromatic residue also hydrophobically contacted with Ala354, Ser355, Ala356, Glu411, and Tyr523 at the S1 subsite. Moreover, the C2 tyrosine of ERKIKVYL hydrophobically interacted with the stabilizing residues Glu162 in the S1’ subsite, Glu281, His353, and Lys511 in the S2 subsite (Table 4). This highlighted the importance of a hydrophobic, aromatic residue at the C2 position of the peptide to facilitate peptide binding to the Zn2+ site and the S1, S2 and S1’ sites (Sitanggang et al. 2021).
N3 position
The amino acid residue at the N3 position was found to exhibit hydrophobic-aliphatic as the dominant property for both medium (48%) and long (38%) peptides followed by basic (24% in medium, 19% in long peptides) and neutral (19%) in long peptides (Fig. 18a). Moreover, the hydrophobic properties of the amino acid residue were the same for both the medium and long peptides considered. However, the properties of reactive amino acid residue tend to be similar with a higher degree of hydrophobic-aliphatic properties in medium (39%) and long peptides (36%); this is followed by hydrophilic-basic properties of medium (25%) and hydrophobic-aromatic properties of long peptides (23%) at the N3 position (Fig. 18b). Overall, the hydrophobicity of the aliphatic chain at the N3 terminal is higher for the non-reactive amino acid residue than in the reactive. Whereas the hydrophobicity of the aromatic chain at the N3 terminal of reactive amino acid is higher than that of the non-reactive amino acid residue. In all the properties considered, none of the short peptides exhibited hydrophobicity, hydrophilic, nor polar neutral properties for both reactive and non-reactive amino acid at the N3 position.
In terms of amino acid profile in the peptides, glycine (25%), leucine (17%), lysine (17%), proline (16%), and arginine (14%) are more dominant at the N3 terminal as presented in Fig. 19a. Following the length of the medium peptides, glycine (18%), leucine (12%), lysine (12%), and arginine (9%) were the dominant amino acid residue at the N3 terminal. The most significant amino acid in the long peptides shares a similar trend with approximately 10% each for glutamine, histidine, glutamic acid, valine, and proline. However, the frequently observed in the reactive amino acids were tyrosine (25%) followed by lysine (23%), glutamine (21%), leucine (19%), and proline (17%), as shown in Fig. 19b. The most significant reactive amino acids in the long peptides at the N3 position are glutamine (18%), tyrosine (14%), and 9% each for glycine, proline, phenylalanine, and lysine. Also, for the medium peptides, the most dominant reactive amino acids include lysine (14%), leucine (14%), tyrosine (11%), and arginine (8%). Overall, in terms of peptides length, 18% of the medium peptides contained glycine as the most predominant amino acid residue at the N3 terminal, while 18% of glutamine was the most significant reactive amino acid from the long peptides. One similar characteristic is that in both instances, methionine and tryptophan were absent, with only the medium and long peptides. Tyrosine, lysine, proline and phenylalanine, leucine, and glutamine seem to be the most significantly abundant reactive amino acid at the N3 terminal.
ACE inhibitory peptide structure–activity relationship for reactive N3 amino acid
Peptides LLYQQPV and YLYEIAR possessing an N3 tyrosine were bound to the active site of ACE in a similar orientation and binding affinity (− 11.3 and − 11.0 kcal/mol, respectively). Both peptides could chelate Zn2+ while their N3 tyrosine either forms hydrogen bond or hydrophobic interaction with the stabilizing residues His353 and Tyr523 in the S1 and S2 subsites (Table 5; Fig. 20a). The aromatic side chain of tyrosine could more readily interact with the hydrophobic and aromatic hotspots surrounding Zn2+ using their delocalized π-electron cloud. Peptides FDKLPGFGD and ERKIKVYL did not bind close to the ACE active site. The N3 lysine of the former hydrophobically interacted with Tyr62, Asn66, and Trp357 (Table 5; Fig. 20b) whereas the latter with Asn66, Ala356, and Trp357 (Table 5). The positive charge of lysine may hinder its ability to interact in the hydrophobic active site of ACE. Besides, the N3 glutamine residue of FPQYLQY was predicted to hydrophobically interact with Val518, Pro519, and Arg522 (Table 5; Fig. 20c). The polar side chain of glutamine may be attracted to the positively charged Arg522 which is the major ligand of Cl− ion involved in Cl− activation of ACE. Next, the presence of a BCAA at the N3 position may favor peptide interaction with the catalytically essential Zn2+. For instance, the N1 and N3 leucine residues in peptide LHLPLPL could stabilize the Zn-peptide chelate complex.
