Abstract
Objective
In this study, we aimed to assess the association between four variants in three genes whose association has been reported in adults but not in children. We evaluated the relationship of the GCKR (rs780094), GCKR (rs1260333), FADS (rs174547), and MLXIPL (rs3812316) polymorphisms with serum lipid levels in Iranian children.
Design
This cross-sectional study was conducted in a subpopulation of the CASPIAN III study. During this study, 550 frozen whole blood samples were selected randomly. Using the recorded information of selected cases, those with and without abnormal lipid levels were determined. Allelic and genotypic frequencies of GCKR (rs780094), GCKR (rs1260333), MLXIPL (rs3812316), and FADS (rs174547) polymorphisms were determined and compared in dyslipidemic and normal children. The association between the studied polymorphisms and lipid profiles was determined using logistic regression analysis.
Results
Prevalence of hypercholesterolemia, hypertriglyceridemia, high low-density lipoprotein cholesterol (LDL-C), and low high-density lipoprotein cholesterol (HDL-C) were 24.9, 34.5, 19.0, and 40.7%, respectively. Significant correlations were found between GCKR (rs780094) and GCKR (rs1260333) polymorphisms and cholesterol and triglyceride levels, between FADS (rs174547) polymorphism and level of triglyceride, and also between MLXIPL (rs3812316) and levels of HDL-C.
Conclusions
The results of this population-based study provide evidence for a relationship between lipid regulatory gene polymorphisms including GCKR (rs780094), GCKR (rs1260333), FADS (rs174547), and MLXIPL (rs3812316) with dyslipidemia in an Iranian population. These results could provide baseline information on as well as further insight into the genetic makeup of lipid profiles in Iranian children, which could be used for preventative strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dyslipidemia is an important risk factor for future cardiovascular disease in children, which could continue/persist into adulthood and increase the rate of morbidity and mortality in these individuals as adults [1]. It is considered as an emerging health problem among children and adolescents due to the high prevalence of the disease. Evidence has reported that unfavorable serum lipid levels are present in approximately one in five children and adolescents aged 8 to 17 years [2]. Results of a systematic review study in Iranian children have indicated that dyslipidemia is a common cardiovascular risk factor with a prevalence of 48%, 3–50%, 5–20%, and 5–88% for hypercholesterolemia, hypertriglyceridemia, elevated low-density lipoprotein cholesterol (LDL-C), and low high-density lipoprotein cholesterol (HDL-C), respectively [3].
Dyslipidemia is a multifactorial trait. Evidence has indicated that multiple genetic and environmental factors and their interaction contribute to the occurrence of different lipid traits [4, 5]. The role of genetic factors and genotype-phenotype association in determining plasma lipoprotein levels has been described in adults [6, 7]. However, there are few studies in this field concerning children and adolescents and there has been no genome-wide association study (GWAS) for lipid levels in children and adolescents. A small number of recent post-GWAS studies have investigated the genetic contribution of GWAS-associated polymorphisms for lipid levels in children. These documented that lipid-related polymorphisms in adults are also associated with blood lipid levels in early life [8, 9]. Christie et al. have demonstrated that lipid-lowering preventative strategies during childhood could be effective in genetically predisposed pediatric populations [9]. In addition, recent studies suggested that for the prevention of subclinical atherosclerosis and its progression to clinical cardiovascular disease (CVD) and related complications, identification of lipid metabolism-related polymorphisms in early life could be helpful [10].
It is also well established that the genetic determinants of disorders (including dyslipidemia) are different in various populations and ethnic groups, the characteristics of dyslipidemia and its dominant components varying in different populations [11].
Dyslipidemia is considered a common metabolic disorder in the Iranian population and the most common variation in children is hypertriglyceridemia and low HDL-C levels [3].
Given the importance of proper management of metabolic risk factors including dyslipidemia for prevention of CVD and related complications, we aimed to assess the association between four variants in three genes whose association has been reported in adults but not in children. We evaluated the relationship of the GCKR (rs780094), GCKR (rs1260333), FADS (rs174547), and MLXIPL (rs3812316) polymorphisms with serum lipid levels in Iranian children. The glucokinase regulatory (GCKR) gene encodes glucokinase regulator protein which regulates pancreatic beta cells and hepatocyte activity and plays a key role in triglyceride and glucose homeostasis [12].
