Abstract
The pathogenicity of four Fusarium oxysporum isolates collected from symptomatic oil palm (Elaeis guineensis Jacq.) from Ghana were tested for the first time to develop a new pathogenicity assay for Fusarium oxysporum f. sp. elaeidis (FOE) infection in oil palm seedlings. All four FOE isolates used for pathogenicity assay were pathogenic to oil palm seedlings within a relatively short time compared to other pathogenicity studies, for which infection/symptoms in oil palm seedlings was time consuming. FOE and “presumed-FOE” (i.e. Fusarium isolates collected from symptomatic oil palm trees whose pathogenicity is not confirmed) were characterised based on partial sequences of a housekeeping gene EF-1α and three Secreted In Xylem genes (SIX8, SIX9 and SIX11). All the phylogenetic trees generated for EF-1α, SIX8, SIX9 and SIX11 showed some variation between FOE, and “presumed-FOE”, but could not cluster isolates based on geographical location. Phylogenetic trees for EF-1α and SIX (SIX9 and SIX11) genes clustered both FOE and “presumed-FOE” from FUSARIUM-ID from GenBank, but SIX8 could not.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pathogen Fusarium oxysporum f. sp. elaeidis (FOE) is a soil borne fungus and causes fusarium wilt disease in the world’s highest oil producing crop, the oil palm (Elaeis guineensis Jacq.) (Corley 2009; Hansen et al. 2015). The pathogen is present in African countries including Ghana, Democratic Republic of Congo and Cameroon (Prendergast 1957; Aderungboye 1982; Corley and Tinker 2003) where oil palm is produced. In Ghana, management of the pathogen such as removal and burning of infected oil palm trees is the only method of control over years in various oil palm plantations.
Although the pathogenicity of Fusarium oxysporum isolates collected from oil palm showing symptoms of fusarium wilt disease have been successfully studied in oil palm seedlings, these pathogenicity tests were extremely time consuming and laborious (Flood et al. 1993; Mepsted et al. 1994a; Rusli et al. 2015). Molecular methods have a strong impact on characterising or fingerprinting isolates of plant pathogens and in the last two decades, use of molecular techniques have expanded due to their sensitivity and accuracy. Tools that have been used for identifying variation in a wide range of F. oxysporum f. sp. include Inter-Simple Sequence Repeat (ISSR) (Thangavelu et al. 2012; Baysal et al. 2013; Liu et al. 2014; Hannachi et al. 2015) or Simple Sequence Repeat (SSR) markers (Bogale et al. 2005; Datta et al. 2011; Datta and Lal 2013). In addition, efforts have been made to use other gene of interest such as housekeeping genes (Baayen et al. 2000; Bogale et al. 2005, 2009; Silva et al. 2014). More so, other studies have been conducted to associate pathogenicity with the presence or absence of effector proteins such as Secreted In Xylem (SIX) genes within other F. oxysporum ff. spp. (Sasaki et al. 2015; Taylor et al. 2016). These molecular tools have been applied to identify and study the relationship or genetic variation of different strains of F. oxysporum (Baayen et al. 2000; Bogale et al. 2005; Silva et al. 2014; Sasaki et al. 2015; Taylor et al. 2016).
The primary objective of this research was to develop pathogenicity assay that would reduce the amount of time necessary for symptom development in oil palm (Elaeis guineensis. Jacq.) using four FOE isolates. Although many studies have tried to understand the association between pathogenicity either the presence or absence of SIX (Secreted In Xylem) genes in F. oxysporum ff. spp. (Sasaki et al. 2015; Taylor et al. 2016), in this present study, the authors considered only to determine the presence or absence of SIX (1–14) in the four FOE isolates used for the pathogenicity test and“presumed-FOE” (i.e. Fusarium isolates collected from symptomatic oil palm trees whose pathogenicity is not confirmed). Phylogenetic trees were also generated to characterise Fusarium oxysporum f. sp. elaeidis (FOE) and “presumed-FOE” isolates via sequencing genes based on three SIX (8, 9 and 11) genes and EF-1α.