The N3 leucine of RMLGQTPWK could also form hydrogen bonds with His353 and hydrophobic interactions with Gln281, Glu384, Lys511, and His513 which were the stabilizing residues of ACE S2 subsites (Fig. 20d). These examples suggested the importance of leucine or BCAA as supporting residue at the N3 position. On the other hand, the N3 proline residue from LGPVRGPFP and YIPIQY only hydrophobically interacted with Ala356 and Trp357. N3 proline may introduce a proline kink to disrupt the hotspot orientation in the ACE active site. Peptides YPFPGPI and NIFYCP also contained an aromatic residue at the N-terminal antepenultimate position. The presence of hydrophobic, aromatic, or BCAA residues adjacent to each other seemed to promote peptide interaction with the Zn-coordinate complex in the active site. For example, the two proline residues accompanied by the N3 phenylalanine of YPFPGPI could induce structural changes in the ACE active site (Li et al. 2018a). The N3 phenylalanine was then predicted to form hydrophobic interactions with Ser355, Ala356, Phe391, His410, and one of the Zn2+ ligands, His387 (Table 5; Fig. 20e). In the case of NIFYCP, the N3 phenylalanine sandwiched between BCAA isoleucine and aromatic tyrosine could form a hydrogen bond with Zn2+ and hydrophobic interactions with the S1 hotspot Glu384 and Zn2+ ligands His387 and Glu411 (Table 5; Fig. 20f). This could be attributed to the hydrophobic shield protection provided by the N2 BCAA and the conducive hydrophobic environment provided by the aromatic tyrosine. Overall, the presence of hydrophobic, aromatic, and BCAA residues at the N3 position seemed more favorable to allow peptide binding to the ACE active site compared to the charged residue.
C3 position
The results indicated that the C3 terminal amino acid residues of the inhibitor peptides mainly possess hydrophobic-aliphatic side chains (56% in medium peptides, and 57% in long peptides (Fig. 21a). Polar neutrality is an important property as well at the C3 terminal of medium and long peptides with a frequency of 22% and 12%, respectively. Sequent to this is the hydrophilic-basic with the fourth-highest property in long peptides (12%), whereas the medium peptides exhibited the lowest of all the properties covered. A similar pattern was also observed in the C3 reactive amino acid residues with hydrophobicity as the most prominent property by the long (70%) and medium peptides (54%) (Fig. 21b). Overall, the degree of hydrophobicity at the aliphatic chain of the reactive amino acid residue is higher when compared with the non-reactive amino acid residue. Moreover, one of the common characteristics between the non-reactive and reactive amino acids in the C3 position is that both do not exhibit properties for the short peptides with only the medium and long peptides exhibiting hydrophobic (aliphatic, aromatic) hydrophilic (acidic, basic), and polar neutral properties.
Based on the amino acid residues, proline (38%), glycine (21%), and glutamine (20%) are the most prominent at the C3 terminal as presented in Fig. 22a. However, for the reactive amino acid, proline (50%), alanine (26%), and glutamine (17%) are the most dominant at the C3 position (Fig. 22b). Moreover, in terms of the length of the peptides, 19% of the long peptides contain proline as the highest amino acid residue while 26% of the long peptides contain proline as the highest reactive amino acid in the C3 terminal. This is jointly followed by 18.8% of the medium peptides containing proline, while 24% of the medium peptides contain proline based on the reactive amino acids. This indicated that the presence of proline at the C3 position of the reactive amino acid is more prominent in the medium peptides. Moreover, for both reactive and non-reactive amino acid residues, the most common characteristic is the presence of proline which is more prominent in medium and long peptides. Several ACE inhibitory peptides have been found to contain two to twelve amino acid residues, according to past research. Peptides containing proline at the C-terminus have IC50 values as high as 182 µM (Byun and Kim 2002). Also, alanine in both reactive (26%) and non-reactive amino acids (14%) are not noticeable in long peptides with none found among the short and medium peptides. Cysteine and asparagine are both not found in both the reactive and non-reactive amino acids in the C3 terminal irrespective of the length of the peptides sequence.