The MLX interacting protein-like MLXIPL (formerly known as ChREBP) regulates the expression of pyruvate kinase, which binds and activates glycolytic pyruvate into lipogenesis through the conversion of dietary carbohydrate to store fat in the liver [13]. The fatty acid desaturase 1 (FADS1) gene encodes fatty acid desaturase enzymes which regulates the desaturation of fatty acids [14].
Materials and methods
This cross-sectional study was conducted in a subpopulation of the third survey of the school-based surveillance system entitled Childhood and Adolescence Surveillance and Prevention of Adult Noncommunicable Disease (CASPIAN III) Study.
Briefly, the CASPIAN III study is a school-based nationwide health survey which was carried out in 27 provinces of Iran in 2009–2010. The protocol of the study was approved by ethics committees and other relevant Iranian national regulatory organizations. Details of the study including data collection, sampling, and measurements have been published [15].
During the main survey, 5528 students aged 10–18 years were selected by multistage random cluster sampling from rural and urban regions of the 27 provinces. Written informed consent and oral assent were obtained from parents and children, respectively. A team of trained health care staff recorded the demographic characteristics of each participant using validated questionnaires. Measurements including weight, height, waist circumference, and blood pressure were performed by the trained team and physicians applying standard protocols and using calibrated instruments. Body mass index (BMI) was calculated (body weight in kg/height in m2). Fasting venous blood sample was obtained from the students for biochemical measurements including plasma levels of glucose and lipid profile variables including total cholesterol (TC), triglycerides (TG), HDL-C, and LDL-C. The lipid profile was measured by autoanalyzer using Pars Azmoon diagnostic kits (Tehran, Iran).
Lipids were measured using auto-analyzers. HDL-C was measured after precipitation of non-HDL-C with dextran sulfate-magnesium chloride. The biochemical measurement in each county was performed in the central provincial laboratory with the standards of the National Reference Laboratory collaborating with the World Health Organization [15].
The protocol of the current study as a substudy of the CASPIAN III study was approved by the regional ethics committee of the Isfahan University of Medical Sciences with the research project number 193058.
In this study, 550 frozen whole blood samples were selected randomly. Using the recorded information of selected cases, those with abnormal lipid levels were determined.
Dyslipidemia was defined as follows for each lipid profile [16]:
-
1.
Low HDL-C, < 40 mg/dL
-
2.
Hypercholesterolemia, ≥ 170 mg/dL
-
3.
Hypertriglyceridemia, ≥ 100 mg/dL
-
4.
High LDL-C, ≥ 110 mg/dL
Allelic and genotypic frequencies of GCKR (rs780094), GCKR (rs1260333), MLXIPL (rs3812316), and FADS (rs174547) polymorphisms were determined in all selected cases and compared in dyslipidemic and normal children.
Molecular study
Genomic deoxyribonucleic acid (DNA) was extracted from peripheral blood samples using the YATA DNA extraction kit (YATA, Iran) according to the manufacturer’s protocol. The GCKR (rs780094), GCKR (rs1260333), MLXIPL (rs3812316), and FADS (rs174547) polymorphisms were identified using the NCBI data bank. Primers of the four polymorphisms were designed by Beacon Designer 8.1(PREMIER Biosoft International, USA) to flank the desired regions. The primers were synthesized by Bioneer (South Korea).
Genotyping was performed by real-time PCR and high-resolution melt analysis (HRM) assay using a Rotor-Gene 6000 instrument (Corbett Life Science, Australia).
Using a Type-it HRM kit (Qiagen, Germany), the amplicons were generated according to the following program: one cycle at 95 °C for 15 min; 40 cycles at 95 °C for 15 s, 60.0 °C for 15 s, 72 °C for 15 s, one cycle of 95 °C for 1 s, 72 °C for 90 s, and a melt from 70 to 95 °C rising to 0.1 °C/s. The amplification mixture of a total volume of 25 μL included 12.5 μL of HRM PCR master mix, 1.75 μL of 10 μM primer mix, 2 μL of genomic DNA as a template, and 8.25 μL of RNase-free water. For each genotype reaction, we included sequence-proven major and minor allele homozygote and heterozygote controls.
