Abstract
Cancer deaths and other dangerous symptoms associated with it make the search for therapies to combat it a necessity. The use of drugs has been implicated in some debilitating side effects, this makes scientists to look for safer, cheaper, and more effective alternatives. One of such ways is the use of plants/fruits that are rich in bioactive components. Citrullus lanatus is one plant that is grown in almost every part of the world and rich in bioactive secondary metabolites. Kirsten rat sarcoma virus is a very popular oncogene in human tumours that has been targeted in the search for anticancer drugs. This work therefore screened the phytochemicals in Citrullus lanatus against a KRAS receptor via molecular docking. The hit compounds and the co-crystallized ligand of the receptor were subjected to molecular dynamic simulation to determine their stability, modelled and optimized at the DFT/B3LYP/6-311G(d,p) level of theory and also screened for their drug-likeness and ADMET properties. Extra-precision docking revealed that ( ±)-Taxiforin (-7.23 kcalmol−1), (E)-Coniferin (-6.31 kcalmol−1) and Isorhamnetin (-5.71 kcalmol−1) displayed better docking scores than the reference compound (-5.07 kcalmol−1). Molecular dynamics simulation reveals the stability of KRAS-ligand complexes. The molecules have sites to interact with biological systems, as revealed by the DFT results. Pharmacokinetics and drug-likeness revealed that the molecules are promising and safe. Citrullus lanatus should be encouraged for consumption, while the hit compounds should be subjected to further pre-clinical and clinical investigations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The second greatest cause of death worldwide is cancer. It is a complex disease linked to several cellular and genetic abnormalities [1]. The multifactorial nature of this disease has made its treatment an uphill task, despite this, the therapy of cancer has advanced. One of such advances is chemotherapy, chemotherapy drugs are powerful enough to prevent cancer from spreading, thereby making it less severe. Most of these chemotherapeutic medicines do, however, have disturbing side effects that are partially attributable to off-target administration and/or poor absorption, which shorten the plasma half-life and decrease their efficacy [2]. Another challenge is that the current drugs are expensive for patients from low income and developing countries like Nigeria.
However, in recent years it has been clear that several oncogenes are constitutively active in cancer because of genetic changes such as mutations, proliferation, rearrangement, etc. Oncogene addiction is the term for how frequently these mutant oncogenes’ roles in the promotion of cancer development are vital. The development of specific anticancer medicines has been greatly facilitated by this insight. The most effective illustration is protein kinase inhibitors, which have shown clinical advantages in a variety of cancer [3]. Kirsten rat sarcoma virus (KRAS) is the most popular oncogene in human tumours [4, 5], along with other RAS isoforms [6]. Despite decades of research into RAS targeted medicines, there are still no clinically effective RAS inhibitors. Noteworthy is the encouraging progress that has been made in recent years in researching the therapeutic potential of RAS inhibition [7].
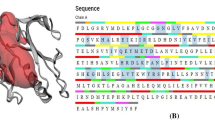
KRAS, also known as the RAS-like GTPases is a member of the RAS superfamily. There are four domains in the KRAS protein. The N-terminal first domain is the same in all three RAS variants, while the second domain has a significantly lower level of sequence identity. The KRAS protein's signalling activity depends on both sections, which together make up the G-domain [8]. The hypothesised downstream effectors and GTPase-activating proteins interact with the KRAS protein's GTP-binding pocket, a part of the protein's G-domain. Additionally, the C-terminal hypervariable region of the KRAS protein controls plasma membrane anchoring and posttranslational modifications. This area is crucial in controlling the bioactivity of the RAS protein. KRAS protein changes from an inactive to an active version owing to the binding of both GTP (guanosine triphosphate) and GDP (guanosine diphosphate), respectively [9]. Under normal physiological and biochemical circumstances, guanine nucleotide exchange factors such as GTPase-activating proteins or Son of Sevenless control the transition between these two states through a variety of mechanisms that include catalysing the swapping of GDP for GTP, increasing inherent GTPase activity, or speeding up RAS-mediated GTP hydrolysis. The same applies to KRAS, which is primarily GDP-bound. KRAS undergoes conformational changes upon binding to GTP, which are known to have two main effects: 1) changing KRAS contacts with GAPs, which increase the RAS protein's GTPase activity by about 100,000-fold; and 2) affecting KRAS connections with GEFs and encouraging the release of GTP3 [10].
Additionally, KRAS accounts for 86% of all RAS mutations and is the isoform of the RAS protein that is most often altered. KRAS-4B is the predominant isoform in human malignancies. It is found in 30%—40% of colon cancers, 90% of pancreatic cancers and 15% to 20% of lung cancers [4]. Additionally, it can be found in breast cancer, myeloid leukaemia, liver cancer, endometrial cancer, bladder cancer, cervical cancer and malignancies of the biliary tract [8]. The 12, 13, or 61 codons in the KRAS gene tend to have the most mutations. Codons 63, 117, 119, and 146 may also experience the changes, albeit less frequently [8]. In more detail, the glycine 12 (G12) mutation interferes with GAP binding and GAP-stimulated GTP hydrolysis, causing RAS activation. Changes at position 13 reduce GAP binding and hydrolysis by interfering sterically with the arginine. Contrarily, glutamine 61 plays a direct part in catalysis by situating the water molecule that is attacking and aiding in stabilising the hydrolysis reaction's transition state [11]. Due to decreased GTPase activity, the nucleotide state of KRAS depends heavily on relative nucleotide affinity and concentration. Due to the advantage this offers GTP over GDP and the rise in the percentage of active GTP-bound RAS, the activated state builds up. Additionally, it has been observed that mutations at residues 12, 13, and 61 reduce, albeit to varying degrees, the affinity of RAF for the RAS-binding domain [12]. However, for more than three decades, it was difficult to produce effective medications to stop RAS-driven oncogenesis, and RAS was thought to be "undruggable." However, the FDA recently approved sotorasib [12]. There are several KRAS inhibitory small compounds undergoing preclinical or clinical trials, and the results are extremely promising.
