Abstract
Bacterial canker disease caused by Pseudomonas syringae pv. syringae is one of the major limiting factors in the growing and productivity of Prunus species in Iran. A total of 293 bacterial strains were purified from the surface and internal tissues of aerial parts of almond (Prunus dulcis) and apricot (Prunus armenica) trees in East Azerbaijan province, Iran. Based on 16 S rRNA gene sequencing of selected 113 strains, these strains belong to 15 different genera with Pseudomonas, Pantoea, and Lysinibacillus being most abundant. Most genera included strains that were either isolated from both the surface (epiphytes) and internal tissues (endophytes). However, strains of Rouxiella, Escherichia, and Curtobacterium were only isolated from internal tissues and strains of Arthrobacter, Massilia, Microbacterium, Paenibacillus and Kocuria were only isolated from the surface. Eighteen of the strains showed antagonistic activity under in vitro conditions against Pseudomonas syringae pv. syringae Pss-170 strain, the causal agent of apricot canker disease. Most of the antagonistic strains belonged to Pseudomonas fluorescens, as confirmed by sequencing a fragment of the citrate synthase (cts) gene. All antagonistic strains were evaluated for their ability to produce auxin, gibberellin, siderophore, protease, ACC-deaminase, and hydrogen cyanide, as well as phosphate solubilization. Each strain was found to have three or more properties related to plant growth promotion. This study revealed plant growth promoting and biocontrol properties of bacterial strains isolated from almond and apricot trees, which can be further tested for their ability to control bacterial canker disease in the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants live in association with a diverse array of microorganisms, especially bacteria, on leaf surfaces, referred to as the phyllosphere or phylloplane. Bacteria living epiphytically on healthy host plant species can develop large populations with their taxonomic composition depending both on the plant genotype and on environmental factors (Thapa et al. 2017).
Endophytic bacteria have been defined as bacteria living inside plants for at least part of their life cycle, interacting with cells of the host, taking up secreted metabolites, and releasing plant-growth-promoting (PGP) compounds without causing negative effects on their host (Schulz and Boyle 2006).
Many studies have shown that endophytic bacteria can have the capacity to control phytopathogens via production of compounds such as antibiotics, siderophores, and enzymes, and enhance plant growth through nitrogen fixation and protection of plants from a series of abiotic stresses including drought, low temperature, and salinity (Ali et al. 2014; Bent and Chanway 1998; Sheibani-Tezerji et al. 2015; Subramanian et al. 2015).
Almond and apricot are one of the most important cropped and consumed fruits in the world, including Iran. Few studies so far have investigated epiphytic and endophytic bacteria of the aerial parts of almond and apricot for their biocontrol and plant growth promoting potential. Such bacteria could be useful to control bacterial canker disease of stone fruit. This disease is one of the most destructive diseases of Prunus species including plums, cherries, peaches, nectarines, apricots, and almonds (Wenneker 2013; Popović et al., 2021). The disease can be caused by either Pseudomonas syringae pv. syringae (Pss), Ps pv. morsprunorum, Ps pv. avii, or Ps pv. persicae. Pseudomonas syringae pv. syringae belongs to genomospecies I (Young 1991) and is unique in its ability to cause disease in over 180 species belonging to both mono- and di-cotyledonous plants including fruit trees, vegetables, ornamentals, and other annual and perennial species (Bradbury 1986; Gardan et al. 1999; Young et al. 1996). Bacterial canker of stone fruit trees caused by Pss is also known as twig blight, blossom blight, gummosis, dieback and spur blight, has a worldwide distribution with causing important economic losses (Kennelly et al. 2007; Kotan and Sahin 2002; Vicente et al. 2004; Wenneker et al. 2012). Pseudomonas syringae pv. syringae infections on stone fruit trees usually start from blossoms, where the pathogen starts to colonize and then reaches a large population size and from where bacteria enter into plant host tissues. When the infection progresses, blossom infections lead to wood invasion and canker formation. Dormant buds are an overwintering site for the bacterial canker pathogens. The ability of Pss to colonize host trees both epiphytically and endophytically limits effective disease management. Also, the absence of effective and specific chemical or biological control measures and poor knowledge of host resistance have made it almost impossible to control bacterial canker disease (Kennelly et al. 2007).
The aim of the present study was to characterize endophytic and epiphytic bacteria associated with aerial parts (e.g. stem, bud, and blossom) of apparently healthy and diseased almond and apricot trees in East Azerbaijan province, Iran, using culture-dependent approaches to evaluate their biological control and plant growth promoting potential.
Materials and methods
Plant sampling, bacterial isolation and identification
Stem, bud, and blossom tissues of 27 almond and 32 apricot trees belonging to different cultivars were collected in March and April 2015 from 13 geographic areas within East Azerbaijan Province, Iran. Trees were either symptomless or symptomatic (canker, oozing on woody tissues, blast of blossoms, and spur dieback). Plant samples were placed in paper bags and immediately brought to the laboratory for further analyses.
Epiphytic bacteria were isolated without surface sterilization of plant material while endophytic bacteria were isolated after surface sterilization. At first, the samples were washed with tap water. For epiphytic isolation, 5 g of healthy and infected tissues were suspended in 20 ml of 0.1 M potassium phosphate buffer (PB) for 10 min on a shaker at 150 rpm. No disinfectant was used. For isolation of endophytic strains, 5 g of healthy and infected tissues were surface-sterilized in 0.5% (for bud and blossom tissues) or 5% (for twig and branch tissues) sodium hypochlorite for 1 and 5 min, respectively, followed by rinsing three times in sterile-double distilled water (DDW). One hundred microliters of the final wash were spread on nutrient agar NA to check sterility. Then, sterilized tissue crushed into pieces of 1 cm were then suspended in 20 ml of 0.01 M magnesium buffer (MB) for 120 min on a shaker at 150 rpm. One hundred microliters of the final suspensions were streaked on Nutrient agar (NA) medium (Merck, Germany) and King’s medium B agar (Biolife, Italy) amended with cyclohexamide (KBC) with three replications. The plates were incubated at 25–28 °C for 3–7 days and observed daily for the growth of bacterial colonies. After incubation, the bacterial population was estimated by counting bacterial colonies.
Pure cultures of randomly selected bacterial colonies with different morphology and pigmentation were obtained by colony subculturing on NA medium and were preliminarily classified based on Gram reaction. Strains were suspended in DDW and maintained at 4 °C for short-term storage. For long-term storage, all bacterial strains were grown in Luria Bertani (LB) (QUELAB, USA) broth medium for 24 h, and maintained in 15% sterile glycerol at − 70 °C.