Using the instrument software, the results of HRM were analyzed by comparing the melting curve shape between studied samples and known controls.
Statistical analysis
Our data were analyzed using IBM SPSS/PC statistical software version 21. The Hardy-Weinberg equation was tested to compare the observed genotype frequencies to those expected by χ2 analysis. Quantitative variables are presented as mean (±SD). Mean levels of cholesterol, TG, LDL-C, and HDL-C in different genotypes of the studied polymorphisms were compared using one-way analysis of variance (ANOVA). The chi-square test was used to compare the genotype and allele frequency in participants with and without dyslipidemia. The association between studied polymorphisms (as independent variables) and dyslipidemia (as a dependent variable) was determined using logistic regression analysis. Variables such as age, sex, low physical activity, prolonged screen time, and familial history of non-communicable diseases were introduced into multivariate models to assess their role as confounders in the analysis of association, while for the covariate control procedures, interaction terms were assessed within the models. Measures of association were estimated as odds ratio and 95% confidence interval (OR, 95% CI). A P value of < 0.05 was considered significant.
Results
In this study, 528 students with a mean age of 15.0 (2.2) were enrolled; 51.7% of the studied population were girls. Prevalence of hypercholesterolemia, hypertriglyceridemia, high LDL-C, and low HDL-C was 24.9, 34.5, 19.0, and 40.7%, respectively. The prevalence of dyslipidemia in the studied population according to gender is presented in Fig. 1. Prevalence of dyslipidemia was not significantly different between boys and girls (P = 0.189 for cholesterol, P = 0.407 for triglyceride, P = 0.537 for LDL-C, and P = 0.348 for HDL-C). Frequency of lifestyle-related factors including prolonged screen time and low physical activity and familial history of non-communicable diseases in the studied children with and without different types of dyslipidemia are presented in Table 1.
Familial history of non-communicable diseases was significantly higher in all types of dyslipidemic children except low HDL-C (P = 0.017 for cholesterol, P = 0.020 for triglyceride, and P = 0.022 for LDL-C).
Prolonged screen time (P = 0.529 for cholesterol, P = 0.357 for triglyceride, P = 0.338 for LDL-C, and P = 0.120 for HDL-C) and low physical activity (P = 0.346 for cholesterol, P = 0.267 for triglyceride, P = 0.497 for LDL-C, and P = 0.579 for HDL-C) were not different between children with and without dyslipidemia.
Pairwise comparisons of the studied polymorphism genotype and allele frequency in participants with and without dyslipidemia are presented in Table 2. Frequency of the GCKR (rs780094) gg allele was significantly lower in participants with hypercholesterolemia and hypertriglyceridemia (P = 0.03 for hypercholesterolemia and P = 0.009 for hypertriglyceridemia). Frequencies of the GCKR (rs1260333) cc allele and FADS (rs174547) tt allele were significantly lower in students with hypertriglyceridemia (P = 0.02 for GCKR (rs1260333) cc allele and P = 0.01 for FADS (rs174547) tt allele). Frequency of the MLXIPL (rs3812316) cc allele was significantly higher in participants with hypertriglyceridemia (P = 0.02).
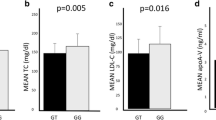
Mean levels of TC, TG, LDL-C, and HDL-C in different genotypes are presented in Table 3. Mean level of cholesterol was significantly lower in participants with the gg and cc alleles of the GCKR (rs780094) (P = 0.011) and GCKR (rs1260333) (P = 0.01) polymorphisms, respectively.
Mean level of TG was significantly lower in participants with the gg and tt alleles of the GCKR (rs780094) (P = 0.006) and FADS (rs174547) (P < 0.001) polymorphisms, respectively. Mean level of TG was significantly higher in participants with the cc allele of the MLXIPL (rs3812316) polymorphism (P = 0.032). Mean level of HDL-C was significantly lower in participants with the cc allele of the MLXIPL (rs3812316) polymorphism (P = 0.018). Mean level of LDL-C was significantly lower in participants with the gg allele of the GCKR (rs780094) polymorphisms (P = 0.004).
