Abstract
Spiroplasma citri, the causal agent of citrus stubborn disease (CSD), causes significant losses in citrus crops. An efficient pathogen detection system is critical for epidemiology studies, particularly when a large sample size is involved. In this study, we report the development of an immunomolecular assay, immunocapture real-time polymerase chain reaction (IC-qPCR), targeting the spiralin gene for direct detection of S. citri without DNA isolation. This method can use either plant sample extracts or media in which S. citri was cultivated. The IC-qPCR protocol demonstrated a limit of detection for pure S. citri culture at a Ct value of 36.523 with a 103-fold dilution factor, making it equally sensitive as qPCR, which exhibited signal disappearance at a 10–3 dilution (Ct value of 37.484). In contrast, the immunological double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) test produced positive results up to a 10–2 dilution only. For S. citri-infected citrus samples, the established IC-qPCR protocol had a limit of detection at 36.46 Ct with a 1/64-fold dilution factor, matching the sensitivity of qPCR, where signal disappearance occurred at a 1/64 dilution (Ct value of 37.21). On the other hand, the immunological DAS-ELISA test yielded positive results only up to a 1/16 dilution, with optical density (OD) values of 0.364 and 0.113 for 1/16 and 1/32 dilutions, respectively. The IC-qPCR assay shows no cross-reaction for any other highly related spiroplasma species and bacteria affecting citrus trees including Candidatus liberibacter, Xylella fastidiosa, and Xanthomonas campestris pv. citri. Therefore, IC-qPCR assay provides an alternative quick and very sensitive method to screening S. citri, with the advantage of not requiring any concentration or DNA purification steps while still allowing an accurate diagnosis of CSD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spiroplasma citri, the causal agent of citrus stubborn disease (CSD), is a phloem‐restricted pathogenic mollicute, which belongs to the Spiroplasmataceae family (Mollicutes). In the field, diseased citrus trees have compressed and stunted appearances, and yield poor‐quality fruits with little market value (Sagouti et al. 2022). Despite the lack of extensive studies on the effects of CSD on citrus commercial production, it is well-acknowledged that the damage varies with the severity of the infection (Mello et al. 2010). There has been a reported 19–100% decrease in orange cumulative yield as a result of CSD, as well as a 6–30 and 12% decrease in fruit weight and size, respectively. The economic damages caused by CSD might be significant (Kyriakou et al. 1996; Mello et al. 2010; Pehrson et al. 1988; Yokomi et al. 2010). Extensive sampling carried out in Morocco between 2020 and 2021 revealed the dominance of S. citri in all citrus-growing regions of the country (Sagouti et al. 2023).
The diagnosis of S. citri is commonly conducted by grafting onto indicator plants, employing electron microscopy, cultivating in specific liquid media, observing pathogen cells under dark-field microscopy, using antibodies–antigen‐based methods, and applying standard polymerase chain reaction (PCR) and real-time PCR (qPCR) using specific primer sets (reviewed in Sagouti et al. 2022). However, these methods are notably time-consuming, and the cultivation in artificial media is susceptible to contamination by unintended microorganisms (Rangel et al. 2005).
Furthermore, using the tree set of primers developed by Yokomi et al. (2008) (P58, Spiralin, and P89), limitations were noted in identifying Moroccan isolates of S. citri using primers targeting the spiralin gene (Sagouti et al. 2023).
Immunomolecular assays are considered to be many orders of magnitude more sensitive than conventional enzyme-linked immunosorbent assay (ELISA) or PCR, increasing sensitivity considerably and enabling a wide range of diagnostic applications (Peroni et al. 2008). Therefore, we developed immunocapture qPCR (IC-qPCR) for CSD detection as an alternative to cultivation and purification of S. citri. The primary goal of this study is to develop an IC-qPCR approach that is extremely sensitive, specific, and reliable for detecting S. citri in infected plants and culture medium by comparing it with currently used detection techniques. Primers targeting the spiralin gene were designed with an emphasis on detecting Moroccan isolates of S. citri.
Material and methods
Plant material, bacterial cultivation, and DNA purification
Ten S. citri-infected and ten healthy citrus samples that were previously collected from citrus Moroccan orchards (Sagouti et al. 2022) were coded and ground (three samples per plant; 30 in total). For experimental analysis, petioles plus the main vein of leaf samples (5 g) were placed in a mesh bag containing 4 mL of commercial extraction buffer (PBS) provided by the supplier (Agdia EMEA, France). The Homogenizer HOMEX (Bioreba, Swiss) instrument was used for crushing plant material. Each sample’s ground material was collected in a 1.5 mL tube and centrifuged at 5000 rpm for 5 min. The resulting supernatant was used for the following tests.