Hypersensitive reaction and pathogenicity test
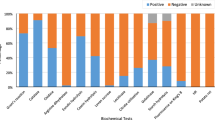
Hypersensitive reaction (HR) was evaluated on tobacco, Nicotiana tabacum, leaves using both Gram negative and Gram-positive bacterial suspensions in DDW from 48-h-old cultures on NA medium at a concentration of approx. 1 × 107 CFU/ml. Bacterial suspensions were injected using a sterile needleless syringe. DDW was used as a negative control. The appearance of necrosis in the injected sites after 48 h was considered as a positive HR reaction.
Pathogenicity tests were performed using cut, one year-old, green apricot shoots. Bacterial strains were grown for 24 h on NA medium at 28 °C and suspended in DDW at a concentration of approx. 1 × 107 CFU/ml. One ml of bacterial suspensions was injected into the shoots at three sites of leaf germination (Little et al. 1998). DDW was used as a negative control. The inoculated tissues were maintained in high moisture conditions at 28 °C for 14 days. The presence of black necrotic lesions was recorded as positive pathogenic reaction.
Molecular characterization of bacterial strains
After extracting the genomic DNA of bacterial strains by boiling for 8 and 15 min at 98 °C for Gram positive and Gram negative strains, respectively, 16 S rRNA oligonucleotide primers 16 S-F (5´- CCAGCAGCCGCGGTAATACG-3´)/16S-R (5´- ATCGGYTACCTTGTTACGACTTC-3´) (Lu et al. 2000) provided by Eton Bioscience Inc. (USA) were used to amplify an approximately 1000 bp-long fragment corresponding to an internal region of the 16 S rRNA gene. For further identification of strains in the genera Pseudomonas and Pantoea/Erwinia, amplification of the citrate synthase (cts) and gyrase (gyrB) genes, respectively, was performed using oligonucleotide primers cts-Fs (5´- CCCGTCGAGCTGCCAATWCTGA-3´)/ cts − Rs (5´- ATCTCGCACGGSGTRTTGAACATC-3´) (Sarkar and Guttman 2004) and gyrB3 (5´-GCGTAAGCGCCCGGGTATGTA − 3´) /gyrB4 (5´-CCGTCGACGTCCGCATCGGTCAT − 3´) (Deletoile et al. 2009) as described in the original papers.
Phylogenetic analyses
PCR products of the 16 S rRNA, cts, and gyrB genes were Sanger-sequenced by Eton Bioscience Inc. (USA). Phylogenetic analysis by Bayesian Inference (BI) was performed using MrBayes v.3.2.2, and the phylogenetic tree was visualized using the program FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The best model of nucleotide substitution was selected under the Akaike Information Criterion (AIC) (Akaike 1974) implemented in MrModeltest v.2.3 (Nylander 2004).
Screening of bacterial strains for antagonistic activity
Antagonistic activity of purified strains was evaluated against the Pseudomonas syringae pv. syringae 170 (Pss-170) strain, a causal agent of apricot canker disease (Vasebi et al. 2019) using a dual culture procedure. All bacterial strains were grown in Tryptic soy broth (TSB) medium for 24 h at 28 °C on a shaker at 150 rpm. One hundred microliters of the Pss-170 strain were added to the Petri dishes (9 cm diameter) of Tryptic soy agar (TSA) medium (MilliporeSigma, USA), spread with glass spreader to produce a lawn of bacteria and maintained at room temperature under a laminar flow hood for 15 min. Then, 5 µl of each bacterial strain were placed on the pathogen-inoculated Petri dishes. All Petri dishes were maintained at 28 °C for 48 h. Strains surrounded by an inhibition zone without visible growth of pathogen were selected for a complementary dual culture assay. In the complementary dual culture assay, a suspension of antagonistic strains at an optical density at 600 nm (OD600) of 0.1 was used against the pathogen at three different optical densities (0.01, 0.1, and 1) on TSA medium. Cultures were incubated at 28 °C for 48 h and the diameter of inhibition zones of every strain against the Pss-170 strain was measured. The experiment was repeated twice with three replications at all three concentrations for each antagonist. DDW was used as a negative control.
Plant growth promoting properties of bacterial strains
Siderophore production
Qualitative assay. Chrome azurol S (CAS) agar medium was used for evaluation of siderophore production according to Schwyn and Neilands (1987). Ten microliters of 24-h-old pure bacterial suspensions grown on LB were cultured on the CAS agar medium and incubated at 28 °C for up to 4 days. Formation of a yellowish orange halo surrounding inoculated colonies indicated siderophore production. Experiments were performed in triplicate.
Quantitative assay. The CAS- shuttle assay was used for quantitative estimation of siderophore production according to Schwyn and Neilands (1987). Bacterial strains were grown in succinate medium and incubated at 28 °C for 24 h on a rotator shaking incubator at 120 rpm. After incubation, cultures were centrifuged at 5000 g at 4 °C for 10 min. Then, the supernatant was filtered using a 0.22 μm filter and the cell- free filtrate was mixed with CAS solution. The equal mixture of CAS solution and uninoculated succinate medium was used as negative control. Color absorbance was determined 20 min after incubation at 630 nm using a spectrophotometer. The percentage of siderophore was estimated using the formula:
[(Ar - As) / Ar] *100.
Ar = the absorbance of the negative control.
As = the absorbance of each treatment.
Phosphate solubilization
Qualitative assay. Qualitative estimation of phosphate solubilization was determined according to Jasim et al. (2014). Ten microliters of 24-h-old bacterial cultures grown in LB medium were sub-cultured on Pikovskaya (PKV) agar (Sigma, USA) medium for 7 days in 28 °C. Formation of a transparent halo around colonies indicated solubilization of phosphate. Experiments were performed in triplicate.
Quantitative assay. Quantitative estimation of phosphate solubilization was done by the spectrophotometric method described by Ruchi et al. (2012). Seventy microliters of a 24-h-old bacterial suspension grown in LB broth medium were cultured in 10 ml PKV broth medium for 7 days at 28 °C on a shaking incubator at 120 rpm. Bacterial suspensions were centrifuged at 4 °C for 20 min at 5000 g. Five milliliters ammonium molybdate reagent (7.5 g of ammonium molybdate, 171 ml of HCl, total volume was made up to 500 ml) was added to 5 ml bacterial supernatant and kept at room temperature for 30 min. Absorbance was measured at 470 nm using a UV-VIS spectrophotometer. A corresponding amount of soluble phosphorous of each strain was calculated from a standard curve of potassium dihydrogen phosphate KH2PO4 in the range of 0-1000 µg/ml.
