Abstract
Strawberry is economically important. Anthracnose caused by Colletotrichum spp. has been a serious threat to this crop globally. To detect and distinguish latent Colletotrichum spp. infection on asymptomatic plants at an affordable cost is crucial for this disease control and the sustainability of strawberry industry. Loop-mediated isothermal amplification (LAMP) assay is superior for the higher sensitiveness, efficiency, and lower cost than other methods for pathogen diagnosis. In this study, six previously reported barcodes were evaluated against DNA templates from strawberry fungal pathogens including six Colletotrichum species and five beyond this genus, and further verified in 40 Colletotrichum isolates. Based on the discernibility revealed, a LAMP assay was developed for the diagnosis of Colletotrichum spp. by designing six primers recognizing the conserved regions in β-tubulin 2 gene from 13 Colletotrichum species of C. gloeosporioides and C. acutatum complexes. The specificity and accuracy of LAMP assay was tested against six Colletotrichum species of two complexes, with a detection sensitivity of 100 pg/μL genomic DNA. Current LAMP assay beginning with a 10 min plant lysis enabled a direct and quick diagnosis of Colletotrichum spp. in strawberry within one hour. Followed by PCR using primers specific to ApMat, Marker 2, and Marker 1, this assay could specifically differentiate Colletotrichum species, at least for the dominant species C. fructicola and C. siamense in China, realizing a diagnosis of latent infection at a species level. Current LAMP-PCR combined protocol allows for a sensitive and efficient detection and differentiation of latent Colletotrichum infection on strawberry, which will be useful for disease management and monitoring pathogen population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern strawberry (Fragaria × ananassa Duchase) is of great economic importance and widely cultivated (Zhao et al. 2012; Edger et al. 2019). Anthracnose caused by Colletotrichum spp. is a serious threat to strawberry production worldwide (Buddie et al. 1999; Martínez-Culebras et al. 2000). All the tissues of strawberry plant could be infected with this disease. Sunken necrosis in strawberry leaves, petioles, and stolons often occurs after a rainfall in the field if no fungicides have been applied in advance. Plant wilting due to anthracnose crown rot constantly causes great losses in nursery and production field after transplanting (Freeman and Katan 1997). Many farmers share their experiences that, during seedling propagation about 90% fungicides are used for anthracnose controlling in the season of high temperature and humidity in Shanghai, where typhoons and heavy rainfalls frequently occur in summer. A broad range of hosts and wide adaptation to temperature in Colletotrichum spp. increase the difficulty for disease management (da Silva et al. 2020). Anthracnose has long been one of the crucial limiting factors for strawberry production, especially hampering soil conservation and ecological cultivation.
Colletotrichum genus was among the top eight important groups of fungal pathogens in the world (Dean et al. 2012). The genetic limits among strawberry anthracnose pathogens of worldwide origins had long been clarified as three clades of C. acutatum, C. fragariae, and C. gloeosporioides (Martínez-Culebras et al. 2000, 2003). Many biologists contributed to understanding Colletotrichum systematics related to strawberry (Cannon et al. 2012), although some synonymies proposed in their work were later considered to be inaccurate (Damm et al. 2012). The emergency of molecular data and multi-loci phylogeny finally revolutionized Colletotrichum species identification.
Currently, a total of 14 species complexes have been proposed in Colletotrichum genus (Damm et al. 2019). Of them, C. gloeosporioides, C. acutatum and C. boninense species complexes including at least 17 species have been reported in relation to strawberry anthracnose: seven species of C. acutatum complex including C. acutatum, C. foriniae, C. godetiae, C. miaoliense, C. nymphaeae, C. salicis, and C. simmondsii, eight of C. gloeosporioides complex including C. aenigma, C. changpingense, C. fructicola, C. gloeosporioides, C. kahawae, C. siamense, C. theobromicola (syn. C. fragariae), and C. viniferum, as well as two species C. boninense and C. karstii of C. boninense complex (Weir et al. 2012; Han et al. 2016; Wang et al. 2019; Chung et al. 2020). The former two complexes C. gloeosporioides and C. acutatum were widely reported in relation to strawberry anthracnose co-existing in North America and East Asia.
The spread of anthracnose in strawberry field is very rapid under appropriate weather conditions, aggravated by ordinary agricultural practice such as transplanting, watering, and trimming old leaves (Ren et al. 2008). To detect and eliminate asymptomatic latent infection is vital for controlling the damage of anthracnose and avoiding a severe outbreak of this disease in strawberry field. Polymerase chain reaction (PCR) has been successfully used for identifying C. acutatum in strawberry (Parikka and Lemmetty 2004). Specific detection of Colletotrichum spp. has been developed for some species (Tao et al. 2013; Azevedo-Nogueira et al. 2021). A barcode named Marker 2 was reported to differentiate among C. fructicola, C. siamense, and C. aenigma virulent to strawberry (Gan et al. 2017). A DNA macroarray was developed for the diagnosis of latent infection of C. acutatum in strawberry plants, based on a PCR using primers specific to MAT1-2 gene (encoding a factor determining mating type) and a subsequent microtube hybridization (MTH) (Furuta et al. 2017). In another report, a loop-mediated isothermal amplification (LAMP) assay was developed for detecting C. gloeosporioides sensu lato in strawberry (Wu et al. 2019). Up to date, there is no method reported for detecting both C. acutatum and C. gloeosporioides species aggregates simultaneously.