The use of plant-derived bioactive chemicals in the treatment of human cancer via a variety of pathways has been documented in literature. Plant sources have always been taken into consideration while searching for novel treatments for a variety of human diseases.
African native Citrullus lanatus, also known as watermelon, is a vine-like flowering plant in the Cucurbitaceae family [13]. C. lanatus has been extensively researched for the treatment of numerous human conditions such as cancer [14], diabetes [15], infectious diseases [16], and cardiovascular diseases [17] among others due to its wide variety of structurally complex phytochemicals. Its therapeutic activity has primarily been attributed to the presence of phenolic antioxidants and specific phytochemicals, such as carotenoid lycopene, which are responsible for this activity [15].
Due to its potent oxygen-scavenging and antioxidant characteristics, lycopene, a red carotenoid pigment present in watermelon, has reportedly been linked to a decreased chance of developing some cancers, primarily of the prostate [18]. According to Wink et al., 2001 it can display both pro- and antioxidant activities [19]. Consequently, it is also recognised that watermelon's β-carotene content may have an anticancer effect [20]. Despite the widely reported health benefit and anticancer properties of C. lanatus fruit, its role in cancer treatment has not been extensively established. Herein, this study seeks to evaluate the bioactive compounds of watermelon as potential noncovalent therapeutic agents against KRAS via computational approach.
2 Materials and Methods
2.1 Ligand Identification and Preparation
The bioactive compounds of plant of interest “Citrullus lanatus” were identified from the watermelon molecules online database (https://watermelon.naturalproducts.net). The compounds (18 of them) were downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and presented (Table 1).
2.2 Protein Retrieval
The crystallographic structure of the human KRAS protein (target protein), 6GJ5 (resolution = 1.50 Å) was retrieved in its 3-diemtional form from the RCSB protein Data Bank (https://www.rcsb.org/). Its co-crystallized ligand (F0N) was downloaded and used as the reference compound.
2.3 Molecular Docking
The identified bioactive compounds of Citrullus lanatus were docked into the binding pocket (x = -21.53, y = -26.76, z = -16.42) of the target protein (KRAS; 6GJ5) after the preparation of the protein and ligands by engaging the Protein preparation Wizard and LigPrep tools respectively, in the Maestro/Schrodinger suite 2022–3 [21].
2.4 MMGBSA Calculation
A quantum mechanics algorithm, MMGBSA helps in eradicating false-positive docking results gotten from molecular docking. It was used to obtain the binding free energy of the docked complexes using the Prime module in Maestro interface [22, 23] using the equation below (Eq. 1).
2.5 Molecular Dynamic Simulation
The complexes of the molecules with higher docking scores than F0N were solvated with the TIP3P water model using the System Builder on Desmond package in Maestro 13.1 using default parameters. This explicit TIP3P model has been used successfully in solvating complexes [24, 25] with the ionic strength set using sodium chloride atoms (0.15 M) before MD simulation was run for 200 ns. The NPT ensemble was employed at 300 K and pressure of 1.013 bar while the isotropic coupling and coulombic cut-off was set at 9.0 Å while the simulation was allowed to run after the systems relaxed. The data generated were analyzed.
2.6 Quantum Chemical Calculations
Density functional theory (DFT) calculation of the best three (3) compounds were done using Gaussian 09 software [26]. The compounds.sdf structures were downloaded from PUBCHEM, viewed on Gaussview was used to verify the downloaded structure and to ensure that all the molecules were well modelled, after which frequency calculations were performed to ensure no imaginary frequencies are in the molecules before geometry optimization, using the B3LYP hybrid functional [27] and 6–311 + + G(d,p) basis set. The molecular parameters computed are frontier molecular orbitals energy (FMOs) which includes the EHOMO and ELUMO. Other important molecular properties such as described earlier [28] were calculated according to Koopman’s theorem [29, 30] using Eq. (2–11).
2.7 Pharmacokinetics and Drug-Likeness Prediction
The bioactive compounds with higher binding affinity than F0N were subjected to pharmacokinetics screening on admetSAR server (http://lmmd.ecust.edu.cn/) for absorption, distribution, metabolism, excretion and toxicity prediction [28, 31]. The compounds were further screened for drug-likeness using the SwissADME server (http://www.swissadme.ch) [32].
3 Results and Discussion
3.1 Molecular Docking
To determine the binding affinity and interaction pattern of the hit compounds against the KRAS target protein, a molecular docking study and analysis were performed on the 18 identified compounds and F0N (the co-crystallized ligand which was also used as the standard drug) using Schrodinger suite 2022–3. According to the results (Table 2), 3 of the bioactive compounds (( ±)-Taxifolin, (E)-Coniferin and Isorhamnetin) had higher binding scores than F0N, based on the XP docking scores. The binding score ranged from -7.23 to -0.91 kcal/mol. ( ±)-Taxifolin had the highest binding affinity (-7.23 kcal/mol) while Spermine had the lowest binding affinity (-0.91 kcal/mol). The standard inhibitor (F0N) had a binding affinity of -5.07 kcal/mol. The order of the molecule according to their binding scores was ranked using the XP score on Table 2.