The genomic DNA was directly extracted from 200 µL of sample grinding liquid for each plant sample using the MagListoTM 5 M Genomic DNA Extraction Kit (Bioneer, Korea). The DNA was then stored at − 20 °C until use. The DNA concentrations in the extracts were assessed using a NanoDrop ND-1000 UV–Vis Spectrophotometer (NanoDrop Technologies, USA), and they were subsequently adjusted to reach a final DNA concentration of 20 ng/µL.
S. citri MK16 isolate was obtained from infected citrus plants from Moroccan citrus orchards (Sagouti et al. 2022). Petioles were excised, surface disinfected, then chopped to small pieces to optimize the osmotic release of S. citri, and soaked for 30 min in 10 mL of R2 liquid medium (Whitcomb 1983). The suspension was passed through 0.45-µm filters and incubated at 32 °C. The obtained pure cultures were used in the PCR-based assays after DNA extraction, as described below.
Primer design
Specific primers targeting conserved sequences in the spiralin gene of Moroccan S. citri isolates (Sagouti et al. 2022) retrieved from GenBank (Accession nos. U13998, OP204695, OP204695, and OP204697) were designed using Geneious software (Table 1). The specificity of the designed primers was assessed using the Primer-BLAST algorithm (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) to identify potential unspecific signals resulting from partial primer annealing and/or primer dimers.
Real-time PCR assay conditions
The qPCR was performed in QuantStudio 5 real-time PCR system (Applied Biosystems, USA). The mixture consists of a 20 µL final volume containing 2 µL of total DNA extract with 20 ng DNA/µL, 0.6 µL of 10 mM of each primer, and 10 µL of Master Mix PowerUp SYBR Green (Invitrogen, USA). Negative controls consisted of the total DNA extracts of healthy citrus leaves. Positive controls were the extracts of the S. citri citrus-infected leaves. All the samples were analyzed in two replicate tubes in the same PCR run. Reactions were performed for 40 cycles, following cycling conditions: 50 °C for 2 min, incubation at 95 °C for 2 min, 40 cycles of 95 °C for 15 s and 59 °C for 15 s, and extension at 72 °C for 2 min. Fluorescence was measured at the end of each cycle. Melting curve steps were added at the end of qPCR as follows: 60 °C–95 °C at melt rates of 0.5 °C/10 s. The results were analyzed by the QuantStudio 5 series software (Applied Biosystems, USA). To check a positive sample, the qPCR amplicons were separated by electrophoresis, and the products were analyzed by 2% agarose gel electrophoresis with 1xTAE buffer stained with SYBR Safe.
The validation of this new qPCR test was further carried out by comparing its results with those obtained by the test developed by Yokomi et al. (2008) and double-antibody sandwich ELISA (DAS-ELISA). The test was carried out on ten citrus leaf samples collected from plants infected with S. citri kept in a greenhouse. qPCR assays have been run using DNA-binding fluorophore SYBR Green with specific primers that amplify a fragment of putative p58 adhesine-like (Yokomi et al. 2008) and the primer pair designed in this study (Table 1).
DAS-ELISA conditions
For the DAS-ELISA analysis assay, 100 µL of the supernatant from the plant sap of the same samples previously tested by qPCR was used. The test was carried out according to the manufacturer’s instruction (Agdia EMEA, France) and as it was described by Sagouti et al. (2023). The test is based on the use of polyclonal antibody alkaline phosphatase conjugate. Each test plate had two replicates of each sample as well as two wells for positive and negative controls (buffer only and healthy control sample). Absorbance was measured after 1 and 2 h of incubation with the p-nitrophenyl phosphate substrate at 37 °C using a BioTek 800 TS absorbance reader that detects optical density (OD) at 405 nm. The ELISA test is considered positive if the average OD value from each of the duplicate sample wells is ≥ 2 × OD of that in the negative control of healthy (HC) plant extracts. The ELISA test is considered doubtful if the average OD value from each of the duplicate sample wells is between 1.5 and 2 × OD of that in the HC. The ELISA test is considered negative if the average OD value from the duplicate sample is < 1.5 × OD of that in the HC.
