Abstract
DNA origami have been established as versatile templates to fabricate plasmonic nanostructures in predefined shapes and multiple dimensions. Limited to the size of DNA origami, which are approximate to 100 nm, it is hard to assemble more intricate plasmonic nanostructures in large scale. Herein, we used rectangular DNA origami as the template to anchor two 30-nm gold nanoparticles (AuNPs) which induced dimers nanostructures. Transmission electron microscopy (TEM) images showed the assembly of AuNPs with high yields. Using the linkers to organize the DNA origami templates into nanoribbons, chains of AuNPs were obtained, which was validated by the TEM images. Furthermore, we observed a significant Raman signal enhancement from molecules covalently attached to the AuNP-dimers and AuNP-chains. Our method opens up the prospects of high-ordered plasmonic nanostructures with tailored optical properties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Plasmonic nanostructures have induced more attention because of their potential applications including biosensing [1,2,3], harvesting energy [4,5,6,7,8] and sub-diffraction limit imaging [9,10,11,12]. The typical method for the assembly of plasmonic nanostructures is top-down lithography. Although it can fabricate plasmonic nanostructures with versatile shapes and novel behaviors, there still exist some problems such as the cost for mass production or making structures in multiple dimensions [13, 14]. Bottom-up approaches, such as soft templates, offer new opportunities to fabricate plasmonic nanostructures [15,16,17,18].
DNA origami technology is a milestone during the history of DNA nanotechnology. It broke through the traditional rules that folded a long single-stranded DNA (ssDNA) by hundreds of staples to create thousands of nanostructures with custom-designed shapes or precisely controlled motions in multiple dimensions [19,20,21,22]. DNA origami have been proved as templates for the assembly of plasmonic nanostructures [23,24,25,26].
Because the local fields are enhanced by the oscillations of conduction electrons around metal nanoparticles (NPs), these plasmonic nanostructures illustrate novel optical behaviors such as surface-enhanced Raman scattering (SERS), ultraviolet–visible spectrometry and circular dichroism [27,28,29,30,31,32]. The high precise addressability of DNA origami makes them excellent tools to study the interaction between metal NPs. A typical example is a 40 × 45 nm2 DNA origami template to organize two 40-nm AuNPs into dimers with sub-5-nm gaps. The strong optical coupling between the AuNP-dimers induced enhanced SERS signals during the measurements of an external analyte and ssDNA oligos attached to the NPs [33]. Also, dynamic DNA origami were employed to organize two gold nanorods (AuNRs). The movement of DNA origami regulated the CD performance of the AuNRs [34].
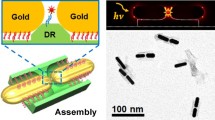
To study the more novel optical behavior of the plasmonic nanostructures, more functional templates and the precise and high-yield placement of NPs onto the template are of vital importance. Limited to the size of DNA origami, which are approximate to 100 nm, it is hard to assemble more intricate plasmonic nanostructures in large scale. Herein, we firstly employed rectangular DNA origami as the template for the programmed positioning of two 30-nm AuNPs. TEM images showed the high yields of AuNPs’ assembly on the rectangular DNA origami. Then, the connecting strands were used to link the DNA origami templates together, which generated chains of AuNPs. Furthermore, we observed a significant Raman signal enhancement from molecules covalently attached to the AuNP-dimers and AuNP-chains.
2 Experimental
2.1 Chemicals and reagents
All DNA strands were purchased from Sangon (PAGE purification). Thiol-modified DNA was purchased from Takara (high-performance liquid chromatography purification). M13mp18 ssDNA was purchased from New England Biolabs which was used as received. 30-nm gold nanoparticles were obtained by BBI. All chemicals were supplied by Sigma.
2.2 Preparation of AuNPs–DNA conjugates
We added 1 µL 100 nM thiol-modified DNA to 100 µL 30 nm AuNP solution in a 0.5 × TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0) and then incubated the buffer for 4 h at 37 °C. Then, we placed 10 µL of 3 M NaCl into the reaction solution slowly for four times in 2 h, and the final concentration of NaCl was reached at 300 mM. Then, the solution was kept at 37 °C for 12 h. We purified the DNA-functionalized AuNPs by repeated centrifugation (7000 rpm, 10 min). Each time, the supernatant was carefully removed and then the AuNPs were resuspended in a 0.5 × TBE buffer to get rid of excess thiol-modified DNA. The concentration of AuNPs was characterized by UV–Vis spectroscopy.
2.3 Assembly of rectangular DNA origami
A rectangular DNA origami template was obtained in a one-pot construction, in which 2 nM of M13mp18 DNA was incubated with 10 nM of staple strands in a 1 × TAE/Mg2+ (40 mM Tris-HCl, 20 mM boric acid, 2 mM EDTA and 12.5 mM magnesium acetate, pH 8.0) buffer. The mixture was annealed from 95 to 20 °C with slowly decreasing the temperature at a rate of 1 °C min−1. Then, the prepared DNA origami were filtered by the 100 kDa (MWCO) centrifuge filters for four times to remove the extra strands.