Protease production
Skimmed milk agar (SMA) medium was used for determining the protease production according to Sgroy et al. (2009) with some modification. Ten microliters of 24-h-old bacterial cultures grown on LB medium were inoculated on SMA medium and incubated at 28 °C for 4 days. Formation of transparent halos around colonies indicated protease production. Experiments were performed in triplicate.
Hydrogen cyanide (HCN) production
Production of hydrogen cyanide in strains was determined using the method of Alstrom and Burns (1989). Fifty microliters of 24-h-old bacterial cultures grown on LB medium were streaked on NA medium. Whatman paper soaked in picric acid solution including 0.5% picric acid and 2% Na2CO3 and placed inside the inoculated Petri dishes’ lids. Dishes were sealed with Parafilm and inversely incubated at 28 °C for 7 days. A change in color of the paper from yellow to orange or red indicated HCN production. Experiments were performed in triplicate.
Gibberellic acid (GA) production
Gibberellic acid production was estimated by the method of Holbrook et al. (1961) with slight modifications. Ten microliters of 24-h-old bacterial cultures grown in LB medium were inoculated into Jenson broth media: sucrose, 20 g/l; K2HPO4, 1 g/l; MgSO4, 0.5 g/l; NaCl, 0.5 g/l; FeSO4, 0.1 g/l; Na2MoO4, 0.005 g/l; CaCO3, 2 g/l) and incubated for 7 days at 28 °C with shaking at 200 rpm. The cultures were then centrifuged at 5000 g for 15 min. Two milliliters zinc acetate was added to 15 ml of the supernatant transferred to a separating funnel, kept for 2 min, and then 2 ml of potassium ferrocyanide solution (10.6% in distilled water) was added and centrifuged at 2000 g for 15 min. Five milliliters of the supernatant was added to 5 ml of 30% HCl and the mixture was incubated at 20 °C for 75 min. Five milliliters of 5% HCI was used as blank. Jenson broth medium without bacterial inoculant was used as negative control. Absorbance was measured at 254 nm in a UV-VIS spectrophotometer. Concentration of gibberellins produced by each strain was calculated by a preparing standard curve by using pure gibberellic acid (Merck, Frankfurt, Germany) in the range of 0-1000 µg/ml.
Indole acetic acid (IAA) production
Production of auxin indole-3-acetic acid by bacteria was tested using LB medium and Salkowski reagent (Rahman et al. 2010). Briefly, bacterial strains were grown in LB medium containing 0.2% (v/v) of sterile L-tryptophan and without L-tryptophan and incubated at 28 °C with shaking at 180 rpm. After growth for 7 days, the cultures were harvested by centrifugation at 5000 g for 10 min. One ml of supernatant was mixed with 2 ml Salkowski’s reagent (150 ml H2SO4, 250 ml distilled water, 7.5 ml FeCl3.6H2O 0.5 m) and incubated at room temperature in the dark for 30 min. The intensity of pink color of the mixture indicating IAA production was read at 530 nm using a spectrophotometer pre-calibrated with the same media. Concentration of indole acetic acid was estimated by preparing a standard curve using pure IAA (Merck, Frankfurt, Germany) in the range of 0-300 µg/ml.
1-aminocyclopropane-1-carboxylate (ACC) production
ACC-deaminase activity was determined according to the method of Glick et al. (1995). Ten microliters of 24-h-old bacterial cultures grown in LB medium were inoculated on NFb medium containing 1-aminocyclopropane-1-carboxylate (5.0 g/l) as unique nitrogen source. Plates were incubated for 4 days at 28 °C to allow colony formation. Colonies were re-inoculated and incubated at 28 °C for 4 days. Newly formed colonies on NFb + ACC medium were considered positive for ACC-deaminase activity.
Statistical analysis
The MSTATC software was used for data analysis, and the comparison of means was carried out using the Duncan test at the 5% probability level for plant growth promotion and biocontrol assays. Graphs were plotted using Excel software.
Results
Strain isolation and characterization
A total of 2867 and 125 bacterial colonies were grown on NA and KBC media, respectively. Two hundred ninety-three of 2992 morphologically different bacterial colonies including 150 Gram negative and 143 Gram positive strains were purified. About 52% and 48% of the purified strains were isolated from almond and apricot trees, respectively. 44% and 56% of the purified strains were isolated endophytically and epiphytically, respectively. One hundred thirteen of 293 purified strains including 81 Gram negative and 32 Gram positive strains were randomly selected for further identification. Among the selected strains, 51% and 49% were isolated from almond and apricot and 44% and 56% were isolated endophytically and epiphytically, respectively. Except for five isolates (Pss-26, Pss-82, Pss-170, Pss-174, and Pss-176) that were later identified as Pss (Vasebi et al. 2019), none of the 288 isolates showed an HR on tobacco leaves or pathogenicity on apricot twigs.
Based on 16 S rRNA sequencing followed by BLAST searches at NCBI, we found that these epiphytic and endophytic bacteria associated with almond and apricot trees belonged to four bacterial classes including Gammaproteobacteria (70.4%), Betaproteobacteria (0.9%), Bacilli (25.1%), and Actinobacteria (3.6%).
Within the Gammaproteobacteria, the four families Pseudomonadaceae, Enterobacteriaceae, Xanthomonadaceae, and Moraxellaceae were found. Bacteria belonged to the following genera: Pseudomonas (26%), Pantoea (26%), Erwinia (9.8%), Stenotrophomonas (5%), Acinetobacter (1.8%), Rouxiella (0.9%), and Escherichia (0.9%). Nine tenths percent of bacteria were identified as members of the genus Massilia, which belongs to the family Oxalobacteraceae within the Betaproteobacteria. Within the Bacilli, the genera Bacillus (9%), Lysinibacillus (14.3%), and Paenibacillus (1.8%) in the Bacillaceae family were identified. Within Actinomycetales, members in the genera Curtobacterium (0.9%) and Microbacterium (0.9%) in the family Microbacteriaceae, and Kocuria (0.9%) and Arthrobacter (0.9%) in the family Micrococcaceae were identified (Fig. 1). Strains isolated from healthy trees just belonged to the four Pseudomonas (31%), Lysinibacillus (31%), Pantoea (25%), and Bacillus (13%) genera. While 16 genera belonged to four bacterial classes including Pseudomonas (26.9%), Pantoea (24.8%), Erwinia (11.4%), Lysinibacillus (11.4%), Bacillus (8.2%), Stenotrophomonas (5.1%), Paenibacillus (2%), Acinetobacter (2%), Rouxiella (1%), Escherichia (1%), Massilia (1%), Curtobacterium (1%), Microbacterium (1%), Kocuria (1%), and Arthrobacter (1%) were isolated from diseased trees. The 16 S rRNA gene sequences obtained in this study were deposited in GenBank under the accession numbers listed in Table 1.