Furthermore, the visualization of the target protein with the co-crystalized ligand revealed the presence of the following vital amino acid restudies at the binding pocket of KRAS; LYS 5, LEU 6, VAL 7, GLU 37, SER 39, ASP 54, LEU 56, MET 67, GLN 70, TYR 71, THR 74 and GLY 75. These amino acids are important in the intramolecular and intermolecular complex interactions such as including pi-pi stacking, hydrophobic, hydrogen bonding and so on. The 2D interactions of the screened ligands-target complex (Table 3) showed that the docked ligands interact with several of the vital amino acids of KRAS binding pocket with majority having a good number of hydrogen bonds which support the efficacy as potential therapeutic agents by donating or accepting hydrogen atom through the formation of hydrogen bond. ( ±)-Taxifolin interacted via hydrogen bonding with LEU 6, GLU 37, ASP 54 and GLN 70 and pi-cation interaction with LYS 5. (E)-Coniferin interacted via hydrogen bonding with LEU 6, ASP 54, GLN 70 and THR 74. Isorhamnetin interacted via hydrogen bonding with LEU 6, ASP 54 and GLN 70 while also bonding with LYS 5 via pi-cation interaction. The co-ligand used as a reference (F0N) interacted with the amino acid residues via hydrogen bonding with ASP 54 and THR 74.
3.2 MMGBSA
Following the docking analysis, the ∆Gbind (Table 2) was used to calculate the binding energy of the screened bioactive compounds. Several researchers rely on MMGBSA result as a reliable method for determining the binding position of docked complexes [33]. When compared to the standard ligand, F0N (-49.88 kcalmol−1), other components have lower binding energy, with Kauralexin A2 having the least (-2.26 kcalmol−1). The top three bioactive components, ( ±)-Taxifolin (-35.13 kcalmol−1), (E)-Coniferin (45.85 kcalmol−1) and Isorhamnetin (-29.77 kcalmol−1) also have decent binding energies. As a result, the ∆Gbind results point to the fact that Citrullus lanatus compounds' have good promise as potential KRAS inhibitors.
3.3 Molecular Dynamic Simulation
The trajectories and coordinates generated from the 200 ns molecular dynamics were analyzed to provide insights into the behavior of the compounds within the binding pocket of KRAS and their resultant impact on the conformational dynamics of the protein. The behavior of a small molecule within its target binding pocket is crucial to understanding the binding affinity and structural complementarity which is underscored by the strength of their interactions [34] and possible mutational effects on the protein [35]. The behavior of the compounds was determined through estimating the root mean square deviations (RMSD) of the compounds over the simulation period and their surface area available for solvent interaction through SASA [36]. The compounds presented average RMSD values of 1.96 ± 0.85 Å, 2.06 ± 0.76 Å, 0.91 ± 0.50 Å and 0.52 ± 0.17 Å for E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference compound respectively. All systems showed RMSD values < 2.5 Å, which is in line with acceptable deviations [37]. As observed in Fig. 1B, the reference compound (F0N) exhibited the highest stability followed by Taxifolin, Isorhamnetin and E-Coniferin. Isorhamnetin became relatively stable after 50 ns after undergoing significant positional change due to the flexibility of both compound and protein which is not catered for in molecular docking. This change possibly resulted in conformational complementarity between Isorhamnetin and KRAS thus inducing the stability after 50 ns. E-Coniferin, however, exhibited consistent fluctuations relative to the other compounds during the simulation period. Generally, the compounds stabilised within the allosteric binding pocket of KRAS with RMSD values less then 2.5 Å. The solvent accessibility surface area of the compounds available for solvent interaction also showed the compounds resided within the hydrophobic core of the protein over the simulation period. The compounds exhibited average SASA values of 212.89 ± 54.03 A2, 135.73 ± 19.18 A2, 131.43 ± 15.54 A2, and 152.11 ± 20.28 A2 for E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference compound (F0N) respectively. As observed in Fig. 1C E-coniferin which had hitherto showed relatively high instability, showed the highest surface area available for solvent interaction suggesting its binding did not penetrate deeper into the hydrophobic pocket compared to the other compounds. Isorhamnetin and ( ±)-Taxifolin were least exposed to solvent interaction than the reference compound.
Comparative plots of the deviations of the compounds from their initial position over the simulation period (RMSD) and the solvent accessibility surface area of the compound. A Shows the solvent surface of KRAS. Insert (A1) shows the compounds within the allosteric binding pocket. B RMDS plots of the compounds over 200 ns period. C SASA plots of the compounds over 200 ns simulation period
The interactions that condition the behaviour of these compounds were monitored over the simulation period through snapshots to ascertain any changes in interaction types that occurred during the docking process. Interestingly, the interaction types remained the same as observed in the Fig. 2 and Table 3. Collectively, the behaviour of the compounds suggest they possess the potential to bind firmly to KRAS. Probing further, the impact of the compounds on the conformation dynamics of KRAS were determined through the RMSD, root mean square fluctuations (RMSF), radius of gyration (RoG) and SASA.