Both real-time PCR and digital PCR (dPCR) allow for accurate quantitation of pathogen molecules and have been widely used in disease diagnosis (Yang et al. 2015; Quan et al. 2018). But these methods rely on special instruments, and the cost for diagnosis often is too expensive to be practically applied in strawberry industry. Loop-mediated isothermal amplification (LAMP) is a rapid nucleic acid amplification technique at a constant temperature with a relatively low cost (Notomi et al. 2000). A LAMP assay is more sensitive and specific than PCR (Hara-Kudo et al. 2007). Hydroxy naphthol blue added into a LAMP reaction tube provides a convenient and fast visual observation of DNA amplification via color changes with a low risk of contamination (Goto et al. 2009). LAMP enables the feasibility of point-of-care testing (POCT) on sites such as strawberry field without a need of special devices. Generally, both LAMP and conventional PCR are affordable for strawberry farmers.
Molecular diagnosis of anthracnose is very important for the sustainable development of strawberry industry. The basic question for strawberry health disease managers, government quarantine personnel and strawberry breeders is to know what Colletotrichum species infect strawberry, and what the range of fungal targets is. To meet this need, the present study intended to evaluate previously published barcodes for differentiating Colletotrichum spp. pathogenic to strawberry. Based on the discernibility revealed, a direct LAMP was developed to identify latent Colletotrichum infection on strawberry, and the LAMP combined with PCR further enabled a definitive resolution in species-specific diagnosis for the dominant species.
Materials and methods
Fungal isolates
This work was based on two previous efforts for characterizing Colletotrichum spp. from diseased strawberry in central and eastern China (Han et al. 2016; Zhang et al. 2020). Six isolates of Colletotrichum spp. representing distinct species were utilized as standard strains (Supplementary Table S1), including five strains of C. gloeosporioides species complex i.e., GQH124 (C. aenigma) and GQHZJ19 (C. siamense) (Zhang et al. 2020), CGMCC3.17371 (C. fructicola) (Ren et al. 2008), Jsh-7-1 (C. gloeosporioides), and Nj-2 (C. murrayae) (Han et al. 2016), as well as one of C. acutatum complex Lch5 (C. nymphaeae) (Han et al. 2016). Five additional pathogenic fungi beyond Colletotrichum genus were used as negative controls in evaluating the discernibility of reported markers. These pathogens consisted of an isolate of Nectria pseudotrichia originating from Guangxi Province caused necrosis and brown to red crown rot (Zhang et al. 2018), an isolate of Fusarium oxysporum triggered wilting from Qingpu Shanghai (ITS rDNA barcode identical to MT453296), an isolate of Pestalotiopsis clavispora) (ITS rDNA barcode-MF399814) from northern Anhui Province causing red leaf disease (Ning et al. 2019), and an isolate of Botrytis cinerea from strawberry fruits causing gray mold (ITS rDNA barcode identical to MH860108). The genomic DNA of Podosphaera aphanis (isolate NWAU1, 2017 purified from Shaanxi) causing strawberry powdery mildew, was a gift from Dr. Jiayue Feng of Northwest A&F University (ITS rDNA barcode-KU207048).
Furthermore, a total of 40 isolates belonging to C. gloeosporioides species aggregate (including 20 C. fructicola, 18 C. siamense, and two C. aenigma, Supplementary Table S2) were used to validate the applicability of candidate barcodes for detecting Colletotrichum spp. These single-spore isolates were previously purified from diseased strawberry in the downstream regions of Yangtze River surrounding Shanghai during 2016 to 2018 (Zhang et al. 2020). All filamentous fungi were maintained on potato dextrose agar (PDA, Lot# 0091369, Becton, Dickinson and Company, USA) plates at 28 ℃.
DNA extraction and PCR amplification
Fungal DNA was extracted from about 100 mg fresh mycelia using the Quick-DNATM Fungal/Bacterial Miniprep Kit (ZYMO Research, USA, Cat. No. D6005) following the manufacturer’s instruction with minor adaption. Specifically, fungal cells were homogenized at 55Hz for 7 min followed by a centrifugation at 10,000 x g for 2 min. When evaluating markers for Colletotrichum spp., strawberry genomic DNA was used as a negative control and purified from leaf tissues using a CTAB method as described previously (Liu et al. 2014). The quality of genomic DNA was evidenced on 1% agarose gel electrophoresis and quantified via a NanoDrop-1000 spectrophotometer (Beckman, USA). All DNA samples were diluted to 10 ng/μL and stored at -20 ℃ until use. For LAMP or PCR detection of Colletotrichum spp. in strawberry tissues either symptomatic or asymptomatic, a simple DNA extraction was accomplished in around 10 min using the lysis buffers of T5 Direct PCR Kit (Plant) (TsingKe Biological Technology, Beijing, China). Briefly, a piece of fresh tissue (leaf blade, petiole or crown) in an appropriate parameter of 2-3 mm was immersed into 50 μL buffer A at 95 ℃ for 10 min, and then mixed with 150 μL buffer B. The diluted lysate was directly used for PCR or LAMP assay.
Amplification with the fungal ITS rDNA (White et al. 1990) and/or strawberry Actin (gene 18570-v1.0-hybrid, FvH4_1g23490) specific primers (Yang et al. 2021) was performed to confirm the quality of DNA template. Six reported barcodes for Colletotrichum spp. were evaluated including β-tubulin 2 (TUB2) (O’Donnell and Cigelnik 1997), Glutamine synthetase (GS) (Prihastuti et al. 2009), Marker1 and Marker 2 (Gan et al. 2017), Virulent-strain specific (Suzuki et al. 2008), and the Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene (ApMat) (Silva et al. 2012). All primer pairs used were shown in Supplementary Table S3.