Phylogenetic tree of partial cts gene sequences of Pseudomnas spp. isolated from almond and apricot trees constructed by Bayesian inference using the GTR + I + G model. The scale bar represents the average number of substitutions per site, and posterior probability values are shown at the nodes obtained for 100,000,000 replicates
A phylogenetic tree using the Bayesian method and evolutionary distances were calculated based on the obtained partial 16 S rRNA gene sequences and sequences of selected bacterial reference strains downloaded from NCBI (Fig. 2). Phylogenetic trees based on the partial sequences of the cts and gyrB genes, respectively, revealed relationships between the isolated members of the genera Pseudomonas spp. and Pantoea spp./Erwinia spp. and selected reference strains (Figs. 3 and 4). Phylogenetic trees based on cts gene sequencing showed that the most of Pseudomonas strains (57%) isolated from almond and apricot had the highest similarity to the P. fluorescens reference strains, some (34%) to the P. graminis reference strain. Phylogenetic trees based on gyrB gene sequencing showed that the Pantoea spp. and Erwinia spp. strains isolated in this study were identified as P. agglomerans and E. billingiae with the highest similarity to the P. agglomerans, and E. billingiae reference strains.
Phylogenetic tree of partial 16S rRNA gene sequences of bacteria isolated from almond and apricot trees constructed by Bayesian inference using the GTR + I + G model. The scale bar represents the average number of substitutions per site, and posterior probability values are shown at the nodes obtained for 100,000,000 replicates
Phylogenetic tree of partial cts gene sequences of Pseudomnas spp. isolated from almond and apricot trees constructed by Bayesian inference using the GTR + I + G model. The scale bar represents the average number of substitutions per site, and posterior probability values are shown at the nodes obtained for 100,000,000 replicates
Phylogenetic tree of partial gyrB gene sequences of Pantoea spp./Erwinia spp. isolated from almond and apricot trees constructed by Bayesian inference using the GTR + I + G model. The scale bar represents the average number of substitutions per site, and posterior probability values are shown at the nodes obtained for 100,000,000 replicates
Antagonistic activity of strains
Thirty five of the 113 sequenced isolates produced an inhibition zone against the Pss-170 strain in the primary dual culture test. Eighteen of these isolates also produced an inhibition zone against the Pss-170 strain in the complementary dual culture test. Of these 18 isolates, 13 were identified as P. fluorescens with the highest similarity to the antagonistic P. fluorescens A506 strain based on 16 S rRNA and cts genes sequencing (Figs. 2 and 3), four isolates were identified as members of the genus Lysinibacillus sp., and one isolate as Paenibacillus sp. based on the 16 S rRNA sequence analysis (Fig. 2). Lysinibacillus strains showed high similarity to Lysinibacillus fusiformis strains based on 16 S rRNA gene sequencing. Isolate 185-2 belongs to the Paenibacillus genus showed high 16 S rRNA gene sequence similarity to Paenibacillus polymyxa DBB1709.
Isolates showed the highest inhibition when plated at an OD600 of 0.1 and when the pathogen was plated at an OD600 of 0.01 and 0.1 with inhibition zone diameters ranging from 8.5 to 13.5 mm and 6.5 to 12 mm, respectively. Only the five P. fluorescens strains (11 − 1, 190-2, 35 − 1, 136-2, 159-5, and 199-3 isolates) inhibited the Pss-170 when the pathogen was plated at an OD600 of 1 with an inhibition zone diameter range from 6.6 to 8.5 mm. Among the P. fluorescens strains, 190-2 and 146-k-1 isolates showed the greatest inhibitory effect on the pathogen with an inhibition zone diameter of 13.5 and 12 mm at an OD600 of 0.01 and 0.1, respectively (Fig. 5).
In spite of Pseudomonas isolate 11 − 1, all other isolates with antagonism against the Pss-170 strain were isolated from apparently infected trees with oozing and canker symptoms, including 11 almond and seven apricot trees from seven geographic areas. Most of the antagonistic isolates, approximately 72%, were endophytes isolated from nine almond and four apricot trees (Table 1).
Evaluation of plant growth promoting properties
All 18 isolates with antagonistic activity against the Pss-170 strain were selected for evaluation of their plant growth promoting ability including siderophore, protease, HCN, GA, IAA, and ACC production, and phosphate solubilization.
Results from the quantitative biosynthesis assay of GA showed differences among isolates. While all strains had some GA production, the highest and lowest one was detected in P. fluorescens 120-3 with 13.7 µg/ml of GA and P. fluorescens 35 − 4 with 1.8 µg/ml of GA, respectively (Fig. 6).
Phylogenetic tree of partial cts gene sequences of Pseudomnas spp. isolated from almond and apricot trees constructed by Bayesian inference using the GTR + I + G model. The scale bar represents the average number of substitutions per site, and posterior probability values are shown at the nodes obtained for 100,000,000 replicates
In the qualitative siderophore production assay, colonies of four isolates had positive results developing yellow to orange haloes on CAS agar (Supplementary Fig. 1 and Table 2). Quantitative siderophore production abilities of these bacteria ranged from 21.2 µg/ml by P. fluorescens 11 − 1 to 0.07 µg/ml by P. fluorescens 159-5, 136-2, 119-3, and 120-3 (Fig. 7).
In the quantitative IAA assay, the highest production rate was observed by P. fluorescens 120-3 with 103 µg/ml of IAA and the lowest production was found in P. fluorescens 188-1 and 199-3 with 1.2 µg/ml of IAA (Fig. 8).
Isolates were screened qualitatively and quantitatively for their ability to solubilize phosphate. All isolates were found to be able to solubilize insoluble phosphate by producing phosphatase enzyme based on the formation of a transparent halo around their colonies (Supplementary Fig. 2 and Table 2). Quantitatively phosphate solubilizing abilities of these bacteria ranged between 17 µg/ml by Lysinibacillus sp. 44-k-1 to 513 µg/ml by P. fluorescens 35 − 1 (Fig. 9).