Proteins often adjust to external stimuli including the binding of small molecules through conformational changes. Disrupting the native conformation of a protein beyond a certain threshold interferes with the basal functionalities of the protein [38]. As such, we determined the impact of the binding of the compounds on the conformational changes of the protein over the 200 ns simulation period. Firstly, the RMSD which is informative of the stability of the protein was determined through computing the deviations of the C-α atoms from the starting positions. Average RMSD values of 2.30 ± 0.62 Å, 1.80 ± 0.47 Å, 1.93 ± 0.58 Å, 1.90 ± 0.44 Å, and 1.78 ± 0.51 Å were computed for the unbound KRAS (Apo), E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference (F0N) complexes respectively. As observed in the graphical format presented in Fig. 3A, the binding of the compounds induced conformational stability on the KRAS protein (reduced RMSD values compared to the Apo). This observation suggests the compounds have the propensity to interfere with the basal conformation of the protein. The reference compound (F0N) was however observed to induce the highest stability. The RMSF metric which reflects the flexibility of the protein showed average values of 1.27 ± 1.03 Å, 0.94 ± 0.71 Å, 1.07 ± 0.86 Å, 1.19 ± 0.88 Å and 1.05 ± 0.73 Å for Apo, E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference complexes respectively (Fig. 3B). The higher the figure the more flexible the protein and vice versa. As presented the compounds’ binding resulted in reduction in KRAS flexibility as evidenced by the higher unbound KRAS values (1.27 ± 1.03 Å). The RoG values which determine the compactness of the protein corroborated the reduced flexibility of KRAS by the compounds wherein they exhibited 15.51 ± 0.15 Å, 15.31 ± 0.11 Å, 15.36 ± 0.12 Å, 15.37 ± 0.10 Å, and 15.32 ± 0.13 Å for Apo, E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference complexes respectively (Fig. 3C). Reduction in the radius of gyration of the atoms of the systems indicated increase in compactness and vice versa. This thus suggest that, aside E-coniferin, the reference compound induced more compactness than the other compounds. The solvent accessibility surface area (SASA) analysis of the protein also showed similar trend wherein the compounds induced a reduction in the surface area of the protein available for solvent interaction. Average SASA values of 8561.91 ± 319.70 A2, 8434.56 ± 285.70 A2, 8451.33 ± 339.91 A2, 8254.77 ± 397.76 A2and 8175.47 ± 278.62 A2 were computed for Apo, E-coniferin, Isorhamnetin, ( ±)-Taxifolin and the reference complexes respectively (Fig. 3D). The compounds’ ability to influence the conformational dynamics of KRAS suggests that they have the potentials of inhibiting its activity upon modification and optimizations.
Comparative plots of the compounds in complex with KRAS. A Shows the RMSD plots of the C-α atoms of KRAS in complex with the compounds. B Shows plots of the residual fluctuations of KRAS in complex with the compounds. C Shows the RoG plots of the C-α atoms of KRAS over the simulation period. D Shows SASA plots over the simulation period
3.4 Quantum Chemical Calculations
DFT is important in elucidating the electronic structure of molecules. In the absence of protein–ligand interactions, electronic structures from DFT could explain the potential interactions of ligands with biological cells [39, 40]. The results of all the electronic properties computed for the leads compounds (( ±)-Taxifolin, (E)-Coniferin and Isorhamnetin) and F0N are shown in Table 4. Their corresponding HOMO and LUMO maps are shown in Table 5, while their electrostatic potential maps are shown in Figs. 4, 5, 6, and 7.
The frontier molecular orbitals energies (EHOMO and ELUMO) play important part in describing chemical reactions [41, 42]. This has been used for explaining the chemically reactivity and stability of drug to target enzyme [43]. From Table 4, isorhamnetin had the higher value of EHOMO (-4.55 eV), therefore, proposing it as the best electron donating compound to among others. ( ±)-Taxifolin had the lowest HOMO value (-6.31 eV), making it the least ready to donate electron. This is also corroborated by their ionization potentials (I). Isorhamnetin needs the lowest I value (4.55 eV) to donate electron, making electron more ready to leave its orbitals compared to other molecules while higher energy would be required to remove electrons from ( ±)-Taxifolin (6.31 eV). The electron donating ability is in the order; Isorhamnetin > F0N > (E)-coniferin > ( ±)-Taxifolin. ( ±)-Taxifolin had the lowest ELUMO (-2.09 eV), indicating it as the best electron acceptor (as seen in its A value).
The energy gap (Eg) of a molecule helps in predicting the stability and reactivity of molecules. Lower energy gap enhances the intra/inter molecular charge movement between HOMO and LUMO thereby increase the reactivity of a molecule and vice versa. In most cases where a molecule had a low EHOMO, its energy gap could be stabilized by its LUMO value. Generally, the hit molecules have lower energy gap than F0N, save for (E)-coniferin. The order of the energy gap is (E)-coniferin (4.87 eV) > F0N (4.79 eV) > ( ±)-Taxifolin (4.22 eV) > Isorhamnetin (2.71 eV). The energy gaps recorded here are within the energy gaps reported for bioactive compounds [44, 45]. Chemical hardness (η) and softness (δ) are consequences of the energy gap. The chemical hardness of the molecule follows (E)-coniferin (2.44 eV) > F0N (2.39 eV) > ( ±)-Taxifolin (2.11 eV) > Isorhamnetin (1.36 eV) while softness follows the reverse trend, isorhamnetin is the softest of all the molecules. Electronegativity (χ) is an important descriptor that explicates the potential of an atom/molecule or functional group to draw electron toward itself [46]. ( ±)-Taxifolin had the highest electronegative value (4.20 eV) while Isorhamnetin had the lowest (3.19 eV).