Immunocapture real-time PCR assay conditions
The IC-qPCR assay developed in the present study was tested on citrus plant samples infected with S. citri and also on pure cultures of S. citri. For plant samples, of samples previously prepared (30 positives and 30 healthy citrus samples) were directly captured by polyclonal antibody following the method described by Saillard et al. (1996). Multiwall plates for qPCR were incubated at 37 °C for 2 h with 100 μL of polyclonal antibody for S. citri diluted 1:500 in a coating buffer (Agdia EMEA, France). After incubation, three washing steps were performed. A total of 50 μL of sap extract was obtained by grinding the petioles in a General Extraction buffer (Agdia EMEA, France). For the cultured sample, 50 µL of a pure colony of S. citri previously isolated on R2 medium (Sagouti et al. 2023) and resuspended in 1 mL of distilled sterilized water was used. After 2 h of incubation at 25 °C, the multiwall was washed 6 times with autoclaved PBS-Tween (available from SEDIAG at 20X concentration), and a 7th time with autoclaved ultrapure water (commercially available DEPC water), dried, and prepared for qPCR as described previously.
Specificity of immunocapture real-time PCR assay
To determine the IC-qPCR specificity, Spiroplasma kunkelii, Spiroplasma melliferum, Spiroplasma phoeniceum, Xylella fastidiosa, and Xanthomonas campestris pv. citri (Table 2) were captured by polyclonal antibody for S. citri. The conditions for carrying out the IC-qPCR test are similar to those described in the previous section.
Sensitivity of immunocapture real-time PCR assay
Sensitivity was tested using tenfold serial dilutions of a culture of S. citri isolate 16MK in sterile DNA-free water, which ranged from undiluted DNA (at a concentration of 20 ng/µL) to a 104-fold dilution. The PCR of the tenfold dilutions was performed 3 times. Also, ½ serial dilution of an S. citri-infected citrus sample (PC4) in healthy plant extract was used. The average of each dilution was used to determine the theoretical sensitivity and reliability of the IC-qPCR assay.
Reproducibility analysis of immunocapture real-time PCR assay
A sap of seven S. citri-infected citrus samples and one negative sample was used as a template to assess the reproducibility of the IC-qPCR assay. The reactions were repeated three times a day and then for 3 consecutive days to assess the intra-assay and inter-assay standard deviations and the repeatability and stability of the method.
Statistical analysis
R software (R 4.3.0) was used for the statistical analysis. For field validation, the results are presented as the mean for three different IC-qPCR reactions for the same ample. Reproducibility between assays was determined by linear regression analysis. T-tests and regression analysis were employed to assess the relationship between the time of sample crushing and the detection efficiency of the IC-qPCR assay.
Results
Real-time PCR assay: standard curve
The melting curve of all amplified S. citri-positive citrus samples (30 samples) with the new pairs of primers (Sp-1F/Sp-1R) produced similar dissociation curve patterns and gave a clear and distinct melt peak. On the other hand, the negative control samples (30 samples) showed a distinctly separate linear curve due to the absence of signals and the lack of primer dimer formation. This parameter holds significant importance in evaluating the specificity of the curves when employing the SYBR Green methodology. The mean Ct varies from 25 to 30 and Tm = 76.5. The qPCR amplicons were separated by electrophoresis, and the products were analyzed under UV to confirm negative and positive samples. Figure 1A shows the 120-bp amplicon corresponding to S. citri-positive citrus samples.
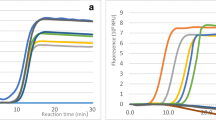
Polymerase chain reaction amplicons produced using Sp-1F/Sp-1R primers with S. citri DNA extracted from plant tissue from the field plants. Lanes 1–16: Moroccan positive samples; lane 17: deionized water control; (M) DNA 100-pb ladder (A); standard curve. Serial dilution 1:10 from 108 to 104 from culture of 16Mk S. citri isolate (B)
The validation of the newly developed qPCR assay in the present study was further carried out by comparing its results with those obtained with a previous assay targeting the P58 gene by testing ten known greenhouse samples infected with S. citri. Our results show a highly significant difference between the two qPCR tests (P = 0.0001308). When comparing the qPCR detection method with that described by Yokomi et al. (2008), the results were superior, with Ct values of 18 and 24 and 25 and 27, respectively (Table 3).