ACE inhibitory peptide structure–activity relationship for reactive C3 amino acid
Based on the molecular docking result, a C3 proline residue of RMLGNTPTK and LGPVRGPFP may be crucial for peptide binding to the Cl− binding site and the active site of ACE. For RMLGNTPTK (− 12.0 kcal/mol), the C-terminal antepenultimate proline was hydrogen-bonded to Arg522 at the Cl− binding site and hydrophobically interacted with His387, His410, and Glu411 at the active site (Table 6; Fig. 8e). For LGPVRGPFP (− 13.1 kcal/mol), its C3 proline hydrophobically interacted with the Zn2+ ligand His383, S1 hotspots Ala354, Gly384, and Tyr523 and the S2 hotspot His353 (Table 6; Fig. 11a). Hence a C3 proline residue could potentially improve peptide interaction with hotspots in both Cl− binding site and the active site of ACE.
Future prospects
In recent times, the application of biotechnology in food-derived bioactive peptides has developed as a potential alternative to pharmaceutical therapies for the prevention and treatment of lifestyle-related illnesses, which are a major global public health concern. Several approaches have been investigated for their synthesis from diverse food sources including dairy foods, grains, and legumes, such as enzyme hydrolysis, fermentation process, or genetic modification using the recombinant DNA transformation. In this study, a total of 173 ACE inhibitory peptides; comprising of 87 short, 44 medium, and 42 long peptides were selected from the UWM BIOPEP database. The binding of these antihypertensive peptides towards a protein structure was succinctly analyzed using the Pepsite2 webservers. The result of this investigation succinctly revealed that the categorization of peptides based on length shows different unique preferences for both the reactive and non-reactive amino acid types as well as their positioning at the C and N terminals with hydrophobicity as the dominant characteristics. To summarize results in the current study, the preferences of amino acids at the C and N terminal ultimate, penultimate and antepenultimate positions were illustrated in Fig. 23. In furtherance to this, the C-terminals (C1, C2, and C3) are more predominant in ACE binding for long-chained peptides, while at the N terminals (N1, N2, and N3), the leucine and isoleucine are branched-chain amino acid (BCAA). Because leucine and isoleucine are BCAA, their bulky side chain forms a hydrophobic shield that protects the Zn-peptide chelate complex from water attack. The hydrophobic fence in turn stabilizes the N1 tyrosine and the disrupted tetrahedral Zn-coordinate complex of ACE. Overall, majority of the reported ACE inhibitory peptides interacted with either the Zn2+ site, Cl− site, and/or the subsites S1, S2 and S1’. Outcomes from the molecular docking study could be used to improve and design more specific domain-targeting peptide inhibitors of ACE. This paper presents an overview of the potential use of various food origins for the green synthesis of antihypertensive peptides using functional ingredients. The outcomes of this study contribute to a better understanding of peptide structural relationships and their functionality in angiotensin-converting enzyme inhibitory effects.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Acharya KR, Sturrock ED, Riordan JF, Ehlers MR (2003) ACE revisited: a new target for structure-based drug design. Nat Rev Drug Discovery 2:891–902. https://doi.org/10.1038/nrd1227
Asoodeh A, Haghighi L, Chamani J, Ansari-Ogholbeyk MA, Mojallal-Tabatabaei Z, Lagzian M (2014) Potential angiotensin I converting enzyme inhibitory peptides from gluten hydrolysate: biochemical characterization and molecular docking study. J Cereal Sci 60:92–98. https://doi.org/10.1016/j.jcs.2014.01.019