The electrophilicity index (ω) measures the ability of a molecule to accept electrons when placed in contact with a nucleophile [30, 47]. In general, in all the electrophilic terms in Table 4, ( ±)-Taxifolin had the highest electrophilicities, followed by Isorhamnetin > (E)-coniferin > F0N. This is indicative that the molecules are better electrophiles than F0N. The descriptors values here show that these molecules allow transfer of electrons between their molecular orbitals and as a result boosting their bioactivity.
The optimized structure, HOMO and LUMO surfaces of all the compounds are shown in Table 5. ( ±)-Taxifolin displayed its HOMO across the entire 3,4-dihydrophenyl ring (including the hydroxyl groups on the ring) while its LUMO is on the other side of the molecule (3,5,7-trihydroxy-2,3-dihydrochromen-4-one). (E)-coniferin had both its HOMO and LUMO map in the same region (methoxyphenoxy ring), only that the HOMO added the methoxy group to itself. Isorhamnetin had its HOMO on the methoxyphenyl ring while the LUMO is spread across almost all the entire molecule, save for the methoxy group. The HOMO of F0N is on the indole ring and the carbonyl and nitrogen atom on dihydroisoindol-1-one ring while its LUMO is on the entire dihydroisoindol-1-one ring.
3.5 Molecular Electrostatic Potential (MEP) Surface Analysis
MEP allows the visualization of the electrostatic potential at the surface of a molecule. It is used for the understanding of the distribution of charges in a molecule and for determining sites for nucleophilic and electrophilic attack reactivity as well as hydrogen-bonding interactions. MEP is represented by a color scheme, red and yellow correspond to regions of high electron density (Electrophilic attack region) and blue corresponding to regions of low electron density (Nucleophilic attack region), with green corresponding to the neutral region. The results of the MEP analysis are shown in Figs. 4, 5, 6, and 7. MEP helps in determining the reactivity of molecules to either positively and/or negatively charged reactants [48].
( ±)-Taxifolin sites for electrophilic attack on the dihydrochromen-4-one carbons and the carbonyl oxygen while the sites for nucleophilic attack are on the hydroxyl hydrogens and the hydroxyphenyl carbons (Fig. 4). (E)-coniferin sites for electrophilic attack on the methoxy and hydroxymethyl oxygen atoms and in the majority of the oxane ring while the site for nucleophilic attack are majorly the hydrogen atoms and the carbon atoms on methoxyphenoxy ring (Fig. 5). Isorhamnetin sites for electrophilic attack are on the carbon and hydrogen atoms in the entire molecule while its sites for nucleophilic attack are on the oxygen atoms in the entire molecule (Fig. 6). The sites for electrophilic attack of F0N are majorly on the carbonyl oxygen in the molecule while its sites for nucleophilic attack are on the nitrogen and hydrogens atoms on the pyrrolidine and indole rings (Fig. 7).
The ligands investigated here are largely electronegative (Figs. 4, 5, 6, and 7), except for F0N which shows more electropositive (blue part). This means that these compounds are likely to favour interactions from the electropositive part of the KRAS protein while F0N is likely to favour interactions from the major electronegative part of the protein [49]. The positive–negative sites of these compounds and the interactions between the opposite ends of the compounds and the protein as well as the free energy of binding of the compounds might be the major contributing factors to the compounds’ stability as seen from the MD simulation.
3.6 Drug Likeness
Oral bioavailability is an important factor in the development of drugs. Lipinski's rule of five is a useful practice for evaluating a molecule's drug-likeness characteristics [50]. Table 6 shows the Lipinski's parameter values for top 3 compounds from molecular docking and F0N. The molecular weights, numbers of H-bond donors (HBD) and H-bond acceptors (HBA) to/from H2O molecules in aqueous solution, the topological polar surface area (TPSA) and the logarithm of the partition coefficient (ilogP) values are reported. The hit compounds and reference compound have zero Lipinski violation.
3.7 Pharmacokinetics
The pharmacokinetics prediction was performed to determine how the hit compounds are absorbed, distributed, metabolised, and eliminated, as well as their potential toxicity. The evaluation results are shown in Table 7. The hit compounds of Citrullus lanatus, ( ±)-Taxiforin, (E)-Coniferin and Isorhamnetin demonstrated low absorption in the intestine via the Caco-2 permeability, which was likely due to their small molecular sizes. Except for (E)-Coniferin, the screen hit compounds demonstrated high intestinal absorption. Pharmacokinetics revealed that ( ±)-Taxiforin, (E)-Coniferin, and Isorhamnetin have no BBB, except for F0N. They, including F0N, are all non-substrates and/or inhibitors of P-glycoprotein (P-GB). These compounds are found primarily in the mitochondria.
Drug metabolism is an important property because it accounts for nearly 50% of drug excretion via Cytochrome P450 enzymes [51]. The therapeutic effects of a drug are highly dependent on metabolism [52, 53]. The most extensively studied CYP450 enzymes, CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4/5, metabolise approximately 90% of all drug molecules [54, 55]. All the compounds are non-inhibitors of CYP2D6, except for F0N. ( ±)-Taxiforin, Isorhamnetin and F0N inhibit CYP1A2 while (E)-Coniferin is a non-inhibitor of CYP1A2. ( ±)-Taxiforin and (E)-Coniferin are non-inhibitors of CYP2C19 while Isorhamnetin and F0N are. Only Isorhamnetin inhibits CYP2C9 while others do not. Only F0N is a CYP2D6 inhibitor. ( ±)-Taxiforin and Isorhamnetin are inhibitors of CYP3A4 while (E)-Coniferin and F0N are not. (E)-Coniferin and F0N are substrates of CYP2C9 while ( ±)-Taxiforin and Isorhamnetin are not. None of all the molecules are substrates of CYP2D6 while all of them are substrates of CYP3A4 except ( ±)-Taxiforin.