Sensitivity of the IC-qPCR assay
To assess the detection sensitivity of the designed IC-qPCR diagnostic and quantification procedure, the limit of detection (LOD) was evaluated against two standards used as reference (titer of crude-infected citrus sample and S. citri fresh culture solution (108 CFU/mL)). To this end, serial dilutions of each standard were prepared, and IC-qPCR was carried out for each dilution (in triplicate), and the LOD was deduced from the lowest dilution providing effective amplification with the three replicas “The last dilution showing 100% response may be accepted as a conservative estimate of the lower limit of detection.”
A standard curve based on decimal dilution of 108 cfu/mL S. citri fresh culture solution was performed, and three replicates for each dilution were made.
To simplify the interpretation of the results, the equations, reliability, and efficiency of the bacterial titer and standard curves are shown in Figure 1B.
The standard curve was reliable, with an R2 value greater than 0.95 (R2 = 0.9899) (Figure 1B).
However, the efficiency of the RT-qPCR amplification, deduced from the slopes, is over 100%.
The sensitivity of the optimized IC-qPCR assay was determined using tenfold serial dilution of a 108 S. citri isolate 16MK solution prepared from fresh culture and ½ serial dilution of an infected citrus sample (PC4) in the healthy plant extract. For pure S. citri culture, the limit of detection of the designed IC-qPCR protocol was found at the 104 CFU/mL concentration with a Ct value of 35,257. This test is more sensitive than qPCR for which the signal disappears at a concentration of 105 (Ct value of 35.184), whereas the immunological DAS-ELISA test gave a positive result up to the concentration of 106; OD values of 0.138 and 0.124 for dilutions 105 and 104, respectively (Table 4). The established IC-qPCR protocol’s limit of detection for the S. citri in the infected citrus sample was determined to be 36.46 Ct using a 1/64-fold dilution factor, which is similar to that of qPCR, with a signal disappearance at a dilution of 1/64 (Ct value of 37.21). In contrast, the immunological DAS-ELISA test only yielded a positive result up to a dilution of 1/16; OD values of 0.364 and 0.113 for dilutions 1/16 and 1/32, respectively (Table 5).
Specificity of the IC-qPCR assay
No fluorescence signal was detected in the DNA samples isolated from the positive controls of other highly related spiroplasma species (S. melliferum, S. kunkelii, and S. phoeniceum), and other bacteria infecting citrus trees including C. liberibacter, X. fastidiosa, and X. campestris pv citri. No cross-reactivity with the DNA of healthy citrus was observed (Fig. 2). The S. citri from the infected citrus gave a marked fluorescent signal (Ct values between 22 and 27) equivalent to that obtained with the qPCR test; Ct values between 22 and 28 (Table 6).
The sensitivity of the IC-qPCR assays tested by tenfold serial dilutions in sterile water of a DNA extract of pure culture of S. citri isolated from an infected citrus tree (isolate 16MK). Agarose gel by electrophoresis detection: Lane 1: pure culture of S. citri isolate 16MK; Lanes 2–4 serial dilutions up to 10–3 of the 16MK isolate; Lane 5: negative PCR control; and Lane 6: positive PCR control. (M) DNA 100-pb ladder (A) and sensitivity of the IC-qPCR based on SYBR Green I (B)
Repeatability of the IC-qPCR assay
Reproducibility is the precision changing any or several of those parameters: day, operator, procedure, materials, and/or equipment. Repeatability and reproducibility were evaluated as the intra-assay. The repeatability of the IC-qPCR test was calculated by performing the test on a sample deposited in 14 different wells, all repeated over five distinct runs. The average Ct value is 26.5, and the standard deviation is 0, 34. Furthermore, using the data from the five runs, the standard deviation values remained below 0, 5 (Table 7), demonstrating the high repeatability and of the procedure.
Detection in greenhouse samples
The reliability of the IC-qPCR assay and the crude plant extract procedure was determined on a total of three replicates of the same citrus leaf samples (10 infected citrus plants and 10 healthy plants) previously tested with qPCR. Among these tested samples, the IC-qPCR assay reported 30 true-positive and 30 true-negative samples (Figs. 3, 4). The accuracy of the whole procedure developed was compared to the standard qPCR detection of S. citri. It was shown that the results were in accordance with the qPCR results (Fig. 4). No significant difference was observed between the results obtained on the 30 samples tested by IC-qPCR and by qPCR (p-value = 0.1386). This finding confirms the results obtained previously indicating that the immunocapture test developed in the present study is as sensitive as qPCR.