Materials and methods
Field sampling
Sampling was done from mature oil palm randomly selected in plantations from two major regions in Ghana (Western and Eastern). In the Eastern Region, the Oil Palm Research Institute (OPRI), and in the Western Region, two different oil palm plantations, namely Norpalm and Benso Oil Palm Plantation (BOPP) and collected samples coded as shown in Table S1. Oil palms showing symptoms of fusarium wilt disease were found at all three sites. In addition to these samples (Table S1) collected from Ghana (West Africa), two other Fusarium isolates, DR CONGO-Z and SURINAME-S (originally from the Democratic Republic of Congo and Suriname respectively) were obtained from the CBS-KNAW culture collection, the Netherlands and two isolates DR CONGO-F3 and Ivory Coast-16F (Table S1) used by Rusli et al. (2015) from the University of Bath-UK. Originally, these two isolates of FOE (F3 and 16F) were from diseased oil palms from the Democratic Republic of Congo (DRC) and Ivory Coast respectively. Isolate 16F was also used by Institut de Recherches pour les Huiles et Oleagineux (IRHO) as an isolate for screening fusarium wilt disease in oil palm (Mepsted et al. 1994a, b).
Plant materials
Oil palm seedlings Tenera or ‘commercial type’ were obtained using the assisted pollination method (Teo 2015). Pre-germinated seeds were transplanted to seed trays (9 mm) filled with a mixture of soil (Levington F2 seed Modular Compost) and perlite at the ratio of 8:1 and maintained in a controlled glasshouse environment (30 °C, 90% RH and 12 h photoperiod), with light [240 μ mol m−2 s−1 photo flux density (PFD)]. The RH in the glasshouse was reduced to 80% after 2 months when the oil palm seedlings were transplanted into Soparco black pots of diameter 13 cm filled a mixture of sand based soil (John Ines No. 3), perlite and vermiculite at a ratio of 8:1:1. Light levels were maintained between 800 and 500 μ mol m−2 s−1 with a day length of 14–17 h; humidity and temperature ranged from 60 to 80% and 20 to 35 °C, respectively. The seedlings were watered at the base at regular intervals when necessary. Conditions in the glasshouse were maintained throughout the experiments.
Pathogenicity test: Preparation of FOE inoculum and inoculation procedures
Spores were collected from BOPP-B5, OPRI-5, NORPALM-N5 and 16F Fusarium oxysporum isolates that were cultured on PDA and incubated at 29 °C for 7 days. Spore suspension was prepared, filtered through Miracloth (CALBIOCHEM) to remove mycelial fragments and the filtrate centrifuged at 670 x g for 10 min with the Centaur 2 (MSE). The sedimented spores were gently resuspended in 50 ml Falcon tubes with sterile distilled water and adjusted to 3 × 106 spores ml−1. Three (3) month-old oil palm seedlings (Fig. 2a) (Tenera or commercial type) were removed from the pot, roots were washed with distilled water followed by wounding the roots with sterile syringe. After wounding the roots, inoculation with 3 × 106 spore ml−1FOE suspension was carried out in two different treatments as described below.
-
1.
The washed roots of oil palm seedlings soaked in FOE suspension, 3X106 spores ml−1 (Fig.2b) overnight with slight wounding around the roots.
-
2.
The washed roots of oil palm seedlings soaked in FOE suspension, 3X106 spores ml−1 (Fig. 2b) overnight without wounding.
The treated oil palm seedlings were assessed (i.e. presence or absence of symptoms) on weekly bases until first symptom observation, followed by testing with primer pairs (Table S2) via PCR. The roots of the symptomatic plants were qualitatively assessed since one of the major symptoms for FOE infection in oil palm is discolouration of roots (Rusli et al. 2015). All oil palm seedlings used as negative controls for pathogenicity test had the roots either wounded or not-wounded, soaked in sterilised distilled water overnight. FOE isolate 16F pathogenicity was confirmed (Flood et al. 1993; Mepsted et al. 1994a, b; Rusli et al. 2015) in oil palm, hence this was selected as the reference (positive control) FOE isolate. Experimental design used was Completely Randomised Design (CRD) and experiment repeated (conducted) three times. Each treatment was replicated three times. Statistical analysis of variance (ANOVA) was done with GenStat 16th Edition.
Root samples with length (0.5–1.0 cm) were collected from both asymptomatic and symptomatic oil palm seedlings and plated on Potato dextrose agar (PDA) purposely for FOE (BOPP-B5, OPRI-5, NORPALM-N5 and 16F) identification and confirmation via PCR. This was done for every repeated pathogenicity test or experiment. The “presumed-FOE” isolates collected from symptomatic oil palm from various sampling sites, were also tested for the presence or absence of SIX (1–14), although not used for inoculation/pathogenicity trials. “Presumed-FOE” isolates were not used for pathogenicity test due to the limited numbers of oil palm seedlings available.