Bahadori M, Hemmateenejad B, Yousefinejad S (2019) Quantitative sequence-activity modeling of ACE peptide originated from milk using ACC–QTMS amino acid indices. Amino Acids 51:1209–1220. https://doi.org/10.1007/s00726-019-02761-y
Balgir PP, Kaur T, Sharma M (2016) Antihypertensive peptides derived from food sources. MOJ Food Process Technol 2:1–6
Barley MH, Turner NJ, Goodacre R (2018) Improved descriptors for the quantitative structure–activity relationship modeling of peptides and proteins. J Chem Inf Model 58:234–243
Brandl M, Weiss MS, Jabs A, Sühnel J, Hilgenfeld R (2001) C-h⋯π-interactions in proteins. J Mol Biol 307:357–377. https://doi.org/10.1006/JMBI.2000.4473
Byun HG, Kim SK (2002) Structure and activity of angiotensin I converting enzyme inhibitory peptides derived from alaskan pollack skin. J Biochem Mol Biol 35:239–243. https://doi.org/10.5483/bmbrep.2002.35.2.239
Ding Q, Sheikh AR, Chen Q, Hu Y, Sun N, Su X, Luo L, Ma H, He R (2021) Understanding the mechanism for the structure-activity relationship of food-derived ACEI peptides. Food Rev Intl. https://doi.org/10.1080/87559129.2021.1936005
Fears KP, Petrovykh DY, Clark TD (2013) Evaluating protocols and analytical methods for peptide adsorption experiments. Biointerphases 8:1–15. https://doi.org/10.1186/1559-4106-8-20
Fitzgerald RJ, Murray BA, Walsh DJ (2004) The Emerging Role of Dairy Proteins and Bioactive Peptides in Nutrition and Health Hypotensive Peptides from Milk Proteins 1:2
Fuchs S, Xiao HD, Hubert C, Michaud A, Campbell DJ, Adams JW, Capecchi MR, Corvol P, Bernstein KE (2008) Angiotensin-converting enzyme C-terminal catalytic domain is the main site of angiotensin I cleavage in vivo. Hypertension 51:267–274. https://doi.org/10.1161/HYPERTENSIONAHA.107.097865
Ghatage T, Goyal SG, Dhar A, Bhat A (2021) Novel therapeutics for the treatment of hypertension and its associated complications: peptide- and nonpeptide-based strategies. Hypertens Res 44:740–755. https://doi.org/10.1038/s41440-021-00643-z
Gobbetti M, Stepaniak L, De Angelis M, Corsetti A, Di Cagno R (2002) Latent bioactive peptides in milk proteins: proteolytic activation and significance in dairy processing. Crit Rev Food Sci Nutr 42:223–239
Gu RZ, Liu WY, Lin F, Jin ZT, Chen L, Yi WX, Lu J, Cai MY (2012) Antioxidant and angiotensin I-converting enzyme inhibitory properties of oligopeptides derived from black-bone silky fowl (Gallus gallus domesticus Brisson) muscle. Food Res Int 49:326–333. https://doi.org/10.1016/j.foodres.2012.07.009
Hemmateenejad B, Yousefinejad S, Mehdipour AR (2011) Novel amino acids indices based on quantum topological molecular similarity and their application to QSAR study of peptides. Amino Acids 40:1169–1183. https://doi.org/10.1007/s00726-010-0741-x
Hernandezledesma B, Quiros A, Amigo L, Recio I (2007) Identification of bioactive peptides after digestion of human milk and infant formula with pepsin and pancreatin. Int Dairy J 17(1):42–49
Ishiguro K, Sameshima Y, Kume T, Ikeda KI, Matsumoto J, Yoshimoto M (2012) Hypotensive effect of a sweetpotato protein digest in spontaneously hypertensive rats and purification of angiotensin I-converting enzyme inhibitory peptides. Food Chem 131:774–779. https://doi.org/10.1016/j.foodchem.2011.09.038
Jabs A, Weiss MS, Hilgenfeld R (1999) Non-proline Cis peptide bonds in proteins. J Mol Biol 286:291–304. https://doi.org/10.1006/JMBI.1998.2459
Jakala P, Vapaatalo H (2010) Antihypertensive peptides from milk proteins. Pharmaceuticals 3:251–272
Jao C-L, Huang S-L, Hsu K-C (2012) Angiotensin I-converting enzyme inhibitory peptides: inhibition mode, bioavailability, and antihypertensive effects. Biomedicine 2:130–136
Kathuria SV, Chan YH, Nobrega RP, Özen A, Matthews CR (2016) Clusters of isoleucine, leucine, and valine side chains define cores of stability in high-energy states of globular proteins: sequence determinants of structure and stability. Protein Sci 25:662–675. https://doi.org/10.1002/pro.2860