Mean level of LDL-C was lower in students with the cc allele of GCKR (rs1260333) (P = 0.05): the P value was on the boundary and the result is marginal and not statistically significant.
The association between lipids and the GCKR (rs780094), GCKR (rs1260333), MLXIPL (rs3812316), and FADS (rs174547) polymorphisms in the studied pediatric population is presented in Table 4. There was a significant correlation between GCKR (rs780094) and TC (P = 0.02, OR = 1.68, CI 95% 1.05–2.57) and TG (P = 0.006, OR = 2.19, CI 95% 1.25–3.85) levels. There was a significant correlation between the FADS (rs174547) polymorphism and level of TG. There was a significant correlation between MLXIPL (rs3812316) and levels of HDL-C (P = 0.009, OR = 2.40, CI 95% 1.24–4.65).
Discussion
In this nationwide study, we investigated the association between four variants in three genes, GCKR (rs780094), GCKR (rs1260333), MLXIPL (rs3812316), and FADS (rs174547) with serum lipid levels of Iranian children. Our findings indicated significant associations between TG level and the GCKR (rs780094) and FADS (rs174547) polymorphisms. Serum TC and HDL-C were significantly associated with GCKR (rs780094) and MLXIPL (rs3812316), respectively. Minor alleles of GCKR (rs780094), GCKR (rs1260333), and FADS (rs174547) and major alleles of MLXIPL (rs3812316) had an effect for/on TG. Minor alleles of GCKR (rs780094) also had an effect on TC.
In the current study, the prevalence of hypercholesterolemia, hypertriglyceridemia, high LDL-C, and low HDL-C was 24.9, 34.5,19.0, and 40.7%, respectively. Hypertriglyceridemia and low HDL-C were the most common types of dyslipidemia in this Iranian pediatric population. The results in our study, which was a substudy of the CASPIAN III study, were similar to those of the main CASPIAN survey and in accordance with the findings of a systematic review in the prevalence of lipid disorders in Iranian children [3]. Though ethnic and genetic backgrounds are associated with different types of dyslipidemia, BMI and dietary habits could also influence the plasma concentration of some lipids such as TG [3].
Results of various studies worldwide have indicated that the prevalence of specific dyslipidemias varies by ethnicity and geographical regions. Among US children, elevated TG and TC were the most common type of dyslipidemia [17]. Low HDL-C and hypercholesterolemia were the most prevalent types of dyslipidemia in Brazilian adolescents [18]. Dyslipidemia among Korean 10–18-year-old children was similar to our results [19]. Hypertriglyceridemia was the most frequent type of dyslipidemia in the pediatric population of Saudi Arabia and Thailand [20, 21].
In the current study, we selected polymorphisms that are associated mainly with TG and HDL-C levels and have not yet been evaluated among the Iranian population.
The results of this study showed that the prevalence of a family history of non-communicable diseases was significantly higher in all types of dyslipidemic children than in non-dyslipidemic ones, whereas the rates of lifestyle-related factors, including prolonged screen time and low physical activity, were not different between children with and without dyslipidemia. It thus seems that genetic factors are the strongest determinants of dyslipidemia in Iranian children. Some evidence has indicated that genetic variation is the most important factor affecting lipid levels in adolescents, presenting a 70–80% heritability rate [22]. The higher heritability of dyslipidemia in children and adolescents than in the adult population is due to the interaction between gene polymorphisms and different environmental factors during adulthood [22].
The role of the GCKR gene in lipid metabolism has not as yet been thoroughly investigated. However, the association of its variants, mainly rs780094 and rs1260326, with plasma TG level has been reported by genome-wide association studies [12, 23, 24].
In this study, two of the abovementioned variants were studied. GCKR polymorphisms were firstly reported by genome-wide association studies regarding their association with TG levels [12]. The results of a number of population-based studies have indicated that the GCKR gene contributes to the risk of dyslipidemia, namely, mainly hypertriglyceridemia and hyperlipidemia [23]. Recently, Surakka et al. have identified genetic variants associated with dyslipidemia by studying 1000 Genomes Imputation in 62,166 samples. They identified 93 loci in this regard. According to their results, GCKR loci were classified as variants with known functional effects on lipid levels of the general population. The loci cause dyslipidemia through Mendelian Inheritance [25].