DNA extraction, PCR and DNA sequencing for FOE or “presumed-FOE” identification
Mycelium (50–100 mg) of FOE and “presumed-FOE” was scraped from the PDA plates using a sterile surgical blade. Tissue disruption was carried out using glass beads and homogenizer (FastPrep®) at a speed of 6.5 m/s for 45 s in the presence of liquid nitrogen. DNA extractions of the cultures were then carried out using DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. Polymerase chain reaction (PCR) of various regions of the template DNA was performed using primer pairs of interest (Table. S2). PCR was carried out in 30 μl volumes consisting of 15 μl of master mix (MangoTaq™ DNA Polymerase), 1 μl (of 10 pmol / ul) each, 12 μl sterile distilled water and 1 μl of template DNA of the isolates of interest. The reaction was performed in a BIO-RAD S1000 Thermal Cycler with the amplification conditions of 95 °C for 2 min for initial denaturation, followed by 35 cycles of denaturation at 95 °C for 2 min, annealing at temperatures suitable for amplification for each primer pair of interest and extension/elongation at 72 °C for 1 min 30 s. The final extension was set at 72 °C for 5 min. PCR products were cleaned using the QIAquick PCR Cleanup kit (Qiagen) following manufacturer’s instruction and DNA concentration was estimated by nano-drop and using 1 kb ladder (Promega). Sequencing reactions were performed by Fisher Scientific or MyGATC. BLAST searches were performed using the GenBank sequence database to confirm the identity of the fungal isolates sequences based on the translation elongation factor-1α (EF-1α) and Secreted In Xylem (SIX-1, 8, 9, 10, 11 and 12) genes. The output from BLAST algorithms was used to query any unknown sequences against the database of all the fungal gene regions. Sequences for EF-1α, SIX8, SIX9 and SIX11 were subsequently submitted to National Centre for Biotechnology Information (NCBI) GenBank for accession number (Table S1) for the eleven isolates of Fusarium oxysporum from symptomatic oil palm.
Phylogenetic analysis
The EF-1α, SIX8, SIX9 and SIX11 partial sequences were manually edited and aligned with the MEGA6 software (Tamura et al. 2013). Phylogenetic analyses were conducted with MEGA6 on the combined selected data set for eleven Fusarium oxysporum isolates collected from symptomatic oil palm with seven reference genes from GenBank (Table S3) to support the EF-1α phylogenetic tree in addition to three known EF-1α FOE sequences host specific to other palms and not oil palm (Table S3). The phylogenetic trees for SIX8, SIX9 and SIX11 were eleven partial sequences of Fusarium oxysporum isolates from symptomatic oil palm with various reference genes from GenBank (Table S3). Six reference genes were used to support SIX8 phylogenetic tree, whereas seven reference genes from GenBank supported both SIX9 and SIX11 phylogenetic trees (Table S3). Unweighted parsimony analyses were performed. Clade stability was assessed by 1000 parsimony bootstrap replications and the grouping statistical method was Unweighted Paired Group Method with Arithmetic Average (UPGMA). Bootstrapping values were shown in the branch nodes of the phylogenetic trees. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was assessed to determine the clade stability.
Results
Sampling
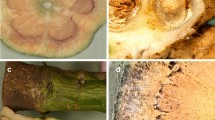
In the study, infected oil palm were identified in the Eastern Region - Oil Palm Research Institute (OPRI), and in the Western Region, two different oil palm plantations, namely Norpalm and Benso Oil Palm Plantation (BOPP). Generally, sampled tissues (Fig. 1a) from the stem or trunk of symptomatic oil palm showed discolouration (Fig. 1b). A cross-section view (Fig. 1e) of symptomatic oil palm leaf frond or branch showed brownish colour (Fig. 1e). Discoloured tissues were absent in both the sampled tissues from stem or trunk and the cross-section view of the leaf frond or branch was also without discouloration in asymptomatic oil palm (Fig. 1c, d).