PCR was performed on an ETC 811 DNA amplifier (Eastwin, Beijing, China) in a 20-μL reaction mixture containing 1 μL DNA template, 3 μM of each primer, 0.5 μL dNTP (2.5 mM), 1x PCR buffer with Mg2+, 1 U Taq DNA polymerase (Biocolor, Shanghai, China). The PCR reaction consisted of an initial denaturation at 94 ℃ for 3 min, followed by 30 cycles of 94 ℃ for 30 s, 60 ℃ for 30 s, 72 ℃ for 30 sec (for strawberry Actin specific primers) or 1 min (for fungal ITS, ApMat, TUB2, GS, Virulent-strain specific, Marker1 and Marker 2 specific primers), then a final extension of 72 ℃ for 5 min. When detecting with a coarse lysate DNA template from fresh strawberry or mycelium tissues, the same PCR system with 35 cycles was performed. PCR products were visualized on 1% (Marker 1 and ApMat specific products) or 1.5% (all the rest barcodes) agarose gel electrophoresis.
Primer design and optimization for LAMP assay to detect both C. gloeosporioides and C. acutatum aggregates
Full length DNA sequences for TUB2 gene of 13 Colletotrichum species related to strawberry anthracnose were collected from GenBank based on a Blast search in NCBI. Sequence alignment revealed the conserved regions in TUB2 gene of C. gloeosporioides and C. acutatum aggregates (Supplementary Fig. S1). A set of six LAMP primers (Table 1) comprising two outer primers F3/B3, two inner primers FIP (F1c-F2)/BIP (B1c-B2), and two loop primers LF/LB were designed in SNAPGENE software following the rules previously described (Notomi et al. 2000).
The components of LAMP assay were largely set as described by Li et al. (2018). The LAMP reaction was initially carried out at 64 ℃ for 60 min in a Digital Dry Bath (MIULAB, Hangzhou, China). DNA of CGMCC3.17371 (C. fructicola) was used as a positive control whereas Botrytis cinerea (BC1) and H2O as negative controls. An optimization of Mg++ concentration ranging from 2 mM to 8 mM revealed that 4 mM MgSO4 provided a sound differentiation, when the color in positive reaction tubes changed from violet to sky blue, whereas a negative one maintained violet. Visual color change was always confirmed by the gel electrophoresis on 2% agarose in 1 x TAE buffer at 95 V for 25 min. LAMP optimization was performed with three replicates in one assay, and independently repeated four times. After optimization, LAMP reaction system in current study was a 25-μL mixture of 2 μL DNA solution (10 ng/μL), 1 μL inner primers FIP/BIP (40 μmol/L), 0.5 μL outer primers F3/B3 (10 μmol/L), 1 μL loop primers LB/LF (40 μmol/L), 2.5 μL dNTPs (10 mmol/L), 1 μLMgSO4 (100 mmol/L), 4 μL Betaine solution (5 mol/L, Sigma, Lot# SLCB2017), 1 μL Bst DNA polymerase Large Fragment (8U/ μL, Vazyme, P701-01), 2.5 μL Hydroxy naphthol blue (HNB) (3 mmol/ L, Sigma, Lot# SHBL8323), and 2.5 μL 10×ThermoPol Buffer. Visual color changes in positive tubes often appeared since 35 min at 64 ℃ and a 50 min bath was enough.
Determining the specificity and sensitivity of LAMP assay
For specificity analysis, the genomic DNAs of six Colletotrichum species and five fungi beyond Colletotrichum genus listed in Supplementary Table S1 were used as templates in LAMP assay adopting the optimal parameters. For sensitivity comparison, LAMP was performed against a serial dilution templates of genomic DNA from C. aenigma, namely, 100 nanogram (ng) /μL, 10 ng/μL, 1 ng/μL, 100 picogram (pg)/μL, 10 pg/μL, 1 pg/μL, 100 femtogram (fg)/μL, and 10 fg/μL. Side-by-side PCR always was carried out with LAMP assay for either specificity or sensitivity test. Both PCR and LAMP reaction products were illustrated by agarose gel electrophoresis. The experiments were independently carried out three times, and each DNA template was tested with three technical replicates one time.
LAMP-PCR combined assay for detecting Colletotrichum spp. in strawberry plants
To validate the detection method in real strawberry, two types of materials including symptomatic plants inoculated with C. fructicola and asymptomatic plants as suspicion of infections from field were used. A direct detection of Colletotrichum spp. in all strawberry tissues started with a 10 min lysis for plant materials using buffers from T5 Direct PCR kit and was followed by a regular LAMP assay as described above. The positive samples revealed by LAMP assay were further examined using a PCR analysis with three barcodes including Marker 1, Marker 2, and ApMat.
To validate the direct LAMP assay for symptomatic plants, five healthy cv. Shanghai Angel plants were inoculated with C. fructicola (CGMCC3.17371). Inoculum was prepared as a conidial suspension from 7 d culture in potato dextrose broth (Lot#0293648, Becton, Dickinson and Company, USA) at 180 rpm under 28 ℃. The resulting conidial suspension was filtered through three layers of lens wiping tissue and adjusted to a concentration of 2×106 per mL by diluting with 0.01% Tween 20 water solution. Plants were inoculated with conidial suspension by misting to runoff using a hand pump sprayer. Control plants were mock-treated with Tween 20 water solution. Plants were immediately placed in a growth incubator (A1000, Conviron, Canada) under 25 ℃ light/23 ℃ dark, and 90-100% relative humidity (100% RH for first two days). The diseased leaf blade, petiole, and crown tissues were sampled at 7-11 days post inoculation. Plant inoculation and sampling were independently repeated twice.