Shang et al. in China have investigated the association between the rs780094 polymorphism and plasma lipid levels in 1026 Chinese children and adolescents aged 7–18 years. They showed that the GCKR (rs780094) polymorphism was associated with hypertriglyceridemia and high LDL-C. In addition, based on their findings, the A-allele of the SNPs could be a genetic risk factor for pediatric dyslipidemia [26].
Horvatovich et al. also suggested a possible association of the GCKR (rs780094) polymorphism with dyslipidemia in children [27]. Lee and colleagues reported that the GCKR variant rs780094 is correlated with hypercholesterolemia and high LDL-C among Korean children [28]. Sparsø et al. demonstrated that the major G allele of GCKR rs780094 has a protective effect on total cholesterol levels in the Danish population [29]. Sotos-Prieto et al. reported that GCKR rs780094 is associated with total cholesterol among European populations and its AA allele was modestly in correlation with total plasma cholesterol levels [30].
Our results showed that the minor alleles of the GCKR (rs780094) polymorphism in Iranian children are associated with hypercholesterolemia and hypertriglyceridemia, this finding being similar to those of the abovementioned studies conducted in Europe, Korea, and China [26, 28, 29].
Another SNP of the GCKR gene which was investigated in the current study was GCKR (rs1260333). The association between GCKR rs1260333 variants with TG levels in children has been reported among European and Chinese children by Waterworth et al. and Shen et al., respectively [31, 32]. In this study, the mean levels of TC and LDL-C were significantly higher in children with a minor allele of the variant and the frequency of minor alleles was significantly higher in children with hypertriglyceridemia; however, logistic regression analysis did not indicate any association with any types of dyslipidemia among Iranian children. The results of logistic regression indicated that it tended to be associated with TG, though the value was not statistically significant. Clearly, in order to obtain more conclusive results, the relation should be investigated in a larger nationwide population.
The exact mechanism of MLXIPL in lipid metabolism has not yet been fully determined [13]. Surakka and colleagues have classified MLXIPL as potential functional variant locus with no known functional impact on lipid levels. MLXIPL has no lipid-related variants listed in the online Mendelian Inheritance in Man database [24]. Recently, several GWAS reported some MLXIPL variants as being associated with plasma TG levels [33]. In the present study, we have evaluated the association of the MLXIPL (rs3812316) polymorphism with dyslipidemia.
The findings of previous studies in this field are not consistent in different populations. Some studies indicated that the G allele of the polymorphism is associated with lower levels of TG in Asians, Indian Asians, and Northern Europeans [34, 35], whereas studies from central Europe and Hungary did not show any such association [36, 37].
In this study, the frequency of the major allele of the MLXIPL (rs3812316) polymorphism was significantly higher in hypertriglyceridemic children. Mean levels of TG and HDL-C were significantly higher and lower in participants with the major allele of the polymorphisms, respectively. However, logistic regression analysis showed a significant correlation between HDL-C and the MLXIPL (rs3812316) polymorphism. Lack of an association between the variants and TG may be due to the small sample size. Our findings regarding the association between the variants and HDL-C have not previously been reported. It is suggested that our results as a novel finding could be used for further studies regarding the role of this gene in the metabolism of HDL-C, especially in the Iranian population, previous studies having indicated that low HDL-C is considered as the most common type of dyslipidemia in the Iranian population [38].
The association of the FADS1 gene variants with serum lipid compositions has been reported by a number of GWAS [14, 39, 40]. We studied the FADS (rs174547) polymorphism because it was investigated in pediatric populations. Standl et al. in Germany have reported that the mean level of TG was significantly higher in children aged 10 years old with minor alleles of FADS (rs174547) polymorphism [41].
In another study in the Netherlands, the KOALA Birth Cohort Study carried out among 2-year-old children, FADS polymorphisms, including rs174547, were associated with plasma TC and HDL-C levels [42].