Pictures showing (a) Drilling process with incremental stem borer in oil palm trunk (b) Drilled core (discoloured/brown/spotted) sample from trunk of diseased oil palm (c) Drilled core (clean) sample from trunk of a healthy oil palm (d) Cross-sectional view of a healthy oil palm lower branch or frond (e) Cross-sectional view of diseased oil palm blocked (brownish part) vascular section lower branch or frond
Pathogenicity test
In the pathogenicity assay for this study, 16F FOE isolate was used as the reference isolate besides Fusarium oxysporum isolates collected from Ghana (OPRI-5, BOPP-B5 and NORP-N5). Oil palm seedlings that had either minor wounds or no wounds on the roots soaked in 3 × 106FOE spores overnight (Fig. 2) before potting also showed symptoms at 4 and 7 months respectively (Fig. 3). However, ANOVA showed there was no significant difference (P > 0.05) among the FOE isolates in the time of first symptoms. The discolouration of the roots as a result of infection by FOE are shown in Fig. 4 b, c. Root discolouration progressed with time in infected plants but did not vary among FOE isolates. Data was not analysed for root discolouration. Roots were tested for the presence of FOE by plating on PDA supplemented with antibiotics (Fig. 5 a, b) and DNA followed by PCR (Table 1).
Identification of SIX genes and EF-1α in four FOE isolates and Fusarium oxysporum isolates from symptomatic oil palm
Secreted In Xylem (SIX8, SIX9 and SIX11) genes were identified within the pathogenic FOE and “presumed-FOE” from Ghana, Ivory Coast, DR Congo and Suriname (Table 2). The SIX gene primers were from published research (Lievens et al. 2009; Taylor et al. 2016). Secreted In Xylem genes (SIX2, SIX3, SIX4, SIX5, SIX6, SIX7, SIX10, SIX13 and SIX14) were not identified within any of the FOE and “presumed-FOE” isolates but SIX1 was present in only FOE (DR Congo-F3) isolate (Table 2) whereas SIX12 was present in FOE OPRI-5 and OPRI-11. EF-1α was identified in all the FOE and “presumed-FOE” isolates based on the similarity with known FOE isolates.
Molecular characterisation of four FOE isolates and Fusarium oxysporum isolates from symptomatic oil palm based on SIX8, SIX9, SIX11 and EF-1α
Characterising the Fusarium oxysporum isolates collected from oil palm, the phylogenetic trees showed SIX gene sequences considerably varied across the FOE or “presumed-FOE”. The phylogenetic trees showed that FOE and “presumed-FOE” isolates could not be clustered based on geographical location using SIX8, SIX9 and SIX11 (Figs. 6, 7 and 8). The SIX9 and SIX11 genes could cluster both FOE and “presumed-FOE” as one group separated from the known FUSARIUM-ID sequences from the GenBank (Figs. 7 and 8), but SIX8 could not (Fig. 6). Characterisation of Fusarium oxysporum isolates from oil palm using housekeeping gene partial sequences of EF-1α, grouped all oil palm FOE and “presumed-FOE” isolates from the known FUSARIUM-ID sequences in the GenBank (Fig. 9).
Phylogenetic tree generated using statistical model Unweighted Pair-Group Method with Arithmetic Average (UPGMA) cluster composite date set from Fusarium oxysporum isolates from symptomatic oil palm and other host from GenBank based on SIX8. Bootstrapping was done for 1000 with bootstrap values shown in the nodes
Phylogenetic tree generated using statistical model Unweighted Pair-Group Method with Arithmetic Average (UPGMA) cluster composite date set from Fusarium oxysporum isolates from symptomatic oil palm and other host from GenBank based on SIX9. Bootstrapping was done for 1000 with bootstrap values shown in the nodes
Phylogenetic tree generated using statistical model Unweighted Pair-Group Method with Arithmetic Average (UPGMA) cluster composite date set from Fusarium oxysporum isolates from symptomatic oil palm and other host from GenBank based on SIX11. Bootstrapping was done for 1000 with bootstrap values shown in the nodes
Phylogenetic tree generated using statistical model Unweighted Pair-Group Method with Arithmetic Average (UPGMA) cluster composite date set from Fusarium oxysporum isolates from symptomatic oil palm and other host from GenBank based on EF-1α. Bootstrapping was done for 1000 with bootstrap values shown in the nodes
Discussion
After testing a new pathogenicity assay in oil palm seedlings which took considerable shorter time for symptoms to show in plants, this study successfully confirmed the pathogenicity of four Fusarium oxysporum f. sp. elaeidis (FOE) isolates based on qualitative assessment and PCR. This current pathogenicity assay used in the current study differed from previous assays used by Rusli et al. (2015). Rusli et al. (2015) applied FOE spore suspension with a sterile syringe onto the soil surface around the base of each palm at age 3 months. Rusli et al. (2015) further reported that the inoculum was then watered with sterile distilled water for 2 weeks. However, symptoms in the current study were rapid compared to Rusli et al. (2015). Some of the isolates such as NORPALM-5 and BOPP-B5 showed mild infections compared to OPRI-5 and 16F. Previous studies have confirmed 16F to be more aggressive than other FOE isolates (Mepsted et al. 1994a, b; Rusli et al. 2015). Effector proteins (Secreted In Xylem-SIX gene) were for the first time studied in FOE and “presumed-FOE” isolates collected from two major regions in Ghana (Western and Eastern) in addition to other FOE isolates from Democratic Republic of Congo, Ivory Coast and Suriname. Characterisation of FOE and “presumed-FOE” isolates based on SIX8, SIX9, SIX11 and EF-1α, was demonstrated via phylogenetic studies which showed non-clustering of FOE and “presumed-FOE” isolates on the bases of geographical location.