To detect latent infection on strawberry, the compound leaves with petioles were randomly sampled at a lower position from asymptomatic plants in a production field located at Zhuanghang trial station of SAAS, where several dead plants had been eliminated previously. For mycelia culture on diseased plant tissues, three pieces of leaf petioles in an approximate size of 3 x 10 mm were detached and twice sterilized using 70% ethanol for 30 s, followed by a culture at 28 ℃ for two days on PDA plate with 25 mg/L chloramphenicol. The positive samples revealed in LAMP assay were further examined using PCR against mycelia DNA for three barcodes including Marker 1, Marker 2, and ApMat. Plant sampling in field was independently performed three times, and similar results were obtained.
Results
PCR analysis of DNA markers for detecting Colletotrichum spp. pathogenic to strawberry
To identify marker(s) fit for molecular detection of Colletotrichum infection in strawberry, the discernibility of several previously reported barcodes was examined for differentiating between fungal species within and beyond Colletotrichum genus. Amplification of six barcodes displayed distinct profiles in strawberry fungal pathogens (Fig. 1). PCR with ApMat specific primers generated a uniform band in C. fructicola, C. siamense, C. murrayae, and C. gloeosporioides sensu stricto, but no band in C. aenigma while a relatively smaller band in C. nymphaeae of C. acutatum species complex. Amplicon of Marker 2 was uniform in C. murrayae, C. siamense and C. gloeosporioides sensu stricto, smaller in C. aenigma, larger in C. fructicola, whereas absent in C. nymphaeae. By contrast, PCR with Glutamine synthetase (GS) specific primers generated uniform bands in all five species of C. gloeosporioides species complex. Similar with Marker 2, GS related amplification was absent in C. nymphaeae of C. acutatum complex. PCR with TUB2 specific primers produced a uniform band in herein six Colletotrichum species. Notably, the primers specific to GS and TUB2 worked with DNA templates of Nectria pseudotrichia and Fusarium oxysporum too, although with obviously smaller bands. Amplification with the Virulent-strain specific primers seemed specific and limited to C. fructicola. Intriguingly, PCR with Marker 1 specific primers generated highly polymorphic profiles in distinct Colletotrichum spp. When we rearranged these PCR products on 1% agarose gel, a DNA ladder could be observed following the order as indicated in Supplementary Fig. S2.
PCR analysis of previous barcodes in differentiating Colletotrichum spp. endangering strawberry. Reported barcodes including the Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene (ApMat), Glutamine synthetase, Marker 1, Marker2, β-tubulin 2 (TUB2), and Virulent-strain specific were amplified against the genomic DNAs of six Colletotrichum species from diseased strawberry. Five fungi pathogenic to strawberry beyond Colletotrichum genus were used as negative controls. Amplification of the fungal ITS rDNA and strawberry Actin was performed to verify the quality of DNA templates. M: DL2000-bp DNA size marker; C.a: C. aenigma, C.f: C. fructicola, C.n: C. nymphaeae, C.s: C. siamense, C.g: C. gloeosporioides, C.m: C. murrayae; B.c: Botrytis cinerea, N.p: Nectria pseudotrichia, P.c: Pestalotiopsis clavispora, F.ox: Fusarium oxysporum, P.a: Podosphaera aphanis; F.a: Fragaria × ananassa cv. Shanghai Angel, H2O: negative control. The approximate size of PCR amplicons in Colletotrichum spp. was indicated at the left side of each panel
Furthermore, the applicability of six barcodes in detecting Colletotrichum spp. was confirmed in a collection of 40 Colletotrichum isolates (Supplementary Table S2) belonging to two dominant species C. fructicola and C. siamense and the sporadically occurred C. aenigma previously isolated from diseased strawberry (Zhang et al. 2020). The results were largely consistent with above analysis in 11 distinct standard species of strawberry pathogens (Supplementary Fig. S3). Accordingly, the performance and discernibility of individual barcode specific primers in resolving Colletotrichum spp. was summarized (Table 2). TUB2 specific primers generated a uniform band in all six Colletotrichum species, enabling a general diagnosis of Colletotrichum spp. infection, at least for C. gloeosporioides and C. acutatum species aggregates. GS specific primers could distinguish C. gloeosporioides species complex from other strawberry pathogens. PCR with Virulent-strain specific primers could be used to specifically recognize C. fructicola. Marker 2 can distinguish C. aenigma and C. fructicola from the rest Colletotrichum species here examined. Marker 1 seems promising in clarifying the genetic limits among six Colletotrichum spp, differentiating anthracnose fungi at a species level. A combined use of these primers in DNA diagnosis could provide a systematic and comprehensive diagnosis, meeting the needs of different levels.
TUB2-based LAMP assay for detecting C. gloeosporioides and C. acutatum species aggregates
TUB2 has been considered as one of the most popular diagnostic markers for Colletotrichum species (Damm et al. 2012). In current study, PCR with TUB2 -specific primers produced uniform bands in Colletotrichum spp. and different bands in Nectria pseudotrichia and Fusarium oxysporum. Since LAMP based DNA amplification requires a set of four or six specific primers that recognize six distinct regions on the template DNA, this method has an extremely higher specificity than PCR (Notomi et al. 2000). Accordingly, TUB2 gene was selected to establish a LAMP assay for a specific detection of both C. gloeosporioides and C. acutatum species aggregates.