In our study, the frequency of minor alleles of FADS (rs174547) was significantly higher in children with hypertriglyceridemia. Mean levels of TG were significantly higher in those with the minor allele of the SNP. There was no significant association between the polymorphism and different lipid compositions.
The present study has some limitations. Though the samples were selected from a population-based study and were a representative sample of Iranian children, the results would be more conclusive in a larger sample size. Moreover, we did not evaluate the sexual maturity of the participants, the impact of sexual maturation on children’s lipid levels having been demonstrated in previous studies [43, 44].
The strength of our study was its novelty in a pediatric population.
Conclusions
The results of this population-based study provide evidence for a relationship between lipid regulatory gene polymorphisms, including GCKR (rs780094), GCKR (rs1260333), FADS (rs174547), and MLXIPL (rs3812316), with dyslipidemia in an Iranian population. The fact that lifestyle-related factors such as screen time and physical activity were not different in dyslipidemic and normal children strongly indicates that genetic factors are important determinants for dyslipidemia in children. Some of our findings such as the association of GCKR (rs780094) with serum TG and TC level and GCKR (rs1260333) with serum TG levels were similar to those of previous studies. However, other findings such as the association of FADS (rs174547) with serum TG and MLXIPL (rs3812316) with HDL-C were novel and have not been previously reported. Our findings regarding the distribution of the studied SNP genotypes and allele frequencies in different types of dyslipidemia were similar to those previously reported.
The obtained data could provide baseline information on as well as further insight into the genetic makeup of lipid profiles in Iranian children, which could be used for preventative strategies. In addition, considering the high prevalence of type 2 diabetes and its increasing trend in low- and middle-income countries (including Iran) and the mutual association between dyslipidemia, type 2 diabetes, and its related cardiometabolic risk factors [45,46,47], understanding the genetic basis of dyslipidemia in our population could be valuable for the prevention of other non-communicable diseases, such as type 2 diabetes.
Given that environmental factors, such as certain dietary components or phenotype such as obesity, could interact with studied SNPs, it is recommended that further studies be designed for the evaluation of the interaction of the SNPs with the abovementioned factors in the development of lipid disorders in children.
References
Juhola J, Magnussen CG, Viikari JS, Kähönen M, Hutri-Kähönen N, Jula A et al (2011) Tracking of serum lipid levels, blood pressure, and body mass index from childhood to adulthood: the Cardiovascular Risk in Young Finns Study. J Pediatr 159(4):584–590
Kit BK, Kuklina E, Carroll MD, Ostchega Y, Freedman DS, Ogden CL (2015) Prevalence of and trends in dyslipidemia and blood pressure among US children and adolescents, 1999–2012. JAMA Pediatr 169(3):272–279
Hovsepian S, Kelishadi R, Djalalinia S, Farzadfar F, Naderimagham S, Qorbani M (2015) Prevalence of dyslipidemia in Iranian children and adolescents: a systematic review. J Res Med Sci: off J Isfahan Univ Med Sci 20(5):503
Ordovas JM (2006) Nutrigenetics, plasma lipids, and cardiovascular risk. J Am Diet Assoc 106(7):1074–1081
Cole CB, Nikpay M, McPherson R (2015) Gene–environment interaction in dyslipidemia. Curr Opin Lipidol 26(2):133–138
Parnell LD (2009) Gene–environment interactions and the impact on obesity and lipid profile phenotypes. Clin Lipidol 4(6):687–690
Talmud PJ, Berglund L, Hawe EM, Waterworth DM, Isasi CR, Deckelbaum RE et al (2001) Age-related effects of genetic variation on lipid levels: The Columbia University BioMarkers Study. Pediatrics 108(3):e50