Specificity of the IC-qPCR assays assessed by testing four Spiroplasma isolates, three Spiroplasma species, and three bacterial species infecting citrus trees. Agarose gel by electrophoresis detection. Lane 1: Spiroplasma melliferum; Lane 2: Spiroplasma kunkelii; Lane 3: Spiroplasma phoeniceum; Lane 4: Xylella fastidiosa; Lane 5: Candidatus liberibacter; Lane 6: Xylella fastidiosa; Lane 7: Xanthomonas campestris pv. citri; and Lanes 7–11: S. citri isolates. Lane 12: deionized water control; (M) DNA 100-pb ladder (A) and specificity of the IC-qPCR based on SYBR Green I (B)
Discussion
Plant diseases significantly reduce plant yield, which results in enormous financial losses. In the face of climate change and globalization, accurate diagnosis and identification of plant diseases are crucial for ensuring food security and halting the spread of invasive pests and pathogens (Balodi et al. 2017). In the field, plant pathogens are often recognized by the characteristic disease symptoms they might induce. To implement efficient management strategies that limit or stop the spread of infections and reduce the detrimental effects of the disease, early detection and identification of pathogens are crucial (Trippa et al. 2023). S. citri is one of the pathogens causing significant losses to citrus production worldwide in arid and semiarid citrus-growing regions including in Morocco (Sagouti et al. 2023, 2022). Several detection methods have been reported for the identification of S. citri (reviewed by Sagouti et al. (2022).
From visual inspection and plant disease identification to high-throughput serological techniques such as ELISA and molecular techniques like PCR, the science of plant disease diagnosis has advanced (Balodi et al. 2017). Due to their low-cost and high-throughput potential, serological methods based primarily on ELISA are the most widely used method for pathogen detection; however, this technique is not particularly reliable and sufficiently sensitive for many pathogens detection during the asymptomatic stage of infection (Trippa et al. 2023), including S. citri (Bové et al. 1987; Saillard et al. 1996). Due to its ease of use and capacity to assess a large number of samples, ELISA has been the most frequently used diagnosis technology in the preliminary sanitary evaluation of propagating material. It can detect S. citri in 95% of symptomatic nursery or field trees in citrus (Bové et al. 1987), but its sensitivity is insufficient to detect S. citri in trees that do not exhibit any symptoms. Early pathogen detection during the establishment of plant diseases is challenging due to the restricted number of spiroplasmas in the phloem (Bové et al. 1987; Saillard et al. 1996). Furthermore, results were inconsistent due to low titer and erratic distribution of S. citri (Saillard et al. 1980). Considering that ELISA has some limitations, immunomolecular assays have been developed for the efficient detection of S. citri. The advantage of this method is that it makes sample preparation simpler while improving the specificity and sensitivity of conventional PCR. Compared to ELISA, IC-PCR is more sensitive in detecting S. citri (El-Banna et al. 2005; Saillard et al. 1996).
A comparative study between molecular and serological methods for detecting S. citri in field samples was conducted in Morocco. The results demonstrated that PCR and qPCR, two molecular methods, are more sensitive than ELISA for detecting S. citri in samples exhibiting symptoms of CSD. It is important to highlight that compared to ELISA and PCR, the qPCR was shown to be the most sensitive in making an accurate diagnosis of S. citri (Sagouti et al. 2023). In this study, an IC-qPCR test was developed to circumvent the time-consuming DNA purification step before running the highly sensitive qPCR test. Primers that target the spiralin gene have been designed (Table 1) to do this by referring to sequences of Moroccan isolates of S. citri. The primary reasons for selecting this gene are that it is responsible for spiralin which is the major protein of S. citri (El-Banna et al. 2005) and its sequence is highly conserved among all spiroplasma strains (Duret et al. 2003). With this as a foundation, the developed method will not only enable the detection of S. citri isolates from Morocco but also those from different countries as well.