There exist at least 15 species belonging to C. gloeosporioides and C. acutatum aggregates reported to be pathogenic to strawberry (Weir et al. 2012; Han et al. 2016; Wang et al. 2019; Chung et al. 2020). TUB2 sequences in GenBank were not available for two species C. changpingense (Jayawardena et al. 2016) and C. miaoliense (Chung et al. 2020). Based on the TUB2 sequences of the rest 13 Colletotrichum species belonging to two aggregates virulent to strawberry (Supplementary Fig. S1), a set of six primers consisting of two outer primers, two inner primers and two loop primers were designed for LAMP assay (Table 1). Side-by-side comparison between PCR and LAMP was always carried out to examine the efficiency of LAMP assay. Typical ladder-like bands were observed on gel electrophoresis of LAMP products for all six Colletotrichum species (Fig. 2). No amplification was present in host strawberry or in fungi beyond Colletotrichum genus. When HNB was used as a visual indicator, color change from violet to sky blue was observed as expected in the tubes of Colletotrichum samples, while the color remained violet in other fungal or healthy strawberry sample tubes. Both gel electrophoresis and visual color change indicated that the LAMP reaction could specifically distinguish Colletotrichum spp. from host DNA and five other fungal pathogens of strawberry.
LAMP assay using six primers specific to β-tubulin 2 (TUB2) (Table 1) allowed a Colletotrichum spp. specific amplification. a Color changes evidencing the products of LAMP using hydroxynaphthol blue (HNB) as a visual indicator. b Detection of LAMP products via 2% agarose gel electrophoresis. The amplification templates were the same genomic DNAs (as in Fig. 1) of 11 fungal isolates infectious to strawberry including six Colletotrichum species (Lanes C.a, C.f, C.n, C.s, C.g, C.m) and five beyond Colletotrichum (Lanes B.c, N.p, P.c, Fox, P.a). Lanes F.a: strawberry, M: DL2000 DNA ladder, H2O: negative control
To reveal the sensitivity of the LAMP assay, a 10-fold serial dilution of the genomic DNA of C. aenigma (strain GQH124) from 100 ng to 10 fg was simultaneously tested in PCR and LAMP assay. Both the visual color changes (Fig. 3a) and gel electrophoresis (Fig. 3b) indicated that LAMP reaction successfully worked with DNA templates at concentrations ranging from 100 ng to 100 pg. Conventional PCR using the primers TUB2-T1 and TUB2-R2 generated a weak band in a reaction with 1 ng DNA template (Fig. 3c). Clearly, the LAMP assay exhibited a lower detection limit, with at least 10 times higher sensitivity in DNA diagnosis for strawberry anthracnose than PCR.
Sensitivity of LAMP assay specific to Colletotrichum spp. pathogenic to strawberry. a Color changes evidencing the LAMP products using HNB. b Agarose gel electrophoresis of LAMP products. c Electrophoresis results of conventional PCR using TUB2-T1 and TUB2-R2 primers (Supplementary Table S3). A gradient dilution of the genomic DNA from C. aenigma, one species causing strawberry anthracnose in Shanghai was used for LAMP and a comparative PCR analysis. Lanes 1-8: 100 ng/μL, 10 ng/μL; 1 ng/μL; 100 pg/μL; 10 pg/μL; 1 pg/μL, 100 fg /μL, 10 fg /μL DNA. M:DL2000 DNA ladder
Direct LAMP detection of Colletotrichum spp. in symptomatic strawberry plants
Different parts from strawberry plants of cv. Shanghai Angel inoculated with C. fructicola were collected at 7-11 days post inoculation together with a control healthy leaf sample (Fig. 4a). The coarse DNA solutions were obtained from the leaf blade, petiole, and crown tissues after a simple pretreatment at 95 ℃ for 10 min in a metal dry bath (Fig. 4b). Then, a LAMP assay was performed using the optimized reaction system as described above. The visual color changes showed that this direct LAMP detection of Colletotrichum infection successfully worked with simply pretreated strawberry samples (Fig. 4c). The LAMP amplification indicated by coloring change was confirmed by gel electrophoresis (data not shown). Color changes in reaction tubes appeared since 35 to 40 min bathing at 64 ℃ in three independent repeats of LAMP assay.
Direct LAMP and PCR detection of Colletotrichum spp. in diseased strawberry. The symptomatic leaf blade (1), petiole (2), and crown (3) tissues were sampled from strawberry plants cv. Shanghai Angel infected with C. fructicola at 7-11 days post inoculation (dpi). The healthy leaf blade tissue (4) was used as a negative control. a The morphology of diseased and healthy strawberry tissues. b Pretreatment of fresh strawberry tissues using lysis buffer A after at 95 ℃ for 10 min. c Direct LAMP assay for the detection of Colletotrichum spp. in strawberry. d Direct PCR detection of Colletotrichum spp. in symptomatic strawberry samples
Comparatively, a direct PCR was performed using TUB2 specific primers to detect Colletotrichum spp. in diseased strawberry. A consistent result was obtained (Fig. 4d), although about 2.5 hr were used for PCR diagnosis. Apparently, no need for a thermal cycler and an electrophoresis apparatus, a DNA diagnosis of Colletotrichum spp. in strawberry samples could be accomplished in one hour via LAMP in a portable dry bath.
LAMP-PCR facilitated a diagnosis of Colletotrichum spp. in asymptomatic strawberry
High quality and timely information on latent infection is crucial for controlling the epidemic of strawberry anthracnose. In this study, we validated the ability of LAMP assay followed by a PCR analysis in detecting and differentiating Colletotrichum spp. in asymptomatic strawberry plants collected from a production field in a SAAS trail station at Zhuanghang Town, Fengxian District, Shanghai.
