Abstract
Orthotospoviruses (genus Orthotospovirus, family Tospoviridae) are amongst the most devastating plant viruses worldwide, causing severe damage to many economically important vegetable crops, such as tomato and sweet pepper. Monitoring virus populations is an important step for estimating virus damage and epidemiology, and gaining insights into the adaptation processes undergone by orthotospoviruses. Here, we studied the orthotospovirus populations infecting vegetable crops in Brazil and the Dominican Republic, including species diversity, genome comparison and phylogenetic analyses. Comparisons of virus populations showed that in Brazil, which is considered a center of orthotospovirus diversity, groundnut rinspot virus (GRSV) is prevalent, infecting 41% of the plants, whereas tomato spotted wilt virus (TSWV) and tomato chlorotic spot virus (TCSV) were present in 4% and 9% of the samples, respectively. In the Dominican Republic, which can be considered an environment with low orthotospovirus diversity, 55% of the samples were infected with TSWV, 11% showed TCSV infection and no GRSV was detected. The occurrence of mixed infection was low in Brazil, at only 5%, but no mixed infection was detected in the Dominican Republic. The low rates of mixed infections may prevent the emergence of genomes resulting from reassortment. Indeed, no reassortant viruses were detected in either country, except for TCSV, recently proposed as representing a reassortant orthotospovirus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Orthotospoviruses (genus Orthostospovirus, family Tospoviridae) are amongst the most devastating plant viruses worldwide, causing severe damage to many economically important vegetable crops, such as tomato and sweet pepper (Pappu et al. 2009; Scholthof et al. 2011). Their importance is not only due to the drastic economic losses caused but also because their control has proven to be difficult, since they have a wide host range and are effectively spread by thrips vectors (Pappu et al. 2009).
Brazil is among the countries with the highest number of reported orthotospovirus species, together with some countries in Asia. Groundnut ringspot virus (GRSV), Tomato spotted wilt virus (TSWV) and Tomato chlorotic spot virus (TCSV) have been the most prevalent orthotospoviruses identified in the country since the early 1990s. They share a similar host range among a high number of plant species and are efficiently transmitted by the thrips species Frankniella occidentalis and F. shultzei (Nagata et al. 1995; Resende et al. 1996, Nagata et al. 2004). Therefore, these virus species are often found in mixed infections in overlapping hosts and vectors. As a consequence, occurrence of genome rearrangements among orthotospovirus species is expected to happen in nature.
In contrast to the situation in Brazil, orthotospoviruses were only recently reported in Central America and the Caribbean region. The Dominican Republic contains large areas of vegetable production, which contribute significantly to the country’s economy. In the Dominican Republic, TSWV was reported for the first time in 2009, simultaneously with the identification of F. occidentalis on the Island. Recently, with the introduction of new virus species, vegetable production has been threatened by orthotospovirus such as TCSV (Martínez et al. 2013; Almeida et al. 2014; Martínez et al. 2018).
Orthotospovirus species are classified mainly using the amino acid sequence of the nucleocapsid (N) protein that is encoded in the S RNA (Plyusnin et al. 2012). New isolates can only be recognized as a new species when their N protein shares less than 90% amino acid sequence identity with members of established species. Although the N protein is used to demarcate new species, phylogenetic comparisons based on the other genome-encoded proteins, such as non-structural protein (NSs), cell-to-cell movement protein (NSm), precursor (GP) to the glycoproteins (Gn and Gc) and L RNA (Kormelink et al. 1994; Adkins et al. 1995; Takeda et al. 2002) are also able to provide useful information on the evolutionary history of orthotospoviruses, as shown by De Oliveira et al. (2012).
According to recent studies, the evolution of orthotospoviruses was mainly driven by the occurrence of intra- and inter-specific viral genome rearrangements for adaptation to different environmental conditions (Margaria et al. 2007; Webster et al. 2011, 2015; Tentchev et al. 2011; Silva et al. 2019). In attempting to develop defense strategies against phytosanitary problems, knowledge of the causal agents and their variability is indispensable, which in turn is crucial for development of efficient tools to elucidate their biological characteristics and their interactions in the environment. Monitoring virus populations in different ecological areas (e.g. Brazil and the Dominican Republic) may shed light on adaptation processes undergone by orthotospovirus species. In addition, the introduction and/or appearance of new species and/or virus variants may drastically affect the behavior of plant varieties and cultivars against virus infection in a determined ecological niche.
Therefore, the aim of this work was to study the orthotospovirus populations infecting vegetable crops in contrasting ecological areas, including species diversity, genome comparisons and their correlation with phylogenetic relationships and virus evolution.
Materials and methods
Field surveys and isolates collection
Isolates of GRSV, TCSV and TSWV found in vegetable crops collected in Brazil and in the Dominican Republic were analyzed based on their genome composition. Sampling was carried out in vegetable-growing regions of the Brazilian states of Goiás, São Paulo and Ceará and in the Distrito Federal. In the Dominican Republic, sampling was conducted in agricultural areas of the provinces La Vega, Monseñor Nouel, San Jose de Ocoa and Santiago.