Kim YE, Yoon S, Yu D et al (1999) Novel angiotensin-I-converting enzyme inhibitory peptides derived from recombinant human as1-casein expressed in Escherichia coli. J Ournal of Dairy Research 66:431–439
Li H, Prairie N, Udenigwe CC, Adebiyi AP, Tappia PS, Aukema HM, Jones PJ, Aluko RE (2011) Blood pressure lowering effect of a pea protein hydrolysate in hypertensive rats and humans. J Agric Food Chem 59:9854–9860
Li M, Xia S, Zhang Y, Li X (2018a) Optimization of ACE inhibitory peptides from black soybean by microwave-assisted enzymatic method and study on its stability. LWT 98:358–365
Li J, Liu Z, Zhao Y, Zhu X, Yu R, Dong S, Wu H (2018b) Novel natural angiotensin converting enzyme (ACE)-inhibitory peptides derived from sea cucumber-modified hydrolysates by adding exogenous proline and a study of their structure⇓activity relationship. Mar Drugs. https://doi.org/10.3390/md16080271
Li M, Fan W, Xu Y (2021) Identification of angiotensin converting enzyme (ACE) inhibitory and antioxidant peptides derived from Pixian broad bean paste. LWT 151:112221. https://doi.org/10.1016/J.LWT.2021.112221
Liu X, Fernandez M, Wouters MA, Heyberger S, Husain A (2001) Arg1098 is critical for the chloride dependence of human angiotensin I-converting enzyme C-domain catalytic activity. J Biol Chem 276:33518–33525. https://doi.org/10.1074/jbc.M101495200
Losacco M, Gallerani R, Gobbetti M, Minervini F, De Leo F (2007) Production of active angiotensin-I converting enzyme inhibitory peptides derived from bovine β-casein by recombinant DNA technologies. Biotechnol J Healthc Nutr Technol 2:1425–1434
Lv GS, Huo GC, Fu XY (2003) Expression of milk-derived antihypertensive peptide in escherichia coli. J Dairy Sci 86:1927–1931
Ma K, Wang Y, Wang M, Wang Z, Wang X, Ju X, He R (2021) Antihypertensive activity of the ACE–renin inhibitory peptide derived from Moringa oleifera protein. Food Funct 2:1–12
Majumder K, Chakrabarti S, Morton JS, Panahi S, Kaufman S, Davidge ST, Wu J (2013) Egg-derived tri-peptide IRW exerts antihypertensive effects in spontaneously hypertensive rats. PLoS ONE. https://doi.org/10.1371/journal.pone.0082829
Masuyer G, Yates CJ, Sturrock ED, Ravi Acharya K (2014) Angiotensin-I converting enzyme (ACE): structure, biological roles, and molecular basis for chloride ion dependence. Biol Chem 395(10):1135–1149
Mei H, Liao Z, Zhou Y, Li SZ (2005) A new set of amino acid descriptors and its application in peptide QSARs. Biopolymers 80:775–786. https://doi.org/10.1002/bip.20296
Meisel H (1998) Overview on milk protein-derived peptides. Int Dairy J 8:363–373
Meisel H (2006) ACE inhibitory peptides. In: Mine Y, Shahidi F (eds) Nutraceutical proteins and peptides in health and disease. CRC Taylor & Francis Group, Boca Raton, pp 269–315
Minkiewicz P, Iwaniak A, Darewicz M (2019) BIOPEP-UWM database of bioactive peptides: current opportunities. Int J Mol Sci 20:5978. https://doi.org/10.3390/ijms20235978
Mojica L, Luna-Vital DA, González De Mejía E (2017) Characterization of peptides from common bean protein isolates and their potential to inhibit markers of type-2 diabetes, hypertension and oxidative stress. J Sci Food Agric 97:401–2410. https://doi.org/10.1002/jsfa.8053
Nakamura Y, Yamamoto N, Sakai K, Takano T (1995) Antihypertensive effects of sour milk and peptides isolated from it that are inhibitors to angiotensin I-converting enzyme. J Dairy Sci 78:1253–1257
Natesh R, Schwager S, Sturrock E (2003) Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 421:551–554
Natesh R, Schwager SL, Evans HR, Sturrock ED, Acharya KR (2004) Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry 43:8718–8724. https://doi.org/10.1021/bi049480n
Nurminen ML, Sipola M et al (2000) Alpha-lactorphin lowers blood pressure measured by radiotelemetry in normotensive and spontaneously hypertensive rats. Life Sci 66:1535–1543