None of the isolates were able to produce protease and hydrogen cyanide. In vitro ACC production indicative of potential plant growth promoting activity was detected for 11 of the 18 isolates (Table 2).
Discussion
Bacterial canker is one of the most dangerous diseases of cultivated Prunus spp. in Iran and the world (Agrios 2005; Ahmadi et al. 2017). One of the causal agents of the disease is the Gram-negative bacterium Pss. Disease management strategies for bacterial canker caused by Pss are important but laborious because of little available knowledge of host resistance, the endophytic nature of the pathogen during some phases of the disease cycle, and the lack of effective systemic chemical bactericides. Copper compounds are the standard bactericides for controlling bacterial canker disease but they are not able to kill the pathogen systemically, they may induce emergence of copper-resistant strains, persist in fruit with harm to consumers, and exhibit phytotoxicity (Kennelly et al. 2007). Therefore, developing alternative control strategies, such as biological control is desirable. Biocontrol using antagonistic bacteria can be an alternative strategy in the management of plant pathogens (Hallmann and Berg 2006). Endophytic bacteria that occupy the internal spaces of plants in vicinity to plant pathogens, are promising biocontrol agents (Berg et al. 2005). Antagonistic bacteria that produce antimicrobial compounds, phytohormones, and siderophores, and that induce systemic resistance can inhibit disease development by plant pathogens (Compant et al. 2010; Zachow et al. 2015).
In the present study, a total of 2992 bacterial strains were isolated from aerial parts of almond and apricot trees of which 113 were identified based on 16 S rRNA gene sequencing. The sequenced strains belonged to 15 bacterial genera including Pseudomonas, Pantoea, Erwinia, Stenotrophomonas, Acinetobacter, Rouxiella, Escherichia, Massilia, Bacillus, Lysinibacillus, Paenibacillus, Curtobacterium, Microbacterium, Kocuria, and Arthrobacter. In many studies, some species in these genera were identified as endophytic bacteria of different plants (Rosenblueth and Martínez-Romero 2006). Two Gram negative genera, Pseudomonas and Pantoea, and two Gram positive genera, Lysinibacillus and Bacillus were the most abundant genera cultured from aerial tissues of almond and apricot trees. According to previous studies, Bacillus, Microbacterium, Pantoea, Pseudomonas, and Stenotrophomonas have been reported as the most commonly isolated bacterial genera, where Bacillus and Pseudomonas are the predominant genera (Chaturvedi et al. 2016).
Both, epiphytic and endophytic strains were isolated for all identified genera with exception in the genera of Rouxiella, Escherichia, and Curtobacterium, for which only endophytes were isolated, and in the genera of Massilia, Paenibacillus, Microbacterium, Kocuria, and Arthrobacter, for which only epiphytes were isolated.
All purified strains were investigated for their antagonistic activity against Pss-170, a strain of the causal agent of apricot canker disease in East Azerbaijan, Iran. Eighteen strains showed antagonistic activity against the pathogen. Antagonistic strains belonged to the genera including Pseudomonas, Lysinibacillus, and Paenibacillus based on 16 S rRNA gene sequencing and were isolated both epiphytically and endophytically. These strains were investigated for their plant growth-promoting characteristics such as IAA, GA, and siderophore production, and phosphate solubilization. They were also tested for their biocontrol potential properties such as protease production and HCN production. Almost 100%, 94%, 78%, and 61% of antagonistic strains had the ability to produce GA, solubilize phosphate, produce siderophore, and ACC, respectively, while none of the strains were able to produce protease and HCN.
Synthesis of plant growth regulators, such as indole acetic acid and gibberellic acid, by some bacteria that live in association with plants have beneficial effects for plants by increasing nutrient availability and promoting plant growth under stressful environments (Duca et al. 2014). IAA and GA are phytohormones known to be produced by plant growth promoting bacteria such as Pseudomonas and Bacillus (Ali et al. 2009; Hussain and Hasnain 2011). Siderophores are low molecular weight bio-molecules secreted by some microorganisms in response to iron starvation. In the present study, both the epiphytic and endophytic antagonistic strains were able to produce siderophores. Siderophore-producing epiphytic and endophytic bacteria are able to compete with phytopathogens for ferrous iron in the rhizosphere as well as inside the host plants and function as a biocontrol agent (van der Lelie et al. 2009). Phosphorus is one of the most important nutrients for plant growth but is usually present in its insoluble form. Many endophytic bacteria with phosphate solubilization activity can enhance phosphorus uptake by plants (Oteino et al. 2015). In agriculture, application of phosphate solubilizing microorganism was reported to facilitate plant growth (Sahu et al. 2016).
One of the key bacterial traits in promoting plant growth and improving plant biomass is the production of the enzyme ACC-deaminase by lowering ethylene accumulation in plants even under stressful conditions such as saline and drought conditions (Gupta and Pandey 2019; Onofre-Lemus et al. 2009). The antagonistic Paenibacillus sp. 185-2 strain was able to produce GA, ACC, and siderophore and was weak in the production of IAA and phosphatase activity. Eastman et al. (2014) reported the presence of genes responsible for plant hormone synthesis, and production of antimicrobials in the P. polymyxa CR1 genome. Paenibacillus species have been isolated from various ecological habitats including soil, air, and rhizosphere. Several studies have shown the antagonistic properties of Paenibacillus species against phytopathogenic bacteria and fungi such as Ralstonia, Agrobacterium, and Fusarium and their ability in plant growth promotion and enhancing yield (Algam et al. 2010; Bosmans et al. 2017; Sato et al. 2014; Yadav 2019). Strains of P. polymyxa have been reported to possess inhibitory activity against plant pathogenic bacterium P. syringae (Eastman et al. 2014; Kwon et al. 2016) with ability in production of HCN, siderophores, phytohormone, and enzymatic activities such as protease and phosphatase production (Gómez-Lama Cabanás et al. 2018).