Because of toxicity, many drug candidates fail before reaching clinical trials [54]. The toxicity of Citrullus lanatus bioactive molecules was determined by testing them for carcinogenicity, hepatotoxicity, human ether-a-go-go inhibition and Ames mutagenesis. None of them is carcinogenic. Except for F0N, none of the compounds were shown to inhibit human ether-a-go-go. Except for ( ±)-Taxiforin, all other compounds were found to be toxic in Ames' test. All the compounds tested positive for high oral toxicity. All compounds were toxic to the liver (hepatotoxic), except for (E)-Coniferin. ( ±)-Taxiforin and F0N were toxic to the kidney and the respiratory system while (E)-Coniferin and Isorhamnetin are not. All compounds were toxic to the reproductive organs.
4 Conclusion
The use of plants for treating diverse diseases has become popular owing to their abundance and safety. The bioactive components of Citrullus lanatus were investigated for their potential anticancer activity via in silico approaches. Molecular docking revealed that ( ±)-Taxiforin, (E)-Coniferin and Isorhamnetin bind strongly to the active site of a KRAS receptor. Molecular dynamins simulation was used to ascertain their stability. The compounds were stable and have favourable interactions with the amino acid residues in the receptor. The reactivity descriptors and frontier molecular orbitals maps revealed that the molecules can interact with biological cells via different nucleophilic and electrophilic sites. Pharmacokinetics screening revealed that the molecules are safe, subject to further clinical and preclinical investigations.
Data Availability
All data generated or analysed during this study are included in this published article and are available from the corresponding author on reasonable request.
References
Kou F, Wu L, Ren X, Yang L (2020) Chromosome abnormalities: new insights into their clinical significance in cancer. Mol Ther- Oncolytics 17:562–570. https://doi.org/10.1016/j.omto.2020.05.010
Muhammad SA, Jaafaru MS, Rabiu S (2023) A meta-analysis on the effectiveness of extracellular vesicles as nanosystems for targeted delivery of anticancer drugs. Mol Pharm 20:1168–1188. https://doi.org/10.1021/acs.molpharmaceut.2c00878
Huang M, Shen A, Ding J, Geng M (2014) Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci 35:41–50. https://doi.org/10.1016/j.tips.2013.11.004
Huang L, Guo Z, Wang F, Fu L (2021) KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 6:1–20. https://doi.org/10.1038/s41392-021-00780-4
Issahaku AR, Salifu EY, Soliman MES (2023) Inside the cracked kernel: establishing the molecular basis of AMG510 and MRTX849 in destabilising KRASG12C mutant switch I and II in cancer treatment. J Biomol Struct Dyn 41:4890–4902. https://doi.org/10.1080/07391102.2022.2074141
Hobbs GA, Der CJ, Rossman KL (2016) RAS isoforms and mutations in cancer at a glance. J Cell Sci 129:1287–1292. https://doi.org/10.1242/jcs.182873
Liu P, Wang Y, Li X (2019) Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B 9:871–879. https://doi.org/10.1016/j.apsb.2019.03.002
Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D (2011) RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11:761–772. https://doi.org/10.1038/nrc3106
Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 130:385. https://doi.org/10.1016/j.cell.2007.07.001
Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic ras mutants. Sci 277:333–338. https://doi.org/10.1126/science.277.5324.333
Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nat 503:548–551. https://doi.org/10.1038/nature12796
FDA Approves LUMAKRASTM (Sotorasib), The First And Only Targeted Treatment For Patients With KRAS G12C-Mutated Locally Advanced Or Metastatic Non-Small Cell Lung Cancer (2018) https://www.amgen.com/newsroom/press-releases/2021/05/fda-approves-lumakras-sotorasib-the-first-and-only-targeted-treatment-for-patients-with-kras-g12cmutated-locally-advanced-or-metastatic-nonsmall-cell-lung-cancer
Mandel H, Levy N, Izkovitch S, Korman SH (2005) Elevated plasma citrulline and arginine due to consumption of Citrullus vulgaris (watermelon). J Inherit Metab Dis 28:467–472. https://doi.org/10.1007/s10545-005-0467-1
Itoh T, Ono A, Kawaguchi K, Teraoka S, Harada M, Sumi K, Ando M, Tsukamasa Y, Ninomiya M, Koketsu M, Hashizume T (2018) Phytol isolated from watermelon (Citrullus lanatus) sprouts induces cell death in human T-lymphoid cell line Jurkat cells via S-phase cell cycle arrest. Food Chem Toxicol 115:425–435. https://doi.org/10.1016/j.fct.2018.03.033
Ajiboye BO, Shonibare MT, Oyinloye BE (2020) Antidiabetic activity of watermelon (Citrullus lanatus) juice in alloxan-induced diabetic rats. J Diabetes Metab Disord 19:343–352. https://doi.org/10.1007/s40200-020-00515-2