Parit R, Jayavel S (2021) Association of ACE inhibitors and angiotensin type II blockers with ACE2 overexpression in COVID-19 comorbidities: a pathway-based analytical study. Eur J Pharmacol 896:173899. https://doi.org/10.1016/j.ejphar.2021.173899
Patel P, Patel M, Pathak Y (2021) Current status of bioactive peptides in clinical studies. In: Bioactive Peptides. CRC Press, pp 403–423
Pihlanto A, Makine S (2013) Antihypertensive properties of plant protein derived peptides, bioactive food peptides in health and disease. In: Hernández-Ledesma B (ed) pp 145–182
Priyanto AD, Doerksen RJ, Chang CI, Sung WC, Widjanarko SB, Kusnadi J, Lin YC, Wang TC, Hsu JL (2015) Screening, discovery, and characterization of angiotensin-I converting enzyme inhibitory peptides derived from proteolytic hydrolysate of bitter melon seed proteins. J Proteomics 128:424–435. https://doi.org/10.1016/J.JPROT.2015.08.018
Rahimi M, Ghaffari SM, Salami M, Mousavy SJ, Niasari-Naslaji A, Jahanbani R, Yousefinejad S, Khalesi M, Moosavi-Movahedi A (2016) ACE- inhibitory and radical scavenging activities of bioactive peptides obtained from camel milk casein hydrolysis with proteinase K. Dairy Sci Technol 96:489–499. https://doi.org/10.1007/s13594-016-0283-4
Rao S, Su Y, Li J et al (2009) Design and expression of recombinant antihypertensive peptide multimer gene in Escherichia coli BL21. J Microb Biot 19:1620–1627
Rawendra RDS et al (2014) Isolation and characterization of a novel angiotensin-converting enzyme-inhibitory tripeptide from enzymatic hydrolysis of soft-shelled turtle (Pelodiscus sinensis) egg white: in vitro, in vivo, and in silico study. J Agric Food Chem 62:12178–12212
Roy F, Boye JI, Simpson BK (2010) Bioactive proteins and peptides in pulse crops: pea, chickpea and lentil. Food Res Int 43:432–442. https://doi.org/10.1016/J.FOODRES.2009.09.002
Ryan JT, Ross RP, Bolton D, Fitzgerald G (2011) Bioactive peptides from muscle sources: meat and fish. Nutrient 3:765–791
Samaei SP, Martini S, Tagliazucchi D, Gianotti A, Babini E (2021) Antioxidant and angiotensin I-converting enzyme (ACE) inhibitory peptides obtained from alcalase protein hydrolysate fractions of hemp (Cannabis sativa L.) bran. J Agric Food Chem 69:9220–9228. https://doi.org/10.1021/acs.jafc.1c01487
Schmidt T, Situ AJ, Ulmer TS (2016) Structural and thermodynamic basis of proline-induced transmembrane complex stabilization OPEN. Nat Publ Group. https://doi.org/10.1038/srep29809
Singh BP, Aluko RE, Hati S, Solanki D (2021) Bioactive peptides in the management of lifestyle-related diseases: current trends and future perspectives. Crit Rev Food Sci Nutr. https://doi.org/10.1080/10408398.2021.1877109
Sitanggang AB, Putri JE, Palupi NS, Hatzakis E, Syamsir E, Budijanto S (2021) Enzymatic preparation of bioactive peptides exhibiting ACE inhibitory activity from soybean and velvet bean: a systematic review. Molecules. https://doi.org/10.3390/molecules26133822
Suetsuna K, Osajima K (1986) The inhibitory activities against angiotensin I-converting enzyme of basic peptides originating from sardine and hair tail meat. Bull Jpn Soc Sci Fish 52:1981–1984
Sun S, Xu X, Sun X, Zhang X, Chen X, Xu N (2019) Preparation and identification of ACE inhibitory peptides from the marine Macroalga Ulva intestinalis. Mar Drugs 17:1–17. https://doi.org/10.3390/md17030179
Surma S, Romańczyk M, Witalińska-Łabuzek J, Czerniuk MR, Łabuzek K, Filipiak KJ (2021) Periodontitis, blood pressure, and the risk and control of arterial hypertension: epidemiological, clinical, and pathophysiological aspects-review of the literature and clinical trials. Infflamm Cardiovasc Dis. https://doi.org/10.1007/s11906-021-01140-x