Four isolates, 44-k-1, 88 − 5, 88 − 9, and 158-k-1, were identified as members of the genus Lysinibacillus with high similarity to L. fusiformis reference strains. These isolates showed the highest inhibition effects against the Pss-170 strain when used at an optical density of 0.01 and when the pathogen was inoculated at an optical density of 0.1. Lysinibacillus is a Gram-positive bacterium that can form dormant endospores under stress conditions which are resistant to heat, chemicals, and ultraviolet light. There are several reports indicating the potential of Lysinibacillus spp. for biocontrol activities against phytopathogens and plant growth promotion like phosphate solubilization and nitrogen fixation and production of higher quantity of IAA, phytohormone, siderophore, HCN, and ACC-deaminase (Naureen et al. 2017; Sahu et al. 2018; Sgroy et al. 2009; Verma et al. 2014; Yadav et al. 2016).
The P. fluorescens (11 − 1, 35 − 4, 69 − 4, 190-2, 35 − 1, 136-2, 119-3, 120-3, 124-1, 146-k-1, 159-5, 188-1, and 199-3 isolates) were found to be the most abundant antagonistic strains. Among all the P. fluorescens antagonistic strains, the 11 − 1, 190-2, 35 − 1, 136-2, 195-5, and 199-3 isolates showed significant inhibition of the Pss-170 strain at all three concentrations of the Pss-170 in the dual-culture assay. All isolated P. fluorescens strains showed high similarity to the P. fluorescens A506 antagonist strain based on molecular identification. This strain was isolated from pear in California and has the ability to reduce the incidence of fire blight in orchards by 50 to 80% (Stockwell et al. 2010). In the present study, the P. fluorescens strains 69 − 4, 190-2, 35 − 1, 124-1, 188-1, and 199-3 showed four properties related to plant growth promotion, including production of ACC and siderophore and phosphate solubilization. The P. fluorescens 120-3 strain showed the highest production of the phytohormones IAA and GA compared to the other strains. The P. fluorescens strains 11 − 1 and 35 − 4 showed the highest ability in production of siderophore. In many studies, strains of P. fluorescens were shown to enhance plant growth promotion and reduce severity of various diseases caused by a range of fungal and bacterial plant pathogens (Gómez-Lama Cabanás et al. 2017; Pujol et al. 2005). This effect is the result of the production of a number of secondary metabolites including antibiotics, siderophores, 2,4-diacetylphloroglucinol, IAA, and hydrogen cyanide as well as ability to solubilize phosphate (Bensidhoum et al. 2016; Couillerot et al. 2009; Duffy and Défago 1999; O’Sullivan and O’Gara 1992; Golanowska et al. 2012) identified the P. fluorescens T660 and T777 strains as antagonistic bacteria against the causal agents of stone fruit canker disease caused by Pss and P. syringae pv. morsprunorum.
In conclusion, this study reported the presence and diversity of culturable epiphytic and endophytic bacteria in almond and apricot trees. Based on our information, this is the first reported study in elucidating the epiphytic and endophytic bacterial diversity associated with aerial parts of almond and apricot trees with plant growth promoting and biocontrol potential based on in vitro assays. The existence of such microorganisms with the ability to promote plant growth and control plant disease suggests that they could be utilized as biocontrol agents in future applications, however, further studies based on in vivo and field conditions are required.
References
Agrios GN (2005) Plant pathology, vol 949. edn, Elsevier Academic Press, p 5
Ahmadi S, Harighi B, Abdollahzadeh J (2017) Phylogenetic relationships of fluorescent pseudomonad isolates associated with bacterial canker of stone fruit trees in Kurdistan province, Iran. Eur J Plant Pathol 150:679–689. https://doi.org/10.1007/s10658-017-1316-4
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19(6):716–723
Algam SAE, Xie G, Li B, Yu S, Su T, Larsen J (2010) Effects of Paenibacillus strains and chitosan on plant growth promotion and control of Ralstonia wilt in tomato. J Plant Pathol 92(3):593–600. https://doi.org/10.4454/jpp.v92i3.303
Ali B, Sabri AN, Ljung K, Hasnain S (2009) Quantification of indole-3-acetic acid from plant associated Bacillus spp. and their phytostimulatory effect on Vigna radiata (L.). World J Microbiol Biotechnol 25(3):519–526. https://doi.org/10.1007/s11274-008-9918-9
Ali S, Charles TC, Glick BR (2014) Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol Biochem 80:160–167. https://doi.org/10.1016/j.plaphy.2014.04.003
Alstrom S, Burns RG (1989) Cyanide production by rhizobacteria as a possible mechanism of plant growth inhibition. Biol Fertil Soils 7:232–238
Bensidhoum L, Nabti E, Tabli N, Kupferschmied P, Weiss A, Rothballer M, Hartmann A (2016) Heavy metal tolerant Pseudomonas protegens isolates from agricultural well water in northeastern Algeria with growth promoting, insecticidal and antifungal activities. Eur J Soil Biol 75:38–46. https://doi.org/10.1016/j.ejsobi.2016.04.006
Bent E, Chanway CP (1998) The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can J Microbiol 44(10):980–988. https://doi.org/10.1139/w98-097
Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J (2005) Endophytic and ectophytic potatoassociated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol 51(2):215–229. https://doi.org/10.1016/j.femsec.2004.08.006
Bosmans L, De Bruijn I, Gerards S, Moerkens R, Van Looveren L, Wittemans L, Van Calenberge B, Paeleman A, Van Kerckhove S, De Mot R, Rozenski J, Rediers H, Raaijmakers JM, Lievens B (2017) Potential for Biocontrol of Hairy Root Disease by a Paenibacillus clade. Front Microbiol 8:447. https://doi.org/10.3389/fmicb.2017.00447
Bradbury JF (1986) Pseudomonas syringae pv. Syringae. Guide to Plant pathogenic Bacteria. CAB International Mycological Institute, Kew, England, pp 175–177
Chaturvedi H, Singh V, Gupta G (2016) Potential of bacterial endophytes as plant growth promoting factors. J Plant Pathol Microbiol 7(9):1–6. https://doi.org/10.4172/2157-7471