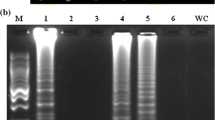
Genomic DNA from the outer leaves at a lower position of asymptomatic plants in field was released at 95 ℃ for 10 min and used in a direct LAMP and PCR detection. Seven positive samples were detected from 20 strawberry plants in LAMP assay, while only four positive samples were detected in conventional PCR (Fig. 5a, b). Apparently, the detection ratio of Colletotrichum spp. in asymptomatic plants was higher for LAMP assay than PCR. In addition, a two days culture at 28 ℃ on PDA for surface-sterilized strawberry samples revealed the fungal mycelium growth on some samples (Fig. 5c). PCR analysis against the mycelium DNA using Colletotrichum TUB2 specific primers suggested that LAMP assay result was consistent with tissue separation and mycelium diagnosis (Fig. 5d). Furthermore, PCR analysis using three additional barcodes including ApMat, Marker1, and Marker 2 distinguished among Colletotrichum species. Six of seven positive samples were infected by C. siamense, and the rest one (No.5) was infected by C. fructicola. Therefore, strawberry plant direct LAMP integrated with PCR developed in this study, not only provided a sensitive and practical diagnosis of latent Colletotrichum infection on strawberry, but also differentiated anthracnose fungi at a species level, supporting a promising chance to monitor Colletotrichum spp. population in field. Finally, the schematic flow of the combined LAMP-PCR assay for latent infection of anthracnose fungi on strawberry developed in this study was summarized in Fig. 6.
LAMP and PCR combined assay resolved the detection and differentiation of latent Colletotrichum infection on strawberry collected from a production field. a Direct LAMP detection of latent Colletotrichum spp. infection on strawberry (an outer petiole at a lower position sampled). b Direct PCR (35 cycles) using primers specific to TUB2 detecting latent infection of Colletotrichum spp. c The fungal colonies associated with the detached leaf petiole samples at 28 ℃ for 2 d. d PCR assay (35 cycles) against additional barcodes (Marker1, Marker 2, ApMat) distinguished latent infection on strawberry at a species level
Schematic flow of the combined LAMP-PCR assay for detecting latent infection of anthracnose fungi on strawberry. The DNA diagnosis consists of seven steps: sampling in the field (Step 1); lysis of fresh strawberry samples at 95 ℃ for 10 min in a digital dry bath (Step 2); a direct LAMP assay for a general detection of Colletotrichum at 64 ℃ for 35-50 min (Step 3); culturing and fungi separation at 28 ℃ for 2 d (Step 4); lysis of fresh fungal mycelium at 95 ℃ for 10 min (Step 5); PCR targeting four barcodes successively (TUB2, ApMat, Marker 2, Marker 1) (Step 6) followed by an electrophoresis analysis on 1-1.5% agarose (Step 7) were performed for a species level diagnosis accomplished in two hours. The experimental details are described in the text
Discussion
The current work found that PCR with the Virulent-strain specific primers (Suzuki et al. 2008) amplified a band unique to C. fructicola (Fig. 1, Supplementary Fig. S3). This pair of primers had been used in classifying Colletotrichum isolates as virulent or less virulent (Gan et al. 2017). Clearly, this barcode might be not relevant to pathogenicity differentiation in Colletotrichum genus. In our previous work, both virulent and less virulent strains were identified in C. fructicola and C. siamense, indicating that a specific species is not closely correlated with its virulence to a certain host (Zhang et al. 2020).
Marker 1 and Marker 2 were developed to distinguish Colletotrichum species pathogenic to strawberry in Japan (Gan et al. 2017). In that work, PCR with Marker 1 specific primers obtained one band of polymorphic sizes in different isolates of certain species, either in C. fructicola, C. siamense, or C. aenigma. However, in this work, a uniform band in each species was observed for Marker 1 specific PCR (Supplementary Fig. S3), at least against 40 Colletotrichum strains identified by a multi-locus phylogeny analysis (Zhang et al. 2020). This barcode indeed holds an ideal discernibility in identifying Colletotrichum at a species level, although the applicability should be tested in more fungal strains reliably identified using multi-gene phylogeny in the future. In addition, current PCR using Marker 1 produced a uniform band in all 19 strains of C. siamense whereas a distinct band in C. murrayae (Supplementary Fig. S3, Fig. 1). Much work is needed to ascertain whether C. murrayae and C. siamense were conspecific as previously suggested (Liu et al. 2016).
It was reported that Marker 2 could distinguish among C. fructicola, C. siamense and C. aenigma, since a common specific band was obtained in each species, and species could be differentiated based on amplicon size (Gan et al. 2017). However, the current work suggested that this barcode from PCR was of same size in C. siamense, C. gloeosporioides sensu stricto, and C. murrayae. Marker 2 also displayed polymorphic weak amplification in 20 C. fructicola strains (Supplementary Fig. S3). These inconsistences might result from the confusion in identifying causal agents for strawberry anthracnose at a species level for a long period, which is common in characterizing many plant pathogens. For example, a primer CgInt related to ITS rDNA was supposed to be specific to C. gloeosporioides (Mills et al. 1992), which has been used for a long time in strawberry anthracnose diagnosis even till 2015 (Rahman et al. 2015). Indeed, the Cg Int primer was found to not match many taxa within C. gloeosporioides complex except for C. fructicola and C. siamense (Weir et al. 2012).