2.4 Assembly of AuNPs onto a DNA origami template to form AuNP-chains
The rectangular origami template was mixed with the AuNPs at a molar ratio of 1:4. The mixture was performed by annealing from 45 to 30 °C four times at a rate of 0.6 °C min−1 and then cooling to 4 °C. Then, the 1 µL 2 µM DNA edge staples were added into the previous mixture overnight.
2.5 TEM characterization
For TEM imaging, a 10-µL sample was dropped on a carbon-coated grid (400 mesh, Ted Pella). During deposition for 15 min, the excess solution drop was wicked from the grid by absorption into filter paper. To get rid of the deposited salt, a droplet of water and filter paper were used to remove the excess water away. The grid was kept in a bake oven at 37 °C for 4 h for drying. The TEM characterization was conducted using a Tecnei G2-20S TWIN system and operated at 200 kV with a bright field mode.
2.6 SERS characterization
For SERS characterization, the samples were incubated on a 5 mM 4-mercaptobenzoic acid (4-MBA, 99%) ethanolic solution for 2 h and dropped onto silicon. The SERS spectra were recorded by a confocal Raman microscope (In Via, Renishaw, England) equipped with a 633-nm He–Ne laser. Resolution grating of 1800 grooves and a slit of 100 μM were used on all measurements. The spectra ranged from 1000 to 1850 cm−1. For all measurements, the experimental parameters were as follows: excitation wavelength 633 nm, objective 20×, laser power 0.08 mW and acquisition time 10 s.
3 Results and discussion
3.1 Strategy for the assembly of 30-nm AuNPs into AuNP-chains
In principle, larger-sized AuNPs may generate stronger plasmonic resonance. Compared to small-sized AuNPs, large-sized AuNPs should be assembled with more ssDNA on their surface which may lower the possibility for precipitation. Hence, more capture strands were needed to resist the charge repulsion between DNA–AuNPs and the templates. The rectangular DNA origami were designed by folding a long ssDNA scaffold with hundreds of staples. Four staples at the central part in each half of the DNA origami were designed with a capture part for the anchoring of two 30-nm AuNPs (Fig S1). The gaps between two AuNPs were set to 10 nm. To make the AuNPs into AuNP-chains, linker A and linker B were prolonged to the staples at each side of the rectangular DNA origami, which could be linked together by hybridization with linker C (Fig. 1).
3.2 Assembly and characterization of AuNP-dimers nanostructure based on rectangular DNA origami
The rectangular DNA origami were folded by M13 (virus ssDNA) with hundreds of staples, carrying capture strands and linkers. AFM images (S1) demonstrated the formation of rectangular DNA origami. The schematic in Fig. 2 illustrated that AuNPs were anchored onto the rectangular DNA origami into dimers by the hybridization of capture strands on the template and ssDNA on AuNPs. The gaps between two AuNPs were set to 10 nm. Typical TEM images in Fig. 2 demonstrated the successful assembly of AuNP-dimers. The yield of AuNP-dimers was 80.6% according to the TEM images. The gaps seem to be 1–2 nm between AuNPs, which were much smaller than 10 nm. This phenomenon may be induced by the shrink during the sample preparation for TEM.
3.3 TEM characterization of the assembly of AuNP-chains
In order to obtain larger plasmonic nanostructures, linkers were employed to organize the rectangular DNA origami template into nanoribbons, which could be validated by the AFM images (Fig. S2). By the addition of the linkers, the AuNP-dimers were organized together into AuNP-chains. Typical TEM images showed that 30-nm AuNPs were linked into chains (Fig. 3 and Fig S3). In fact, there was another way to form AuNPs, which used DNA nanoribbon as the template. But this approach seemed much easier to make the AuNPs to participate. In the TEM images, similar phenomenon of shrink also happened. The gaps between AuNPs in AuNP-chains seem to be 1–2 nm wide according to the TEM images.