Using the recently designed primers (Table 1), we first confirmed the qPCR test before starting the IC-qPCR assay. This test was initially run on thirty known S. citri-infected greenhouse samples and thirty negative samples. With the new primer pairs (Sp-1F/Sp-1R), the melting curves of all thirty amplified S. citri-positive citrus samples generated comparable dissociation curve patterns and yielded a distinct melt peak. However, because there were no signals and no primer dimer synthesis, the thirty samples in the negative control group had a distinct linear curve. This parameter is very important to consider when assessing the curves’ specificity using the SYBR Green approach. The Tm is equivalent to 76.5 °C, and the mean Ct ranges from 25 to 30. The qPCR test was subsequently compared to that targeting the putative adhesion gene P58 known for its high sensitivity in the detection of S. citri (Sagouti et al. 2023; Yokomi et al. 2008). According to our findings, there was a significant difference (P = 0.0001308) between the two qPCR assays by testing ten known greenhouse samples infected with S. citri. The qPCR detection approach yielded better results than the technique published by Yokomi et al. (2008), with Ct values ranging from 18 to 24 and 25 to 27, respectively (Table 2). Our results allowed the validation of the qPCR test developed in the present study. In light of these results, an IC-qPCR test was developed using polyclonal antibody alkaline phosphatase conjugate. This choice is supported by the fact that a wide variety of antigens can be detected using antibody-based PCR tests, such as IC-PCR, which combine the sensitivity of PCR with the versatility of immunoassays, such as ELISA (Peroni et al. 2008).
It is noteworthy to emphasize that this study is the first to report on the use of real-time PCR in conjunction with immunocapture for S. citri detection. The last tests were done with conventional PCR (El-Banna et al. 2005; Saillard et al. 1996). IC-PCR, like ELISA, requires the capture of the bacterial cells by coating the wells with specific polyclonal antibodies as an initial step (Peroni et al. 2008) followed by antigen coating. Therefore in preliminary tests, different temperatures (25 and 31 °C) and incubation times (2 and 4 h) were evaluated to establish the best conditions for IC-qPCR, since the coating conditions may influence the accuracy of pathogen detection. The incubation at 25 °C for 2 h was enough for coating the antigen onto the plate, and this condition was adopted in all further experiments (data not shown). It is crucial to highlight that, rather than letting the antigen incubate overnight at 4 °C, we were able to shorten its incubation period to 2 h at 25 °C. In other words, in addition to avoiding time-consuming DNA purification steps, we were also able to optimize the IC-qPCR test. The successful validation of reducing the antigen incubation time in IC-qPCR from 2 days to 6 h represents a significant advancement in diagnostic methodology, with potential benefits in terms of efficiency, cost savings, and robustness. However, careful attention to quality control and regulatory considerations is essential to ensure that diagnostic accuracy and reliability are not compromised by this change.
Specificity, inclusivity, exclusivity, sensitivity, accuracy, and precision are indicators of an assay’s reliability and robustness. As opposed to assays used for screening and surveying, confirmatory assays for regulatory applications are typically subjected to a more stringent evaluation process in terms of accuracy and detection specificity. However, assay validation may fall short in certain emergency scenarios; recognized analytical specificity, sensitivity, and reproducibility are the minimal requirements (Cardwell et al. 2018). Additionally, a precise, sensitive, and specific diagnosis is required for the effective and affordable control of plant diseases (Balodi et al. 2017). For this reason, we were interested in assessing the sensitivity, specificity, and repeatability of the developed IC-qPCR test in the current study.
The sensitivity was evaluated using tenfold dilutions of S. citri isolate “16MK” culture and ½ serial dilution of an infected citrus sample (PC4) in the healthy plant extract. The results demonstrated that the IC-qPCR test is more sensitive than aPCR and the immunological DAS-ELISA test, which yielded a positive result only up to a dilution of 106 CFU (OD values of 0.138 and 0.124 for dilutions 105 and 104, respectively), and more sensitive of qPCR, for which the signal disappears at a dilution of 104 as well (Table 3). Using a 1/64-fold dilution factor, the estimated limit of detection for the S. citri-diseased citrus sample in the IC-qPCR technique was 36.46 Ct. Comparing this test to the immunological DAS-ELISA test, which only produced a positive result up to a dilution of 1/16 (OD values of 0.364 and 0.113 for dilutions 1/16 and 1/32, respectively), it demonstrates a better level of sensitivity and the same level of signal disappearance as qPCR (Ct value of 37.21) (Table 4).
The cross-reactivity can interfere with the specificity, making the diagnosis useless, due to false-positive results (Peroni et al. 2008) even in the absence of the S. citri. The used polyclonal antibodies are highly specific for S. citri, since it has no cross-reactivity in IC-qPCR with other related spiroplasma species such as S. melliferum, S. kunkelii, and S. phoeniceum and other bacterial species infecting citrus such as C. liberibacter, X. fastidiosa, and X. campestris pv. citri. Furthermore, no cross-reactivity with the DNA of healthy citrus was observed (Table 5).