The oil palm-infecting FOE has not been classified into races (Gordon and Martyn 1997) unlike banana F. oxysporum f. sp. cubense isolates (Li et al. 2015). Some research on the pathogenicity of FOE isolates has been done (Flood et al. 1989; Flood 2006; Cooper 2011; Rusli et al. 2015) but was laborious and time consuming (Mouyna et al. 1996). The pathogenicity test in this present study showed that all four FOE were pathogenic to oil palm and this is congruent with reports (Flood et al. 1993; Rusli et al. 2015). The pathogenicity of 16F Fusarium oxysporum isolate collected from oil palm has been confirmed in a previous study (Rusli et al. 2015) but not the three Fusarium oxysporum isolates (BOPP-B5, NORP-5 and OPRI-5) collected from Ghana. Symptoms for FOE infection in oil palm seedlings showed at different times depending on the method of inoculation, indicating the complex nature of the FOE pathogen. The earliest time symptoms showed in inoculated oil palm seedlings via wounding of the roots and soaking in spore suspension, was 4 months under glasshouse conditions. However, Rusli et al. (2015) reported of severe symptoms at 25 weeks in Tenera (commercial type) oil palm. This shows that the protocol used for this current study, reduced the amount of time necessary for symptom development in oil palm seedlings compared to research conducted by Rusli et al. (2015), where higher concentration of 6 X 106 spores ml−1 was poured directly onto the soil surface around the base of the 3-month-old oil palm kept under glasshouse conditions. Other studies, Corley and Tinker (2003) pathogenicity test was in an open environment and the conditions optimized by providing shade to maintain the best temperature for pathogen invasion but the specific method of inoculation was not stated though plants showed late symptom development compared to the present study. Generally, there was yellowing of the leaves of the infected oil palm seedlings which is one prominent symptom of fusarium wilt disease (Flood et al. 1993; Flood 2006; Rusli et al. 2015).