Tomatsu M, Shimakage A, Shinbo M, Yamada S, Takahashi S (2013) Novel angiotensin I-converting enzyme inhibitory peptides derived from soya milk. Food Chem 136:612–616. https://doi.org/10.1016/j.foodchem.2012.08.080
Tzakos AG, Galanis AS, Spyroulias GA, Cordopatis P, Manessi-Zoupa E, Gerothanassis IP (2003) Structure-function discrimination of the N- and C- catalytic domains of human angiotensin-converting enzyme: implications for Cl- activation and peptide hydrolysis mechanisms. Protein Eng 16:993–1003. https://doi.org/10.1093/protein/gzg122
Vallabha VS, Tiku PK (2014) Antihypertensive peptides derived from soy protein by fermentation. Int J Pept Res Ther 20:161–168
Villegas JM et al (2014) Milk-derived angiotensin-I-converting enzyme-inhibitory peptides generated by Lactobacillus delbrueckii subsp. lactis CRL. Peptidomics 1:22–29
Wang J, Hu J, Cui J, Bai X, Du Y, Miyaguchi Y, Lin B (2008) Purification and identification of a ACE inhibitory peptide from oyster proteins hydrolysate and the antihypertensive effect of hydrolysate in spontaneously hypertensive rats. Food Chem 111:302–308. https://doi.org/10.1016/j.foodchem.2008.03.059
Wu J, Aluko RE, Nakai S (2006) Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure-activity relationship modeling of peptides containing 4–10 amino acid residues. QSAR Comb Sci 25:873–880. https://doi.org/10.1002/qsar.200630005
Xiang L, Qiu Z, Zhao R, Zheng Z, Qiao X (2021) Advancement and prospects of production, transport, functional activity and structure-activity relationship of food-derived angiotensin converting enzyme (ACE) inhibitory peptides. Crit Rev Food Sci Nutr. https://doi.org/10.1080/10408398.2021.1964433
Xie J, Chen X, Wu J, Zhang Y, Zhou Y, Zhang L, Tang Y, Wei D (2018) Antihypertensive effects, molecular docking study, and isothermal titration calorimetry assay of angiotensin I-converting enzyme inhibitory peptides from Chlorella vulgaris. J Agric Food Chem 66:1359–1368
Xu Z, Wu C, Sun-Waterhouse D, Zhao T, Waterhouse GI, Zhao M, Su G (2021) Identification of post-digestion angiotensin-I converting enzyme (ACE) inhibitory peptides from soybean protein Isolate: their production conditions and in silico molecular docking with ACE. Food Chem 345:128855
Xue L, Yin R, Howell K, Zhang P (2021) Activity and bioavailability of food protein-derived angiotensin-I-converting enzyme-inhibitory peptides. Comp Rev Food Sci Food Safe. https://doi.org/10.1111/1541-4337.12711
Yamamoto N (1997) Antihypertensive peptides derived from food proteins. Biopolym Peptide Sci Sect 43:129–134
Yates CJ, Masuyer G, Schwager SL, Akif M, Sturrock ED, Acharya KR (2014) Molecular and thermodynamic mechanisms of the chloride-dependent human angiotensin-I-converting enzyme (ACE). J Biol Chem 289:1798–1814. https://doi.org/10.1074/jbc.M113.512335
Yu J, Mikiashvili N, Bonku R, Smith IN (2021) Allergenicity, antioxidant activity and ACE-inhibitory activity of protease hydrolyzed peanut flour. Food Chem 360:129992. https://doi.org/10.1016/J.FOODCHEM.2021.129992
Author information
Authors and Affiliations
Contributions
OOA writing—original draft preparation, formal analysis, and final editing. PGY investigation, writing—original draft preparation, and formal analysis. CYG conceptualization, supervision, and project administration.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants and/or animals
This research does not involve the use of human participants or animals.
Informed consent
No informed consent is required for this study.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Olalere, O.A., Yap, PG. & Gan, CY. Comprehensive review on some food-derived bioactive peptides with anti-hypertension therapeutic potential for angiotensin-converting enzyme (ACE) inhibition. J Proteins Proteom 14, 129–161 (2023). https://doi.org/10.1007/s42485-023-00106-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-023-00106-8