To evaluate the repeatability of the IC-qPCR technique, seven citrus leaf samples from seven S. citri-diseased plants kept in a greenhouse were used. The outcomes show how repeatable the IC-qPCR test is over three days as well as on a single day. Between samples tested at 1 h intervals on the same day (P > 0.05) and those tested at 1 day intervals for 3 days (P > 0.05), there was no discernible variation in the Ct values (Fig. 3).
Following verification of the IC-qPCR test’s reliability and robustness, we analyzed known S. citri-infected samples that were kept in a greenhouse. The developed procedure’s accuracy was assessed in comparison with the standard qPCR detection of S. citri. The results from both IC-qPCR and qPCR on the 30 tested samples showed no significant difference (p-value = 0.1386) (Fig. 5). This reaffirms the previous findings, indicating that the immunocapture test introduced in this study is equally sensitive as qPCR.
Detection of S. citri in greenhouse samples by the IC-qPCR and qPCR tests developed in the present study. The Ct values reflect S. citri DNA accumulation. Within the boxes, the horizontal line indicates the median value (50% quantile), the box itself delimits the 25% and 75% quantiles, and lines represent the normal range of the values. Boxplots with letters indicate significant differences of DNA accumulation (T-test; p-value = 0.1386): Small letters correspond to the comparisons between S. citri detection by qPCR and IC-qPCR
The method developed in the present study can be used for routine diagnosis of CSD and may aid in the study of S. citri epidemiology. The sensitivity, specificity, and repeatability of the IC-qPCR make it possible to detect S. citri even in asymptomatic samples, which could improve the management of CSD.
References
Balodi R, Bisht S, Ghatak A, Rao KH (2017) Plant disease diagnosis: technological advancements and challenges. Indian Phytopathol 70:275–281. https://doi.org/10.24838/ip.2017.v70.i3.72487
Bové JM, Vignault JC, Saillard C (1987) Spiroplasma citri detection by enzyme- linked immunosorbent assay (ELISA), culture and dot hybridization. J Med Sci 23:729–731
Cardwell K, Dennis G, Flannery AR, Fletcher J, Luster D, Nakhla M, Rice A, Shiel P, Stack J, Walsh C, Levy L (2018) Principles of diagnostic assay validation for plant pathogens: a basic review of concepts. Plant Heal Prog 19:272–278. https://doi.org/10.1094/PHP-06-18-0036-RV
Clark TB, Whitcomb RF, Tully JG, Mouches C, Saillard C, Bové JM, Wroblewski H, Carle P, Rose DL, Henegar RB, Williamson DL (1985) Spiroplasma melliferum, a new species from the honeybee (Apis mellifera). Int J Syst Bacteriol 35:296–408. https://doi.org/10.1099/00207713-35-3-296
Duret S, Berho N, Danet JL, Garnier M, Renaudin J (2003) Spiralin is not essential for helicity, motility, or pathogenicity but is required for efficient transmission of spiroplasma citri by its leafhopper vector circulifer haematoceps. Appl Environ Microbiol 69:6225–6234. https://doi.org/10.1128/AEM.69.10.6225
El-Banna OM, AbouZeid AA, Fawzya IM, Azza GF (2005) Immunocapture polymerase chain reaction (IC-PCR) and nucleic acid hybridization techniques for detection of spiroplasma citri. Int J Virol 1:13–13
Kyriakou A, Eliades G, Ioannou N, Kapari-Isaia T (1996) Effect of stubborn disease on growth, yield and fruit quality of frost washington navel and frost valencia oranges in cyprus. J Hortic Sci 71:461–467. https://doi.org/10.1080/14620316.1996.11515427
Mello AFS, Yokomi RK, Melcher U, Chen JC, Fletcher J (2010) Citrus stubborn severity is associated with spiroplasma citri titer but not with bacterial genotype. Plant Dis 94:75–82. https://doi.org/10.1094/PDIS-94-1-0075
Pehrson JE, Gumpf DJ, Ohr HD (1988) Living with stubborn disease in Central California citrus, in: tenth international organization of citrus virologists conference proceedings (1957–2010), 10(10). pp. 304–306. https://doi.org/10.5070/C55wf251nd