There was an initial thought that SIX genes were unique for F. oxysporum f. sp. lycopersici (FOL). (Fraser-Smith et al. 2014) but in other F. oxysporum f. spp. (Rep et al. 2004; Lievens et al. 2009; Laurence et al. 2015; Rocha et al. 2016; Taylor et al. 2016). The identified SIX genes common to all four FOE used for pathogenicity test were SIX8, SIX9 and SIX11. However, FOE OPRI-5 isolate was detected to have SIX12 which was however, absent in the other three FOE isolates used for pathogenicity test. Taylor et al. (2016) reported similar results for F. oxysporum f. sp. cepae (Foc) in onion, as some SIX genes were either present or absent in both pathogenic and non-pathogenic isolates. This is the first time fourteen SIX (1–14) genes have been studied in four FOE confirmed to be pathogenic to oil palm and “presumed-FOE” isolates from Ghana, Ivory Coast, DR Congo and Suriname. However, conclusions cannot be made for this present study as to whether pathogenicity could be associated with the presence or absence of SIX genes in FOE as this would require further study. The phylogenetic tree generated from SIX genes (SIX9 and SIX11) sequences grouped all FOE and “presumed-FOE” isolates into same cluster with known Fusarium oxysporum f. sp.-ID sequences from GenBank. On the contrary, SIX8 did not cluster FOE and “presumed-FOE” from known Fusarium oxysporum f. sp. sequences-ID, as some isolates (DR Congo-Z, DR Congo-F3 and Suriname-S) clustered widely from the other FOE and “presumed-FOE” in the phylogenetic tree. The Genetic variation in FOE and “presumed-FOE” isolates based on the individual phylogenetic trees for SIX8, SIX9 and SIX11is likely to be because SIX genes reside in the class II transposable elements (TEs) - enriched chromosomal sub-regions as reported (McDonald and Linde 2002; Ma et al. 2010; Schmidt et al. 2013). This characteristics of the SIX genes as reported (Ma et al. 2010; Schmidt et al. 2013) could have resulted in the failure of the clustering of the FOE isolates based on their geographical location where the isolates were sampled. A similar report showed the inability of SIX genes to differentiate Australian F. oxysporum f. sp. canariensis (Foc) isolates from international Foc isolates (Laurence et al. 2015). Laurence et al. (2015) further stated low sequence diversity among the Foc isolates based on the SIX genes and suggested horizontal gene transfer mechanisms as the cause. The diversity within formae speciales of Fusarium has been revealed by multiple studies that reported amino acid and nucleic acid sequences of SIX genes as highly conserved (Rep et al. 2004; Houterman et al. 2008; van der Doe et al. 2008; Thatcher et al. 2012) which is likely to have contributed to the genetic variation among FOE, “presumed-FOE” and known Fusarium oxysporum f. sp. sequences-ID from GenBank based on the SIX8, SIX9 and SIX11.
The ribosomal DNA (rDNA) genes are useful for identification of plant pathogens (Abd-Elsalam et al. 2003). Frequently used loci for studying the genetic variation within Fusarium species complexes is the EF-1α (Geiser et al. 2004; Laurence et al. 2015; Pinaria et al. 2015). The phylogenetic tree for EF-1α grouped all FOE and “presumed-FOE” isolates into a cluster separated from other Fusarium oxysporum f. sp.-ID sequences from GenBank. Similar results were reported for EF-1α phylogeny in F. oxysporum f. sp. canariensis (Laurence et al. 2015) and F. oxysporum f. sp. vanillae (Pinaria et al. 2015). The EF-1α showed genetic variation among the FOE and “presumed-FOE” as isolates from Ghana were mixed with Suriname, Ivory Coast and DR Congo within the same cluster in the phylogenetic tree. This observation is congruent with recent EF-1α phylogeny in F. oxysporum f. sp. vanillae sampled from Mexico (Flores-de la Rosa et al. 2018). Diversity among FOE and “presumed-FOE” based on EF-1α region is likely to be as a result of exchange of genetic material between the FOE and “presumed-FOE” isolates that lead to the switch of clades as reported for F. oxysporum f. sp. lycopersici and F. oxysporum f. sp. radices-lycopersici (Lievens et al. 2009).
In this study, pathogenicity assay developed has drastically reduced the time for symptom development and future researchers can be confident in this improved pathogenicity protocol developed. Different SIX genes have also been identified in both FOE and “presumed-FOE” isolates collected from Ghana, Suriname-S, Ivory-Coast and DR Congo. These isolates obtained from geographic origins where oil palm production is a significant part of their economy could tap into this knowledge and research further to improve on the management and control of fusarium wilt disease in oil palm. Future study should be conducted to verify if the three SIX genes identified within the four FOE isolates could be associated with pathogenicity.