3.4 AuNP-dimers and AuNP-chains for surface-enhanced Raman spectroscopy
The AuNP-dimers and AuNP-chains are typical plasmonic nanostructures which impelled us to consider their potential functionalities. Previous studies showed that proper distances between AuNPs in the plasmonic nanostructures would generate hot spots which may have induced signals of surface-enhanced Raman scattering (SERS). The distance between the capture strands is 10 nm. When 30-nm AuNPs were anchored, the AuNPs were closed to each other with no gaps. According to the TEM images, the gaps were 1–2 nm because there existed the charge repulsion between DNA–AuNPs. AuNPs with these gaps may generate the coupling of plasmons in the AuNPs assemblies (Fig. S4). 4-Mercaptobenzoic acid (4-MBA) was employed as a Raman-active molecule. They can covalently attach to the AuNPs by the strong interaction of an Au–S bond. The typical SERS spectrum is shown in Fig. 4. The frequency of 1578 and 1075 cm−1 in the SERS spectra was clearly attributed to 4-MBA. Using individual AuNPs samples (black curve) as a control, the AuNP-dimers (red curve) and AuNP-chains (blue curve) obtained significant enhanced signals under SERS detection. Taken the peak at 1075 cm−1 for example, the SERS signal intensity increased from 929.6 (AuNP-dimers) to 4963.9 au (AuNP-chains) (Fig. 5), the reason of which may own to the increasing of hot spots between AuNPs.
4 Conclusion
In summary, we have anchored large-sized AuNPs on to the rectangular DNA origami with high yields. By using the linkers to connect the DNA origami templates together, chains of AuNPs were obtained. Thus, prepared AuNP-dimers and AuNP-chains were validated by TEM images which demonstrated their organization and structural uniformity. Furthermore, significant enhanced signals of SERS detection were obtained for the samples of AuNP-dimers and AuNP-chains. This strategy is a simple and timesaving way to construct large-scale plasmonic nanostructures based on AuNPs, which may open up the prospects of high-ordered plasmonic nanostructures with tailored optical properties.
References
F. Peng, Y. Su, Y. Zhong et al., Silicon nanomaterials platform for bioimaging, biosensing, and cancer therapy. Acc. Chem. Res. 47, 612–623 (2014). https://doi.org/10.1021/ar400221g
J. Mcphillips, A. Murphy, M.P. Jonsson et al., High-performance biosensing using arrays of plasmonic nanotubes. ACS Nano 4, 2210–2216 (2010). https://doi.org/10.1021/nn9015828
J. Chao, D. Zhu, Y. Zhang et al., DNA nanotechnology-enabled biosensors. Biosens. Bioelectron. 76, 68–79 (2016). https://doi.org/10.1002/adma.201002767
J. Zhou, Y. Zuo, X. Wan et al., Solution-processed and high-performance organic solar cells using small molecules with a benzodithiophene unit. J. Am. Chem. Soc. 135, 8484–8487 (2013). https://doi.org/10.1021/ja403318y
F. Wang, C. Li, H. Chen et al., Plasmonic harvesting of light energy for Suzuki coupling reactions. J. Am. Chem. Soc. 135, 5588–5601 (2013). https://doi.org/10.1021/ja310501y
Ł. Bujak, T. Ishii, D.K. Sharma et al., Selective turn-on and modulation of resonant energy transfer in single plasmonic hybrid nanostructures. Nanoscale (2017). https://doi.org/10.1039/c6nr08740j
W. Shen, M.F. Bruist, S.D. Goodman et al., A protein-driven DNA device that measures the excess binding energy of proteins that distort DNA. Angew. Chem. Int. Ed. 43, 4750–4752 (2004). https://doi.org/10.1117/12.2051277
M. Lee, J.U. Kim, K.J. Lee et al., Aluminum nanoarrays for plasmon-enhanced light harvesting. ACS Nano 9, 6206–6213 (2015). https://doi.org/10.1021/acsnano.5b01541
K. Dopf, S. Heunisch, P. Schwab et al., Superresolution optical fluctuation imaging (SOFI) aided nanomanipulation of quantum dots using AFM for novel artificial arrangements of chemically functionalized colloidal quantum dots and plasmonic structures. SPIE Photonics Eur 9, 91260N–912609N (2014). https://doi.org/10.1117/12.2051277
Y. Xiong, Z. Liu, C. Sun et al., Two-dimensional imaging by far-field superlens at visible wavelengths. Nano Lett. 7, 3360–3365 (2007). https://doi.org/10.1021/nl0716449
T. Tian, J.-C. Zhang, H.-Z. Lei et al., Synchrotron radiation X-ray fluorescence analysis of Fe, Zn and Cu in mice brain associated with Parkinson’s disease. Nucl. Sci. Tech. 26, 030506 (2015). https://doi.org/10.13538/j.1001-8042/nst.26.030506
N. Fang, H. Lee, C. Sun et al., Sub–diffraction-limited optical imaging with a silver superlens. Science 308, 534–537 (2005). https://doi.org/10.1126/science.1108759