Latsuzbaia A, Jaddoe VW, Hofman A, Franco OH, Felix JF (2016) Associations of genetic variants for adult lipid levels with lipid levels in children. The Generation R Study. J Lipid Res 57(12):2185–2192
Christie S, Robiou-du-Pont S, Anand SS, Morrison KM, McDonald SD, Paré G et al (2017) Genetic contribution to lipid levels in early life based on 158 loci validated in adults: the FAMILY study. Sci Rep 7
Santos IR, Fernandes AP, Sousa MO, Ferreira CN, Gomes KB (2013) Genetic polymorphisms as a risk factor for dyslipidemia in children. J Pediatr Genet 2(2):69–75
Ellman N, Keswell D, Collins M, Tootla M, Goedecke JH (2015) Ethnic differences in the association between lipid metabolism genes and lipid levels in black and white South African women. Atherosclerosis 240(2):311–317
Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H et al (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316(5829):1331–1336
Uyeda K, Repa JJ (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4(2):107–110
Nakayama K, Bayasgalan T, Tazoe F, Yanagisawa Y, Gotoh T, Yamanaka K et al (2010) A single nucleotide polymorphism in the FADS1/FADS2 gene is associated with plasma lipid profiles in two genetically similar Asian ethnic groups with distinctive differences in lifestyle. Hum Genet 127(6):685–690
Kelishadi R, Heshmat R, Motlagh ME, Majdzadeh R, Keramatian K, Qorbani M et al (2012) Methodology and early findings of the third survey of CASPIAN study: a national school-based surveillance of students’ high risk behaviors. Int J Prev Med 3(6):394
Zimmet P, Alberti G, Kaufman F, Tajima N (2007) The metabolic syndrome in children and adolescents. Lancet 369(9579):2059
Li J, Motsko SP, Goehring EL, Tave A, Pezzullo JC, Jones JK (2010) Prevalence of pediatric dyslipidemia: comparison of a population-based claims database to national surveys. Pharmacoepidemiol Drug Saf 19(10):1031–1040
Faria Neto JR, Bento VFR, Baena CP, Olandoski M, Gonçalves LGO, Abreu GA et al (2016) ERICA: prevalence of dyslipidemia in Brazilian adolescents. Rev Saude Publica 50
Yang S, Hwang JS, Park HK, Lee HS, Kim HS, Kim EY et al (2012) Serum lipid concentrations, prevalence of dyslipidemia, and percentage eligible for pharmacological treatment of Korean children and adolescents; data from the Korea National Health and Nutrition Examination Survey IV (2007–2009). PLoS One 7(12):e49253
Al-Shehri SN, Saleh ZA, Salama MM, Hassan YM (2004) Prevalence of hyperlipidemia among Saudi school children in Riyadh. Ann Saudi Med 24(1):6–8
Rerksuppaphol S, Rerksuppaphol L (2011) Prevalence of dyslipidemia in Thai schoolchildren. J Med Assoc Thail 94(6):710
Middelberg RP, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB (2012) Evidence of differential allelic effects between adolescents and adults for plasma high-density lipoprotein. PloSone 7(4):e35605
Johansen CT, Kathiresan S, Hegele RA (2011) Genetic determinants of plasma triglycerides. J Lipid Res 52(2):189–206
Hadarits F, Kisfali P, Mohás M, Maász A, Duga B, Janicsek I et al (2012) Common functional variants of APOA5 and GCKR accumulate gradually in association with triglyceride increase in metabolic syndrome patients. Mol Biol Rep 39(2):1949–1955
Surakka I, Horikoshi M, Mägi R, Sarin A-P, Mahajan A, Lagou V et al (2015) The impact of low-frequency and rare variants on lipid levels. Nat Genet 47(6):589–597
Shang X, Song J, Liu F, Ma J, Wang H (2014) Association between rs780094 polymorphism in GCKR and plasma lipid levels in children and adolescents. Zhonghua Liu Xing Bing Xue Za Zhi 35(6):626–629
Horvatovich K, Bokor S, Polgar N, Kisfali P, Hadarits F, Jaromi L et al (2011) Functional glucokinase regulator gene variants have inverse effects on triglyceride and glucose levels, and decrease the risk of obesity in children. Diabetes Metab 37(5):432–439
Lee H-J, Jang HB, Kim H-J, Ahn Y, Hong K-W, Cho SB et al (2015) The dietary monounsaturated to saturated fatty acid ratio modulates the genetic effects of GCKR on serum lipid levels in children. Clin Chim Acta 450:155–161