Compant S, Clement C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42(5):669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moënne-Loccoz Y (2009) Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol 48(5):505–512. https://doi.org/10.1111/j.1472-765X.2009.02566.x
Deletoile A, Decre D, Courant S, Passet V, Audo J, Grimont P, Arlet G, Brisse S (2009) Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol 47:300–310. https://doi.org/10.1128/JCM.01916-08
Duca D, Lorv J, Patten CL, Rose D, Glick BR (2014) Indole-3-acetic acid in plant–microbe interactions. Anton Leeuw 106:85–125. https://doi.org/10.1007/s10482-013-0095-y
Duffy BK, Défago G (1999) Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 65(6):2429–2438. https://doi.org/10.1128/AEM.65.6.2429-2438
Eastman AW, Weselowski B, Nathoo N, Yuan Z-C (2014) Complete genome sequence of Paenibacillus polymyxa CR1, a plant growth-promoting bacterium isolated from the corn rhizosphere exhibiting potential for biocontrol, biomass degradation, and biofuel production. Genome Announc 2(1):e01218–e01213. https://doi.org/10.1128/genomeA.01218-13
Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont PAD (1999) DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. Int J Syst Bacteriol 49(2):469–478. https://doi.org/10.1099/00207713-49-2-469
Glick B, Karaturovíc D, Newell P (1995) A novel procedure for rapid isolation of plant growth-promoting rhizobacteria. Can J Microbiol 41(6):533–536. https://doi.org/10.1139/m95-070
Golanowska M, Ankiewicz H, Taraszkiewicz A, Kamysz W, Czajkowski R, Krolicka A, Jafra S (2012) Combining antagonistic potential of selected Pseudomonas spp. strains and synthetic peptide CAMEL towards Pseudomonas syringae pv. syringae and P. syringae pv. morsprunorum. J Plant Pathol 94: S1. 69-S1.73. https://doi.org/10.4454/jpp.v94i1sup.012
Gómez-Lama Cabanás C, Ruano-Rosa D, Legarda G, Pizarro-Tobías P, Valverde-Corredor A, Triviño JC, Roca A, Mercado-Blanco J (2018) Bacillales Members from the Olive Rhizosphere are effective Biological Control Agents against the defoliating pathotype of Verticillium dahlia. Agric 8(7):90. https://doi.org/10.3390/agriculture8070090
Gómez-Lama Cabanás C, Sesmero R, Valverde-Corredor A, López-Escudero FJ, Mercado-Blanco J (2017) A split-root system to assess biocontrol effectiveness and defense-related genetic responses in above-ground tissues during the tripartite interaction verticillium dahliae-olive-Pseudomonas fluorescens PICF7 in roots. Plant Soil 417:433–452. https://doi.org/10.1007/s11104-017-3269-y
Gupta S, Pandey S (2019) ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in french bean (Phaseolus vulgaris) plants. Front Microbiol 10:1506. https://doi.org/10.3389/fmicb.2019.01506
Hallmann J, Berg G (2006) Spectrum and population dynamics of bacterial root endophytes. In: Schulz. BJE, Boyle. CJC, Sieber. TN (eds) Microbial Root Endophytes, vol 6. Springer-Verlag, Berlin, pp 15–31
Holbrook AA, Edge WJW, Bailey P (1961) Spectrophotometric method for determination of Gibberellic acid. Advances in Chemistry, In. Gibberellines, vol. 28. Chapter 18, (pp. 159–167), American Chemical Society, Washington, DC. https://doi.org/10.1021/ba-1961-0028
Hussain A, Hasnain S (2011) Interactions of bacterial cytokinins and IAA in the rhizosphere may alter phytostimulatory efficiency of rhizobacteria. World J Microbiol Biotechnol 27:2645–2654. https://doi.org/10.1007/s11274-011-0738-y
Jasim B, Joseph AA, John CJ, Mathew J, Radhakrishnan EK (2014) Isolation and characterization of plant growth promoting endophytic bacteria from the rhizome of Zingiber officinale. 3 Biotech 4:197–204. https://doi.org/10.1007/s13205-013-0143-3
Kennelly MM, Cazorla FM, De Vicente A, Ramos C, Sundin GW (2007) Pseudomonas syringae disease of fruit, progress toward understanding and control. Plant Dis 91(1):4–17. https://doi.org/10.1094/PD-91-0004
Kotan R, Sahin F (2002) First record of bacterial canker caused by Pseudomonas syringae pv. Syringae, on apricot trees in Turkey. Plant Pathol 51(6):798–798. https://doi.org/10.1046/j.1365-3059.2002.00768.x
Kwon YS, Lee DY, Rakwal R, Baek SB, Lee JH, Kwak YS, Seo JS, Chung WS, Bae DW, Kim SG (2016) Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16(1):122–135. https://doi.org/10.1002/pmic.201500196
Little EL, Bostock RM, Kirkpatrick BC (1998) Genetic characterization of Pseudomonas syringae pv. Syringae strains from stone fruits in California. Appl Environm Microbiol 64:3818–3823. https://doi.org/10.1128/AEM.64.10.3818-3823.1998
Lu JJ, Perng Cl, Lee S, Wan CC (2000) Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol 38(6):2076–2080. https://doi.org/10.1128/.38.6.2076-2080.2000
Naureen Z, Rehman NU, Hussain H, Hussain J, Gilani SA, Al Housni SK, Mabood F, Khan AL, Farooq S, Abbas G, Harrasi AA (2017) Exploring the potentials of Lysinibacillus sphaericus ZA9 for plant growth promotion and biocontrol activities against phytopathogenic fungi. Front Microbiol 8:1477. https://doi.org/10.3389/fmicb.2017.01477
Nylander JAA (2004) MrModeltest v2.0. Program distributed by the author. Evolutionary Biology Centre. Uppsala University, Uppsala, Sweden
Onofre-Lemus J, Hernández-Lucas I, Girard L, Caballero-Mellado J (2009) ACC (1-Aminocyclopropane-1-Carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Plant Microbiol 75(20):6581–6590. https://doi.org/10.1128/AEM.01240-09
O’Sullivan DB, O’Gara F (1992) Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev 56(4):662–676
Oteino N, Lally RD, Kiwanuka S, Lioyd A, Ryan D, Germaine KJ, Dowling DN (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol 6:745. https://doi.org/10.3389/fmicb.2015.00745
Popović T, Menković J, Prokić A, Zlatković N, Obradović A (2021) Isolation and characterization of Pseudomonas syringae isolates affecting stone fruits and almond in Montenegro. J Plant Dis Prot 128:391–405. https://doi.org/10.1007/s41348-020-00417-8
Pujol M, Badosa E, Cabrefiga J, Montesinos E (2005) Development of a strain-specific quantitative method for monitoring Pseudomonas fluorescens EPS62e, a novel biocontrol agent of fire blight. FEMS Microbiol Lett 249(2):343–352. https://doi.org/10.1016/j.femsle.2005.06.029