Peroni LA, dos Reis JRR, Coletta-Filho HD, de Souza AA, Machado MA, Stach-Machado DR (2008) Assessment of the diagnostic potential of immmunocapture-PCR and immuno-PCR for citrus variegated chlorosis. J Microbiol Methods 75:302–307. https://doi.org/10.1016/j.mimet.2008.06.024
Rangel B, Krueger RR, Lee RF (2005) Current research on Spiroplasma citri in California, in: Sixteenth international organization of citrus virologists conference proceedings. pp. 439–442. https://doi.org/10.5070/C555n9m8n6
Saglio P, Lhospital M, Laflèche D, Dupont G, Bové JM, Tully JG, Freundt EA (1973) Spiroplasma citri gen. and sp. n.: a mycoplasma—like organism associated with “ stubborn ” disease of citrus. Int J Syst Evol Microbiol 23:191–204. https://doi.org/10.1099/00207713-23-3-191
Sagouti T, Belabess Z, Rhallabi N, Barka EA, Tahiri A, Lahlali R (2022) Citrus stubborn disease: current insights on an enigmatic problem prevailing in citrus orchards. Microorganisms 10:1–23. https://doi.org/10.3390/microorganisms10010183
Sagouti T, Rhallabi N, Polizzi G, Tahiri A, Belabess Z, Barka EA, Lahlali R (2023) Comparison of serological and molecular methods for detection of spiroplasma citri in moroccan citrus-growing areas. Plants 12:1–11. https://doi.org/10.3390/plants12030667
Saillard C, Vignault JC, Bove JM, Raie A, Tully JG, Williamson DL, Fos A, Garnier M, Gadeau A, Carle P, Whitcomb RF (1987) Spiroplasma phoeniceum sp. nov., a new plant-pathogenic species from Syria. Int J Syst Bacteriol 37:106–115. https://doi.org/10.1099/00207713-37-2-106
Saillard C, Garcia-Jurado O, Bové JM, Vignault JC, Moutous G, Fos A, Bonfils J, Nhami A, Vogel R, Viennot-Bourgin G (1980) Application of ELISA to the detection of spiroplasma citri in plants and insects, in: eighth international organization of citrus virologists conference proceedings. pp. 145–152
Saillard CA, Nhami A, Moreno P, Garnier M, Bové JM (1996) Spiroplasma citri detection by immuno-capture PCR, in: thirteenth international organization of citrus virologists conference proceedings (1957–2010). p. 413
Trippa D, Scalenghe R, Basso MF, Panno S, Davino S, Morone C, Giovino A, Oufensou S, Luchi N, Yousefi S, Martinelli F (2023) Next-generation methods for early disease detection in crops. Pest Manag Sci. https://doi.org/10.1002/ps.7733
Whitcomb RF (1983) Culture media for spiroplasmas. In: Razin S, Tully JG (eds) Methods in mycoplasmology. New York
Whitcomb RF, Chen TA, Williamson DL, Lia C, Tully JG, Bové JM, Rose DL, Coan ME, Clark TB (1986) Spiroplasma kunkelii sp.nov.: characterization of the etiological agent of corn stunt disease. Int J Syst Biol 36:170–178
Yokomi RK, Mello AFS, Saponari M, Fletcher J (2008) Polymerase chain reaction-based detection of Spiroplasma citri associated with citrus stubborn disease. Plant Dis 92:253–260
Yokomi RK, Mello AFS, Fletcher J, Saponari M (2010) Estimation of citrus stubborn incidence in citrus groves by real-time PCR, in: seventh international organization of citrus virologists. pp. 131–141. https://doi.org/10.5070/C50ph9p8sx
Acknowledgements
This research was financially supported by the Department of Plant Protection, Ecole Nationale d’Agriculture de Meknes, and the Faculté des Sciences et Techniques de Mohammedia.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethical approval of human or animal rights
All authors are fully aware of this submission and have declared that they have no competing interests. This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
All authors have reviewed the manuscript and approved its submission to Journal of Plant Disease and Protection.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sagouti, T., Rhallabi, N., Tahiri, A. et al. Development and application of an immunocapture real-time PCR for the detection of Spiroplasma citri, the causal agent of citrus stubborn disease. J Plant Dis Prot (2024). https://doi.org/10.1007/s41348-024-00960-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41348-024-00960-8