Sparsø T, Andersen G, Nielsen T, Burgdorf K, Gjesing A, Nielsen A et al (2008) The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia 51(1):70–75
Sotos-Prieto M, Luben R, Khaw K-T, Wareham NJ, Forouhi NG (2014) The association between Mediterranean Diet Score and glucokinase regulatory protein gene variation on the markers of cardiometabolic risk: an analysis in the European Prospective Investigation into Cancer (EPIC)-Norfolk study. Br J Nutr 112(1):122–131
Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S et al (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 30(11):2264–2276
Shen Y, Wu L, Xi B, Liu X, Zhao X, Cheng H et al (2013) GCKR variants increase triglycerides while protecting from insulin resistance in Chinese children. PLoS One 8(1):e55350
Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR et al (2008) Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet 40(2):149–151
Nakayama K, Yanagisawa Y, Ogawa A, Ishizuka Y, Munkhtulga L, Charupoonphol P et al (2011) High prevalence of an anti-hypertriglyceridemic variant of the MLXIPL gene in Central Asia. J Hum Genet 56(12)
Been LF, Nath SK, Ralhan SK, Wander GS, Mehra NK, Singh J et al (2010) Replication of association between a common variant near melanocortin-4 receptor gene and obesity-related traits in Asian Sikhs. Obesity 18(2):425–429
Vrablik M, Ceska R, Adamkova V, Peasey A, Pikhart H, Kubinova R et al (2008) MLXIPL variant in individuals with low and high triglyceridemia in white population in Central Europe. Hum Genet 124(5):553–555
Polgár N, Járomi L, Csöngei V, Maász A, Sipeky C, Sáfrány E et al (2010) Triglyceride level modifying functional variants of GALTN2 and MLXIPL in patients with ischaemic stroke. Eur J Neurol 17(8):1033–1039
Kelishadi R, Motlagh ME, Roomizadeh P, Abtahi S-H, Qorbani M, Taslimi M et al (2013) First report on path analysis for cardiometabolic components in a nationally representative sample of pediatric population in the Middle East and North Africa (MENA): the CASPIAN-III Study. Ann Nutr Metab 62(3):257–265
Lattka E, Illig T, Koletzko B, Heinrich J (2010) Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol 21(1):64–69
Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE et al (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41(1):56–65
Standl M, Lattka E, Stach B, Koletzko S, Bauer C-P, von Berg A et al (2012) FADS1 FADS2 gene cluster, PUFA intake and blood lipids in children: results from the GINIplus andLISAplus studies. PLoS One 7(5):e37780
Moltó-Puigmartí C, Jansen E, Heinrich J, Standl M, Mensink RP, Plat J et al (2013) Genetic variation in FADS genes and plasma cholesterol levels in 2-year-old infants: KOALA Birth Cohort Study. PLoS One 8(5):e61671
Garcés C, de Oya I, Lasunción MA, López-Simón L, Cano B, de Oya M (2010) Sex hormone–binding globulin and lipid profile in pubertal children. Metabolism 59(2):166–171
Aydın B, Winters SJ (2016) Sex hormone-binding globulin in children and adolescents. J Clin Res Pediatr Endocrinol 8(1):1
Niroumand S, Dadgarmoghaddam M, Eghbali B, Abrishami M, Gholoobi A, Taghanaki HRB et al (2016) Cardiovascular disease risk factors profile in individuals with diabetes compared with non-diabetic subjects in north-east of Iran. Iran Red Crescent Med J 18(8):e29382
Khajedaluee M, Dadgarmoghaddam M, Saeedi R, Izadi-Mood Z, Abrishami M (2014) The burden of diabetes in a developing country. Open J Prev Med 4(04):175
Shaw JE, Sicree RA, Zimmet PZ (2010) Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract 87(1):4–14
Funding
This study was supported by Isfahan University of Medical Sciences (research project number: 193058).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Hovsepian, S., Javanmard, S.H., Mansourian, M. et al. Relationship of lipid regulatory gene polymorphisms and dyslipidemia in a pediatric population: the CASPIAN III study. Hormones 17, 97–105 (2018). https://doi.org/10.1007/s42000-018-0020-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42000-018-0020-x