Morimoto R, Yoshioka K, Nakayama M, Nagai E, Okuno Y, Nakashima A, Ogawa T, Suzuki K, Enomoto T, Isegawa Y (2021) Juice of Citrullus lanatus var. citroides (wild watermelon) inhibits the entry and propagation of influenza viruses in vitro and in vivo. Food Sci Nutr 9:544–552. https://doi.org/10.1002/fsn3.2023
Poduri A, Rateri DL, Saha SK, Saha S, Daugherty A (2008) Citrullus lanatus `Sentinel’ (Watermelon) extract reduces atherosclerosis in LDL receptor deficient mice. J Nutr Biochem 23:1–7. https://doi.org/10.1016/j.jnutbio.2012.05.011.Citrullus
Black HS, Boehm F, Edge R, Truscott TG (2020) The benefits and risks of certain dietary carotenoids that exhibit both anti-and pro-oxidative mechanisms—A comprehensive review. Antioxidants 9:1–31. https://doi.org/10.3390/antiox9030264
Knight J, Caseldine C, Boykoff MT (2010) Mechanisms of the antioxidant effects of nitric oxide. Antioxid Redox Signal 176:267–269
Barkur S, Bankapur A, Chidangil S, Mathur D (2017) Effect of infrared light on live blood cells: Role of β-carotene. J Photochem Photobiol B Biol 171:104–116. https://doi.org/10.1016/j.jphotobiol.2017.04.034
Schrödinger Release 2023–3: Maestro, Schrödinger (2023)
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins Struct Funct Bioinforma 55:351–367
Balogun TA, Ipinloju N, Abdullateef OT, Moses SI, Omoboyowa DA, James AC, Saibu OA, Akinyemi WF, Oni EA (2021) Computational Evaluation of Bioactive Compounds from Colocasia affinis Schott as a novel EGFR inhibitor for cancer treatment. Cancer Inform 20. https://doi.org/10.1177/11769351211049244
Florová P, Sklenovský P, Banáš P, Otyepka M (2010) Explicit water models affect the specific solvation and dynamics of unfolded peptides while the conformational behavior and flexibility of folded peptides remain intact. J Chem Ther Comput 6:3569–3579. https://doi.org/10.1021/ct1003687
Issahaku AR, Ibrahim MAA, Mukelabai N, Soliman MES (2023) Intermolecular and dynamic investigation of the mechanism of action of reldesemtiv on fast skeletal muscle troponin complex toward the treatment of impaired muscle function. Protein J 42:263–275. https://doi.org/10.1007/s10930-023-10091-y
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery J, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox D, Gaussian 09, Revision C.01, Gaussian, Inc. (2009)
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652. https://doi.org/10.1063/1.464913
Ipinloju N, Ibrahim A, da Costa RA, Adigun TB, Olubode SO, Abayomi KJ, Aiyelabegan AO, Esan TO, Muhammad SA, Oyeneyin OE (2023) Quantum evaluation and therapeutic activity of (E)-N-(4-methoxyphenyl)-2-(4-(3-oxo-3-phenylprop-1-en-1-yl) phenoxy)acetamide and its modified derivatives against EGFR and VEGFR-2 in the treatment of triple-negative cancer via in silico approach. J Mol Model 29:159. https://doi.org/10.1007/s00894-023-05543-2
Chattaraj PK, Chakraborty A, Giri S (2009) Net electrophilicity. J Phys Chem A 113:10068–10074. https://doi.org/10.1021/jp904674x
Pérez P, Domingo LR, Aizman A, Contreras R (2007) The electrophilicity index in organic chemistry in: theoretical aspects of chemical reactivity. Theor Asp Chem React 19:139–201
Yang H, Lou C, Sun L, Li L, Cai Y, Wang Z, Li W, Liu G, Tang Y (2019) AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 35:1067–1069. https://doi.org/10.1093/bioinformatics/bty707
Daina A, Michielin O, Zoete V (2017) SwissADME : a free web tool to evaluate pharmacokinetics, drug- likeness and medicinal chemistry friendliness of small molecules. Nat Publ Gr 7:42717. https://doi.org/10.1038/srep42717
Tripathi S, Muttineni R, Singh S (2013) Extra precision docking, free energy calculation and molecular dynamics simulation studies of CDK2 inhibitors. J Theor Biol 334:87–100. https://doi.org/10.1016/j.jtbi.2013.05.014
Du X, Li Y, Xia Y, Ai S, Liang J, Sang P, Ji X, Liu S (2016) Insights into protein-ligand interactions: mechanisms, models, and methods. Int J Mol Sci 17(2):144. https://doi.org/10.3390/ijms17020144
Issahaku AR, Aljoundi A, Soliman MES (2022) Establishing the mutational effect on the binding susceptibility of AMG510 to KRAS switch II binding pocket: computational insights. Informatics Med Unlocked 30:100952. https://doi.org/10.1016/j.imu.2022.100952
Durojaye OA, Okoro NO, Odiba AS, Nwanguma BC (2023) MasitinibL shows promise as a drug-like analog of masitinib that elicits comparable SARS-Cov-2 3CLpro inhibition with low kinase preference. Sci Rep 13:6972. https://doi.org/10.1038/s41598-023-33024-2
Barazorda-Ccahuana HL, Valencia DE, Aguilar-Pineda JA, Gómez B (2018) Art v 4 protein structure as a representative template for allergen profilins: homology modeling and molecular dynamics. ACS Omega 3:17254–17260. https://doi.org/10.1021/acsomega.8b02288