References
Abd-Elsalam K, Bahkali A, Moslem M et al (2003) PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr J Biotechnol (4):82–85
Aderungboye FO (1982) Significance of vascular wilt in oil palm plantation in Nigeria: In Pushparajah and Chew, P.S. (ed.) The oil palm and agriculture in the eighties. Incorporated Society of Planters, Kuala Lumpur Press 475–484
Baayen RP, O’Donnell K, Bonants PJM et al (2000) Gene genealogies and AFLP analyses within the Fusarium oxysporum complex identify monophyletic and non-monophyletic Formae specialis causing wilt and rot disease. Phytopathology 90:891–900
Baysal Ö, Karaaslan Ç, Siragusa M et al (2013) Molecular markers reflect differentiation of Fusarium oxysporum forma speciales on tomato and forma on eggplant. Biochem Syst Ecol 47:139–147
Bogale M, Wingfield BD, Wingfield MJ et al (2005) Simple sequence repeat markers for species in the Fusarium oxysporum complex. Mol Ecol Notes 5:622–624
Bogale M, Steenkamp ET, Wingfield MJ et al (2009) Diverse Fusarium solani isolates colonise agricultural environments in Ethiopia. European Journal for Plant Pathology 124:369–378
Cooper RM (2011) Fusarium wilt of oil palm: a continuing threat to south east Asian plantations. The planter, vol 87, Kuala Lumpur, pp 409–418
Corley RHV (2009) How much palm oil do we need? Environmental Science Policy 12:134–139
Corley RHV, Tinker PB (2003) The oil palm, fourth edition. Blackwell publishing, Oxford
Datta J, Lal N (2013) Genetic diversity of fusarium wilt races of pigeon pea in major regions of India. Afr Crop Sci J 3:201–211
Datta S, Choudhary RG, Shamim M et al (2011) Molecular diversity in Indian isolates of Fusarium oxysporum f. sp. lentis inciting wilt disease in lentil (Lens culinaris Medik). Afr J Biotechnol 38:7314–7323
Flood J (2006) A review of Fusarium wilt of oil palm caused by Fusarium oxysporum f. sp. elaeidis. Phytopathology 96:660–662
Flood J, Cooper RM, Lees PE (1989) An investigation of pathogenicity of four isolates of Fusarium oxysporum from South America, Africa and Malaysia to clonal oil palm. J Phytopathol 124:80–88
Flood J, Mepsted R, Velez A et al (1993) Comparison of virulence of isolates of Fusarium oxysporum f. sp. elaeidis from Africa and South America. Plant Pathol 42:168–171
Flores-de la Rosa FR, De Luna E, Adame-Garcia J et al (2018) Phylogenetic position and nucleotide diversity of Fusarium oxysporum f. sp. vanillae worldwide based on translation elongation factor 1α sequences. Plant Pathol 67:1278–1285
Fraser-Smith S, Czislowski E, Meldrum R.A et al (2014) Sequence variation in the putative effector gene SIX8 facilitates molecular differentiation of Fusarium oxysporum f. sp. cubense. Plant Pathology 63:044–1052
Geiser DM, Jiménez-Gasco MM, Kang S et al (2004) FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479
Gordon TR, Martyn RD (1997) The evolutionary biology of Fusarium oxysporum. Annu Rev Phytopathol 35:111–128
Hannachi I, Poli A, Regui S et al (2015) Genetic and phenotypic differences of Fusarium oxysporum f. sp. citri isolated from sweet orange and tangerine. Eur J Plant Pathol 142:269–280
Hansen SB, Padfield R, Syayuti K et al (2015) Trends in global palm oil sustainability research. J Clean Prod 100:140–149
Houterman PM, Cornelissen BJ, Rep M (2008) Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog 5:e1000061. 21
Laurence MH, Summerell BA, Liew ECY (2015) Fusarium oxysporum f. sp. canariensis: evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathol 64:1068–1075
Li WM, Dita M, Wu W et al (2015) Resistance sources to Fusarium oxysporum f. sp. cubense tropical race 4 in banana wild relatives. Plant Pathol 64:1061–1067
Lievens B, van Baarlen P, Verreth C et al (2009) Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici isolates inferred from mating type, elongation factor-1alpha and exopolygalacturonase sequences. Mycol Res 113(10):1181–1191
Liu F, Wei JG, Zhan RL et al (2014) Genetic diversity of Fusarium mangiferae isolated from mango malformation disease in China. Scientia Horticulturae 65:352–356
Ma L-J, van der Does HC, Borkovich KA et al (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373
McDonald BA, Linde C (2002) The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124:163–180
Mepsted R, Flood J, Cooper RM (1994a) Fusarium wilt of oil palm; ‘susceptibility’ of some palms of a ‘resistant’ clone. Oleagineux 49:205–208
Mepsted R, Flood J, Paul T et al (1994b) Virulence and aggressiveness in Fusarium oxysporum f. sp. elaeidis; implications for screening for disease resistance. Oleagineux 49:209–212