LAMP assay is applicable for a fast, sensitive and on-site diagnosis of plant pathogens. In this study, a LAMP assay was successfully established beginning with a 10 min lysis of strawberry tissues in a parameter of 2-3 mm. This method allowed a direct detection of most reported Colletotrichum species of C. gloeosporioides and C. acutatum complexes within one hour. The current LAMP assay could be further improved for the convenience of strawberry farmers and used in practical fields, although we should be more cautious in providing an accurate diagnosis based on LAMP assay, considering the site of sampling tissues, the potential existence of dead spores of Colletotrichum spp., and no differentiation between fungi species. Since the fungal pathogen may localize in an unsampled part of a plant at the initial stage of infection, the possibility of incorrect judgment resulting from sampling limits could not be eliminated in a direct on-site DNA diagnosis. Without an initial culturing of fungi, all DNA diagnosis methods have a risk of confusing between dead and living pathogens. Since different Colletotrichum species might use different infection strategies and require distinct management, it is important to distinguish between pathogen species. For example, the aggressive C. siamense and C. fructicola caused larger lesions on wounded and non-wounded strawberry leaves, while the opportunistic C. boninense, C. karstii, and C. miaoliense only caused lesions on wounded leaves (Chung et al. 2020). Simply, when no aggressive Colletotrichum spp. has been detected, fungicide is not needed in a strawberry field, so long as avoiding wounding caused by agricultural practice. These facts re-enforced the need of establish an on-site LAMP and in lab PCR combined method to ensure a sensitive and comprehensively accurate diagnosis of Colletotrichum spp. on strawberry.
The LAMP-PCR combined method developed in this study combined the advantages of LAMP as a sensitive, affordable, and on-site diagnosis of latent Colletotrichum infection on strawberry, as well as the high discernibility of PCR in lab in distinguishing Colletotrichum species, which might support a definitive pathogen identification for strawberry anthracnose management. This method could detect Colletotrichum spp. of both C. gloeosporioides and C. acutatum species complexes. Using this method we successfully differentiated two dominant species C. fructicola and C. siamense in asymptomatic strawberry. Due to the limited inclusivity of fungal isolates tested in current study, the ability of current method to differentiate other species (C. gloeosporioides, C. murrayae, C. aenigma, C. nymphaeae) seems promising but validation work is needed in the near future.
References
Azevedo-Nogueira F, Gomes S, Lino A, Carvalho T, Martins-Lopes P (2021) Real-time PCR assay for Colletotrichum acutatum sensu stricto quantification in olive fruit samples. Food Chem 339:127858
Buddie AG, Martinez-Culebras P, Bridge PD, García MD, Querol A, Cannon PF et al (1999) Molecular characterization of Colletotrichum strains derived from strawberry. Mycological Research 103:385–394
Cannon PF, Damm U, Johnston PR, Weir BS (2012) Colletotrichum – current status and future directions. Stud Mycol 73:181–213
Chung PC, Wu HY, Wang YW, Ariyawansa HA, Hu HP, Hung TH, Tzean SS, Chung CL (2020) Diversity and pathogenicity of Colletotrichum species causing strawberry anthracnose in Taiwan and description of a new species Colletotrichum miaoliense sp. nov. Sci Rep 10(1):14664
da Silva LL, Moreno HLA, Correia HLN, Santana MF, de Queiroz MV (2020) Colletotrichum: species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl Microbiol Biotechnol 104(5):1891–1904
Damm U, Cannon PF, Woudenberg JHC, Crous PW (2012) The Colletotrichum acutatum species complex. Stud Mycol 73:37–113
Damm U, Sato T, Alizadeh A, Groenewald JZ, Crous PW (2019) The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud Mycol 92:1–46.
Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13(4):414–30
Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, Smith RD, Teresi SJ, Nelson ADL, Wai CM, Alger EI, Bird KA, Yocca AE, Pumplin N, Ou S, Ben-Zvi G, Brodt A, Baruch K, Swale T, Shiue L, Acharya CB, Cole GS, Mower JP, Childs KL, Jiang N, Lyons E, Freeling M, Puzey JR, Knapp SJ (2019) Origin and evolution of the octoploid strawberry genome. Nat Genet 51(3):541–547
Freeman S, Katan T (1997) Identification of Colletotrichum Species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathology 87(5):516–521
Furuta K, Nagashima S, Inukai T, Masuta C (2017) Construction of a system for the strawberry nursery production towards elimination of latent infection of anthracnose fungi by a combination of PCR and microtube hybridization. Plant Pathol J 33(1):80–86
Gan P, Nakata N, Suzuki T, Shirasu K (2017) Markers to differentiate species of anthracnose fungi identify Colletotrichum fructicola as the predominant virulent species in strawberry plants in Chiba Prefecture of Japan. J Gen Plant Pathol 83:14–22
Goto M, Honda E, Ogura A, Nomoto A, Hanaki K (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Bio Techniques 46(3):167–172
Han YC, Zeng XG, Xiang FY, Ren L, Chen FY, Gu YC (2016) Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei. China. Plant Dis 100:996–1006
Hara-Kudo Y, Nemoto J, Ohtsuka K, Segawa Y, Takatori K, Kojima T, Ikedo M (2007) Sensitive and rapid detection of Vero toxin-producing Escherichia coli using loop-mediated isothermal amplification. J Med Microbiol 56(Pt 3):398–406
Jayawardena RS, Huang JK, Jin BC, Yan JY, Li XH, Hyde KD, Bahkali AH, Yin SL, Zhang GZ (2016) An account of Colletotrichum species associated with strawberry anthracnose in China based on morphology and molecular data. Mycosphere 7:1147–1163