Liu H, Dastidar SG, Lei H, Zhang W, Lee MC, Duan Y (2008) Conformational changes in protein function BT- molecular modeling of proteins. Andreas Kukol Ed. Totowa, NJ: Humana Press. pp. 258–275
Ramalingam A, Kuppusamy M, Sambandam S, Medimagh M, Oyeneyin OE, Shanmugasundaram A, Issaoui N, Ojo ND (2022) Synthesis, spectroscopic, topological, hirshfeld surface analysis, and anti-covid-19 molecular docking investigation of isopropyl 1-benzoyl-4-(benzoyloxy)-2,6-diphenyl-1,2,5,6-tetrahydropyridine-3-carboxylate. Heliyon 8:e10831. https://doi.org/10.1016/j.heliyon.2022.e10831
Omoboyowa DA, Singh G, Fatoki JO, Oyeneyin OE (2022) Computational investigation of phytochemicals from Abrus precatorius seeds as modulators of peroxisome proliferator-activated receptor gamma (PPARγ). J Biomol Struct Dyn 41:5568–5582. https://doi.org/10.1080/07391102.2022.2091657
Ojo ND, Krause RW, Obi-Egbedi NO (2020) Electronic and nonlinear optical properties of 3-(((2-substituted-4-nitrophenyl)imino) methyl)phenol. Comput Theor Chem 1192:113050. https://doi.org/10.1016/j.comptc.2020.113050
Kumar A, Sambandam S, Ramalingam A, Krishnamoorthy R, Arumugam D, Oyeneyin OE (2022) Synthesis, molecular docking of 3-(2-chloroethyl)-2,6-diphenylpiperidin-4-one: Hirshfeld surface, spectroscopic and DFT based analyses. J Mol Struct 1262:132993. https://doi.org/10.1016/j.molstruc.2022.132993
Oderinlo OO, Iwegbulam CG, Ekweli OA, Alawode TT, Oyeneyin OE (2022) Acridone alkaloids: in-silico investigation against SARS-CoV-2 main protease. Chem Africa 5:1441–1450. https://doi.org/10.1007/s42250-022-00440-2
Houngue MTAK, Doco RC, Kpotin GA, Kuevi UA, Simplice K, Wilfried K, Atohoun YGS, Mensah JB (2017) DFT study of chemical reactivity of free radicals ABTS° + and DPPH° by Myricetin, Quercetin and Kaempferol. World Sci News 90:177–188
Kamaraj RR (2022) Synthesis, Crystal Structure and Theoretical Investigations of (3-(2-Chlorophenyl)-5-Tosyl- 1,3,3a,4,5,9b-Hexahydroisoxazolo[4,3-c]Quinolin-3a- yl)Methanamine. Biointerface Res Appl Chem 12:8394–8405
Oyeneyin OE, Ojo ND, Ipinloju N, James AC, Agbaffa EB (2022) Investigation of corrosion inhibition potentials of some aminopyridine Schiff bases using density functional theory and monte Carlo simulation. Chem Africa 5:319–332. https://doi.org/10.1007/s42250-021-00304-1
Chattaraj PK, Sarkar U, Roy DR (2006) Electrophilicity index. Chem Rev 106:2065–2091. https://doi.org/10.1021/cr040109f
Holman JB, Shi Z, Fadahunsi AA, Li C, Ding W (2023) Advances on microfluidic paper-based electroanalytical devices. Biotechnol Adv 63:108093
Dorujaye OA, Ejaz U, Uzoeto HO, Fadahunsi AA, Opabunmi AO, Ekpo DE, Sedzro DM, Idris MO (2023) CSC01 shows promise as a potential inhibitor of the oncogenic G13D mutant of KRAS: an in silico approach. Amino Acids 55:1745–1764. https://doi.org/10.1007/s00726-023-03304-2
Lipinski CA (2016) Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv Drug Deliv Rev 101:34–41. https://doi.org/10.1016/j.addr.2016.04.029
Song Y, Li C, Liu G, Liu R, Chen Y, Li W, Cao Z, Zhao B (2021) Drug-Metabolizing cytochrome P450 enzymes have multifarious influences on treatment outcomes. Clin Pharmacokinet 60:585–601. https://doi.org/10.1007/s40262-021-01001-5
Kazmi SR, Jun R, Yu M, Jung C, Na D (2019) In silico approaches and tools for the prediction of drug metabolism and fate: a review. Comput Biol Med 106:54–64. https://doi.org/10.1016/j.compbiomed.2019.01.008
Kazmi F, Sensenhauser C, Greway T (2019) Characterization of the in vitro inhibitory potential of the oligonucleotide imetelstat on human cytochrome P450 enzymes with predictions of in vivo drug-drug interactions. Drug Metab Dispos 47:9–14. https://doi.org/10.1124/dmd.118.084103
Jia Y, Li B, Yang Z, Li F, Zhao Z, Wei C, Yang X, Jin Q, Liu D, Wei X, Yost J, Lund H, Tang J, Robinson KA (2023) Trends of randomized clinical trials citing prior systematic reviews, 2007–2021. JAMA Netw Open 6:E234219. https://doi.org/10.1001/jamanetworkopen.2023.4219
Fatunde OA, Brown S (2020) The role of CYP450 drug metabolism in precision cardio-oncology enzyme. Int J Mol Sci 21:604
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Informed Consent
Not applicable.
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oyeneyin, O.E., Ipinloju, N., da Costa, R.A. et al. In Silico Evaluation of Bioactive Compounds of Citrullus lanatus as Potential Noncovalent KRAS Inhibitors in the Treatment of Human Cancer. Chemistry Africa 7, 2553–2567 (2024). https://doi.org/10.1007/s42250-024-00946-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-024-00946-x