Rahman A, Sitepu IR, Tang SY, Hashidoko Y (2010) Salkowski’s reagent test as a primary screening index for functionalities of Rhizobacteria isolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Biosci Biotechnol Biochem 74:2202–2208. https://doi.org/10.1271/bbb.100360
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact 19(8):827–837. https://doi.org/10.1094/MPMI-19-0827
Ruchi Kapoor R, Kumar A, Patil S, Thapa S, Kaur M (2012) Evaluation of plant growth promoting attributes and lytic enzyme production by fluorescent Pseudomonas diversity associated with Apple and Pear. Int J Sci Res Publ 2(2):1–8
Sahu PK, Shivaprakash MK, Mallesha BC, Subbarayappa CT, Brahmaprakash GP (2018) Effect of bacterial endophytes Lysinibacillus sp. on plant growth and fruit yield of tomato (Solanum lycopersicum). Int J Curr MicrobiolAppl Sci 7(5):2319–7706. https://doi.org/10.20546/ijcmas.2018.705.397
Sahu PK, Lavanya G, Gupta A, Brahmaprakash GP (2016) Fluid bed dried microbial consortium for enhanced plant growth: a step towards next generation bioformulation. Vegetos 29(4):6–10. https://doi.org/10.5958/2229-4473.2016.00093.8
Sarkar SF, Guttman D (2004) Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol 70:1999–2012. https://doi.org/10.1128/aem.70.4.1999-2012.2004
Sato I, Yoshida S, Iwamoto Y, Aino M, Hyakumachi M, Shimizu M, Takahashi H, Ando S, Tsushima S (2014) Suppressive potential of Paenibacillus strains isolated from the tomato phyllosphere against Fusarium crown and root rot of tomato. Microbes Environ 29(2):168–177. https://doi.org/10.1264/jsme2.ME13172
Schulz B, Boyle C (2006) What are endophytes? In: Schulz BJE, Boyle CJC, Sieber TN (eds) Microbial Root Endophytes. Springer-Verlag, Berlin, pp 1–13
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Sgroy V, Cassán F, Masciarelli O, Del Papa MF, Lagares A, Luna V (2009) Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasisregulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl Microbiol Biotechnol 85(2):371–381. https://doi.org/10.1007/s00253-009-2116-3
Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B (2015) Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6(5):e00621–e00615. https://doi.org/10.1128/mBio.00621-15
Stockwell VO, Johnson KB, Sugar D, Loper JE (2010) Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1applied as single strains and mixed inocula. Phytopathol 100(12):1330–1339. https://doi.org/10.1094/PHYTO-03-10-0097
Subramanian P, Mageswari A, Kim K, Lee Y, Sa T (2015) Psychrotolerant endophytic Pseudomonas sp. strains OB155 and OS261 induced chilling resistance in tomato plants (Solanum lycopersicum Mill.) By activation of their antioxidant capacity. Mol Plant-Microbe Interac 28(10):1073–1081. https://doi.org/10.1094/MPMI-01-15-0021-R
Thapa S, Prasanna R, Ranjan K, Velmourougane K, Ramakrishnan B (2017) Nutrients and host attributes modulate the abundance and functional traits of phyllosphere microbiome in rice. Microbiol Res 204:55–64. https://doi.org/10.1016/j.micres.2017.07.007
Van der Lelie D, Taghavi S, Monchy S, Schwender J, Miller L, Ferrier R, Rogers A, Wu X, Zhu W, Weyens N, Vangronsveld J, Newman L (2009) Poplar and its bacterial endophytes: coexistence and harmony. Crit Rev Plant Sci 28(5):346–358. https://doi.org/10.1080/07352680903241204
Vasebi Y, Khakvara R, Faghihi MM, Vinatzer BA (2019) Genomic and pathogenic properties of Pseudomonas syringae pv. Syringae strains isolated from apricot in East Azerbaijan province, Iran. Biocatal Agric Biotechnol 19:101167. https://doi.org/10.1016/j.bcab.2019.101167
Verma P, Yadav AN, Kazy SK, Saxena AK, Suman A (2014) Evaluating the diversity and phylogeny of plant growth promoting bacteria associated with wheat (Triticum aestivum) growing in central zone of India. Int J Curr Microbiol Appl Sci 3(5):432–447
Vicente JG, Alves JP, Russell K, Roberts SJ (2004) Identification and discrimination of Pseudomonas syringae isolates from wild cherry in England. Eur J Plant Pathol 110:337–351. https://doi.org/10.1023/B:EJPP.0000021060.15901.33
Wenneker M, Janse JD, de Bruine A, Vink P, Pham K (2012) Bacterial canker of plum caused by Pseudomonas syringae pathovars, as a serious threat for plum production in the Netherlands. J Plant Pathol 94:S11–S13
Wenneker M, Meijer H, Maas FM, de Bruine A, Vink P, Pham K (2013) Bacterial canker of plum trees (Prunus domestica), caused by Pseudomonas syringae pathovars, in the Netherlands. Acta Hortic 985:235–239. https://doi.org/10.17660/ActaHortic.2013.985.30
Yadav AN (2019) Microbiomes of wheat (Triticum aestivum L.) endowed with multifunctional plant growth promoting attributes. EC Microbiol 15:9: 700–705
Yadav AN, Sachan SG, Verma P, Saxena AK (2016) Bioprospecting of plant growth promoting psychrotrophic Bacilli from the cold desert of north western indian Himalayas. Indian J Exp Biol 54(2):142–150
Young JM (1991) Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. Syringae van Hall 1902. Ann Appl Biol 118(2):283–298. https://doi.org/10.1111/j.1744-7348.1991.tb05629.x
Young JM, Saddler GS, Takikawa Y, Boer SH, Vauterin L, Gardan L, Gvozdyak RI, Stead DE (1996) Names of plant pathogenic bacteria 1864–1995. Rev Plant Pathol 75(9):721–763
Zachow C, Jahanshah G, de Bruijn I, Song C, Ianni F, Pataj Z, Gerhadt H, Pianet I, Lämmerhofer M, Berg G, Gross H, Raaijmakers JM (2015) The novel lipopeptide poaeamide of the endophyte Pseudomonas poae RE*1-1-14 is involved in pathogen suppression and root colonisation. Mol Plant Microbe Interact 28(7):800–810. https://doi.org/10.1094/MPMI-12-14-0406-R
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that there is no known conflict of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vasebi, Y., Khakvar, R. & Vinatzer, B.A. Characterization of culturable epiphytic and endophytic bacteria of Prunus spp. and their potential for plant growth promotion and antagonistic activity against bacterial canker disease. J Plant Pathol 105, 749–766 (2023). https://doi.org/10.1007/s42161-023-01342-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-023-01342-z