Li Q, Shen D, Yu J, Zhao Y, Zhu Y, Dou D (2018) Rapid detection of Pythium aphanidermatum by loop-mediated isothermal amplification. Journal of Nanjing Agricultural University 41(1):79–87 (In Chinese)
Liu F, Wang M, Damm U, Crous PW, Cai L (2016) Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evol Biol 16:81
Liu H, Xie WF, Zhang L, Valpuesta V, Ye ZW, Gao QH, Duan K (2014) Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J Integr Plant Biol 56(4):350–63
Martínez-Culebras PV, Barrio E, García MD, Querol A (2000) Identification of Colletotrichum species responsible for anthracnose of strawberry based on the internal transcribed spacers of the ribosomal region. FEMS Microbiol Lett 189(1):97–101
Martinez-Culebras PV, Querol A, Suarez-Fernandez MB, Garcia-Lopez MD, Barrio E (2003) Phylogenetic relationships among Colletotrichum pathogens of strawberry and design of PCR primers for their identification. J Phytopathol 151:135–143
Mills PR, Sreenivasaprasad S, Brown AE (1992) Detection and differentiation of Colletotrichum gloeosporioides isolates using PCR. FEMS Microbiol Lett 98:137–144
Ning Z, Yi X, Lan W, Huang X, Zhang D, Dong L, Qian X (2019) Isolation and Identification of the Pathogen of Red Leaf Disease in “Sweet Charlie” Strawberry in Anhui Province. Molecular Plant Breeding 17(15):5051–5056 (In Chinese)
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:63e
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
Parikka P, Lemmetty A (2004) Tracing latent infection of Colletotrichum acutatum on strawberry by PCR. Eur J Plant Pathol 110:393–398
Prihastuti H, Cai L, Chen H, McKenzie E, Hyde KJFD (2009) Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers 39:89–109
Quan PL, Sauzade M, Brouzes E (2018) dPCR: A Technology Review. Sensors (Basel) 18(4):1271
Rahman M, Ojiambo P, Louws F (2015) Initial inoculum and spatial dispersal of Colletotrichum gloeosporioides, the causal agent of strawberry anthracnose crown rot. Plant Dis 99(1):80–86
Ren XJ, Liang Y, Lu JP, Yang BR, Dai FM (2008) Identification of Colletotrichum spieces from strawberry in Shanghai. Acta Phytopathol Sinica 3:325–328 (In Chinese)
Silva DN, Talhinhas P, Várzea V, Cai L, Paulo OS, Batista DJM (2012) Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Plant Pathol 104:396–409
Suzuki T, Tanaka-Miwa T, Ito Y, Uematsu S, Hirayama Y, Okayama K (2008) Development of specific PCR primers designed for the detection of Colletotrichum gloeosporioides causing strawberry anthracnose (abstract in Japanese). Jpn J Phytopathol 74:198
Tao G, Hyde KD, Cai L (2013) Species-specific real-time PCR detection of Colletotrichum kahawae. J Appl Microbiol 114(3):828–35
Wang NY, Forcelini BB, Peres NA (2019) Anthracnose fruit and root necrosis of strawberry are caused by a dominant species within the Colletotrichum acutatum species complex in the United States. Phytopathology 109(7):1293–1301
Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology 73:115–180
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. Academic Press New York pp:315-322
Wu JY, Hu XR, Zhang CQ (2019) Molecular detection of QoI resistance in Colletotrichum gloeosporioides causing strawberry anthracnose based on loop-mediated isothermal amplification assay. Plant Dis 103:1319–1325
Yang HC, Haudenshield JS, Hartman GL (2015) Multiplex Real-time PCR Detection and differentiation of Colletotrichum Species Infecting Soybean. Plant Dis 99(11):1559–1568
Yang J, Ding Z, Wang J, Tian S, Duan K, Gao Q (2021) Bet v 1 potential allergens are involved in anthracnose resistance of strawberry varieties. J Berry Res 11:21–32
Zhang LQ, Song LL, Duan K, Zhang L, Gao QH (2018) First report of Nectria pseudotrichia causing crown rot of strawberry in China. Plant Dis 102(8):1655
Zhang L, Song L, Xu X, Zou X, Duan K, Gao Q (2020) Characterization and fungicide sensitivity of Colletotrichum species causing strawberry anthracnose in Eastern China. Plant Dis 104(7):1960–1968
Zhao MZ, Wang J, Wang ZW, Qian YM, Wu WM (2012) Strawberry production and trade in the world. Fruit Grow Friend 6 (In Chinese)
Acknowledgments
This work was funded by Shanghai Municipal Agriculture Commission Project (Grant No. Hu Nong Ke Zhong Zi-2017-2-1). We are grateful to Dr. Jiayue Feng of Northwest A&F University for kindly sharing the genomic DNA of Podosphaera aphanis, Professor Kai Zhao of SAAS for valuable guidance in LAMP assay, as well as Professor Daolong Dou and Dr. Danyu Shen of Nanjing Agricultural University for critical discussion and suggestions. Thanks are due to anonymous reviewers for valuable comments which improved this manuscript.
Funding
This work was funded by Shanghai Agriculture Applied Technology Development Program, China (Grant No. Z201702) to Qinghua Gao.
Author information
Authors and Affiliations
Contributions
K. D. and Q-H. G conceived this work. Y. L. carried out most experiments and data analysis. Y. J. performed pathogen culture work. L-L. S., L-Q. Z., Y-C. H., and Z-Y. N. contributed to purification of all fungal isolates used in this work. W-Z. Y sampled diseased strawberry. Y. L. and K. D. prepared the manuscript. All authors contributed to revising and approved the manuscript.
Corresponding authors
Ethics declarations
Research involving human and animal participants
This research did not involve any human participants or animals.
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Ji, Y., Han, Y. et al. Loop-mediated isothermal amplification and PCR combined assay to detect and distinguish latent Colletotrichum spp. infection on strawberry. J Plant Pathol 103, 887–899 (2021). https://doi.org/10.1007/s42161-021-00873-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-021-00873-7