S. Biswas, J. Duan, D. Nepal et al., Plasmonic resonances in self-assembled reduced symmetry gold nanorod structures. Nano Lett. 13, 2220–2225 (2013). https://doi.org/10.1021/nl4007358
J.A. Fan, Y. He, K. Bao et al., DNA-enabled self-assembly of plasmonic nanoclusters. Nano Lett. 11, 4859–4864 (2011). https://doi.org/10.1021/nl203194m
G. Dai, X. Lu, Z. Chen et al., DNA origami-directed, discrete three-dimensional plasmonic tetrahedron nanoarchitectures with tailored optical chirality. ACS Appl. Mater. Interfaces 6, 5388–5392 (2014). https://doi.org/10.1021/am501599f
C. Helgert, E. Pshenay-Severin, M. Falkner et al., Chiral metamaterial composed of three-dimensional plasmonic nanostructures. Nano Lett. 11, 4400–4404 (2011). https://doi.org/10.1021/nl202565e
A. Dhawan, Y. Du, D. Batchelor et al., Hybrid top-down and bottom-up fabrication approach for wafer-scale plasmonic nanoplatforms. Small 7, 727–731 (2011). https://doi.org/10.1002/smll.201002186
B. Wei, M. Dai, P. Yin, Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012). https://doi.org/10.1038/nature11075
P.W. Rothemund, Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). https://doi.org/10.1038/nature04586
E.S. Andersen, M. Dong, M.M. Nielsen et al., Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009). https://doi.org/10.1038/nature07971
H. Dietz, S.M. Douglas, W.M. Shih, Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009). https://doi.org/10.1126/science.1174251
H. Zhang, J. Chao, D. Pan et al., DNA origami-based shape IDs for single-molecule nanomechanical genotyping. Nat. Commun. 8, 14738 (2017). https://doi.org/10.1038/ncomms14738
A. Kuzyk, R. Schreiber, Z. Fan et al., DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012). https://doi.org/10.1038/nature10889
H. Gu, J. Chao, S.-J. Xiao et al., A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010). https://doi.org/10.1038/nature09026
M.J. Urban, C. Zhou, X. Duan et al., Optically resolving the dynamic walking of a plasmonic walker couple. Nano Lett. 15, 8392–8396 (2015). https://doi.org/10.1021/acs.nanolett.5b04270
X. Lan, Z. Chen, G. Dai et al., Bifacial DNA origami-directed discrete, three-dimensional, anisotropic plasmonic nanoarchitectures with tailored optical chirality. J. Am. Chem. Soc. 135, 11441–11444 (2013). https://doi.org/10.1021/ja404354c
P. KüHler, E.-M. Roller, R. Schreiber et al., Plasmonic DNA-origami nanoantennas for surface-enhanced Raman spectroscopy. Nano Lett. 14, 2914–2919 (2014). https://doi.org/10.1021/nl5009635
V.V. Thacker, L.O. Herrmann, D.O. Sigle et al., DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat. Commun. 5, 3448–3453 (2014). https://doi.org/10.1038/ncomms4448
B. Liu, C. Song, D. Zhu et al., DNA-origami-based assembly of anisotropic plasmonic gold nanostructures. Small 13, 1603991–1603999 (2017). https://doi.org/10.1002/smll.201603991
X. Lan, X. Lu, C. Shen et al., Au nanorod helical superstructures with designed chirality. J. Am. Chem. Soc. 137, 457–462 (2014). https://doi.org/10.1021/ja511333q
C. Zhou, X. Duan, N. Liu, A plasmonic nanorod that walks on DNA origami. Nat. Commun. 6, 9102 (2015). https://doi.org/10.1038/ncomms9102
S. Pal, Z. Deng, H. Wang et al., DNA directed self-assembly of anisotropic plasmonic nanostructures. J. Am. Chem. Soc. 133, 17606–17609 (2011). https://doi.org/10.1021/ja207898r
M. Pilo-Pais, A. Watson, S. Demers et al., Surface-enhanced Raman scattering plasmonic enhancement using DNA origami-based complex metallic nanostructures. Nano Lett. 14, 2099–2104 (2014). https://doi.org/10.1021/nl5003069
Z. Chen, X. Lan, Y.-C. Chiu et al., Strong chiroptical activities in gold nanorod dimers assembled using DNA origami templates. Acs Photonics 2, 392–397 (2015). https://doi.org/10.1021/ph500434f
Author information
Authors and Affiliations
Corresponding authors
Additional information
This work was supported by the National Natural Science Foundation of China (No. 21475064), the Natural Science Foundation of Jiangsu Province (No. BK20151504), Program for Changjiang Scholars and Innovative Research Team in University (No. IRT_15R37), Sci-Tech Support Plan of Jiangsu Province (No. BE2014719), the Priority Academic Program Development of Jiangsu Higher Education Institutions (No. PAPD, YX03001), the Mega-projects of Science and Technology Research (No. AWS13C007) and NUPTSF (No. 214175).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, MZ., Wang, X., Xing, YK. et al. DNA origami-templated assembly of plasmonic nanostructures with enhanced Raman scattering. NUCL SCI TECH 29, 6 (2018). https://doi.org/10.1007/s41365-017-0347-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41365-017-0347-z