Mouyna I, Renard JL, Brygoo Y (1996) DNA polymorphism among Fusarium oxysporum f. sp. elaeidis populations from oil palm, using a repeated and dispersed sequence “palm”. Curr Genet 30:174–180
Pinaria AG, Laurence MH, Burgess LW et al (2015) Phylogeny and origin of Fusarium oxysporum f. sp. vanillae in Indonesia. Plant Pathol 64:1358–1365
Prendergast AG (1957) Observations on the epidemiology of vascular wilt disease of the oil palm. Journal of The West Africa Institute for Oil Palm Research 2:148–175
Rep M, van der Does HC, Meijer M et al (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53:1373–1383
Rocha LO, Laurence MH, Ludowici VA et al (2016) Putative effector genes detected in Fusarium oxysporum from natural ecosystems of Australia. Plant Pathol. https://doi.org/10.1111/ppa.12472
Rusli MH, Idris AS, Cooper RM (2015) Evaluation of Malaysian oil palm progenies for susceptibility, resistance or tolerance to Fusarium oxysporum f. sp. elaeidis and defence-related gene expression in roots. Plant Pathol 64:638–647
Sasaki K, Nakahara K, Tanaka S et al (2015) Detection and quantification of onion isolates of Fusarium oxysporum f. sp. cepae in onion plant. J Gen Plant Pathol 81:232–236
Schmidt SM, Lukasiewicz J, Farrer R et al (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14:119
Silva FP, Marta H, Ricardo H (2014) EF-1α gene and IGS rDNA sequencing of Fusarium oxysporum f. sp. vasinfectum and F. oxysporum f.sp. phaseoli reveals polyphyletic origin of strains. Tropical Plant Pathology 39:64–73
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Taylor A, Vágány V, Jackson AC et al (2016) Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol Plant Pathol 17:1032–1047
Teo TM (2015) Effectiveness of the oil palm pollinating weevil, Elaeidobius kamerunicus, in Malaysia
Thangavelu R, Kumar KM, Ganga DP et al (2012) Genetic diversity of Fusarium oxysporum f. sp. cubense isolates (Foc) of India by inter simple sequence repeats (ISSR) analysis. Mol Biotechnol 51:203–211
Thatcher LF, Gardiner DM, Kazan K et al (2012) A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular Plant Microbe 25:180–190
van der Doe HC, Duyvesteijn RGE, Goltstein PM et al (2008) Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet Biol 45:1257–1264
Acknowledgements
This research was fully funded by Commonwealth Scholarship Commission-UK. We thank Dr. Julie Flood the Global Director of CABI Research for technical advice and support and the late Prof. Richard Cooper former Lecturer at University of Bath – UK for providing us with two isolates of FOE. Further thanks go to Dr. Y. Ndede a Scientist and Frank Dwumfour a Technician at the CSIR-OPRI, Ghana for the immense assistance during the field work. Our appreciation also goes to University of Nottingham – UK, Plant Science School of Biosciences for giving us access to laboratory facilities. We also appreciate the support of bioinformatician Dr. Preeti Panda of Scion-Forest Protection Team. We finally thank all the various oil palm plantation sites in Ghana where sampling was undertaken including CSIR-OPRI, NORPALM, BOPP and TOPP.
Funding
This study was funded by Commonwealth Scholarship Commission – UK with grant number (GHCS-115- 235 2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author Dr. Kwasi Adusei-Fosu currently a Forest Pathologist with Scion (New Zealand Forest Research Institute), received research grant from Commonwealth Scholarship Commission-UK and was supervised by Co-author Prof. Matthew Dickinson, Senior Plant Pathologist, Diagnostician Researcher, and Lecturer at the University of Nottingham – UK.
Also, there is no conflict of interest in existence: Author Dr. Kwasi Adusei-Fosu declares that he has no conflict of interest. Author Prof. Matthew Dickinson also declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. Further, this article does not contain any studies with animals performed by any of the authors.
This article does not contain any studies with human participants or animals performed by authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 32 kb)
Rights and permissions
About this article
Cite this article
Adusei-Fosu, K., Dickinson, M. Development of pathogenicity assay and characterization of Fusarium oxysporum f. sp. elaeidis (FOE) based on Secreted In Xylem genes and EF-1α. J Plant Pathol 101, 1013–1024 (2019). https://doi.org/10.1007/s42161-019-00332-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-019-00332-4