Initially, in both countries, a total of 166 samples of symptomatic leaves showing orthotospovirus-like symptoms were collected from plants of the families Solanaceae, Fabaceae, Asteraceae, Alliaceae and Chenopodiaceae between the years of 2013 and 2016. Samples were collected at random, mainly in open fields, although samples from protected crops were also collected in the Dominican Republic. The main purpose was to collect samples from solanaceous plants, with emphasis on tomato and sweet pepper. Collected samples were maintained at 4 °C for short-term and at −80 °C for long-term storage.
In additional surveys conducted in the Dominican Republic in 2017 and 2018, 62 symptomatic plants were tested for common viruses infecting plant species of the Solanaceae family using immunostrips (Agdia Inc.) for Tobacco mosaic virus (TMV), Cucumber mosaic virus (CMV) and Potato virus Y (PVY). Positive samples were confirmed either by ELISA with species-specific antisera or RT-PCR using virus-specific primers (Table 1).
Enzyme-linked immunosorbent assay (ELISA)
Plates (96-well) were first coated with a capture commercial IgG (Agdia Inc.) for PVY, TMV, CMV or TSWV (100 μL/well) diluted in 0.01 mol/L phosphate buffer saline (PBS, pH 7.4) at 4 °C overnight. After three washes with PBS containing 0.05% Tween 20 (PBST), the wells were blocked with PBST containing 5% skimmed milk powder for 30 min. Crude extracts of infected sweet pepper and tomato leaves diluted at 1:20 (w/v, g/mL) in PBS were added to each well, and the plates were incubated at 37 °C for 1 h. After three washes with PBST, the corresponding commercial conjugate (Agdia Inc.) was added to the wells, and the plates were incubated at 37 °C for 1 h. After four washes with PBST, a p-nitrophenylphosphate solution (Sigma-Aldrich) was added to each well. A positive control (infected tissue) and a negative control (sterilized water) were used, and the results were measured visually based on color intensity.
RT-PCR and sequence analysis
Total RNA of all 228 (166 plus 62) samples collected from 2013 to 2018 were extracted by the Trizol® method (Invitrogen) following the manufacturer’s specifications. The integrity of the extracted RNA was verified by agarose gel electrophoresis for the presence of intact 16S and 18S ribosomal RNA in the samples. Initially, all samples were tested with a pair of primers synthesized from a conserved region of the L RNA of GRSV, TCSV and TSWV, to confirm the infection by orthotospovirus in the samples. Then, the positive samples were evaluated with species-specific primers to separately detect the S, M and L RNA of GRSV, TCSV and TSWV (Table 1). For final orthospovirus species identification, in addition to the conserved L RNA sequences, the N protein genes were amplified and sequences were determined and compared.
Reassortant analysis
For reassortant analyses, a representative isolate of each orthotospovirus species found in each plant species analyzed, collected from the different geographic regions sampled in both countries, was completely sequenced. For cloning and sequencing of the complete genes, cDNA was synthetized with M-MLV reverse transcriptase (Promega) using specific primers designed for N, NSm and L genes (Table 1). PCR was performed with one isolate from each plant species using Platinum Taq polymerase (Invitrogen) in a Gene Amp PCR System 2400–9600 (Perkin Elmer) and the program: 5 min denaturation at 95 °C followed by 35 cycles of 10 s at 95 °C, 10 s annealing at appropriate temperature for each primer, 50 s extension at 72 °C, and a 5 min final extension at 72 °C. Amplified DNA products were resolved on a 1% agarose gel, fragments corresponding in size to each viral gene were gel purified and subsequently cloned into pGEM-T Easy vector (Promega). Positive clones were selected and sequenced.
The RNA sequences representing the S, M and L RNAs segments of the GRSV, TCSV and TSWV isolates were aligned and compared with sequences of other isolates deposited in the GenBank using CLUSTALW algorithm implemented in the program MEGA v6.0. Nucleotide and amino acid sequence identities between TSWV isolates were calculated from the p-distance with MEGA v6.0 as (1 – p-distance).
Phylogenetic analyses of the NSm genes
In order to search for reassortant genomes in virus from samples obtained in Brazil and the Dominican Republic, sequences of NSm genes from TCSV, GRSV and TSWV isolates (an isolate of each orthotospovirus species, from each plant species analyzed, collected in the different geographic regions sampled) were used to construct a neighbor-joining tree with 1000 bootstrap replicates. TSWV sequences were included in this analysis for comparison. The partial amino acid sequences encoded by the S, M and L segments of the three orthotospovirus species were used to generate maximum likelihood (ML) trees. For comparison of the M RNA segment, the amino acid sequences were concatenated. Only orthotospovirus isolates with all open reading frames (ORFs) sequenced (L, M and S RNA segments) were included in this analysis. Multiple alignments were done using the program MUSCLE (Edgar 2004), and the phylogenetic trees were built using the software PhyML v3.2 (Guindon et al. 2010) with 1000 bootstrap replicates implemented in Geneious 8.
Results
Orthotospovirus population and distribution in Brazil and the Dominican Republic
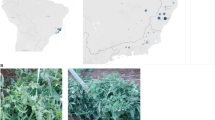
In the field surveys conducted between 2013 and 2018, a total of 228 samples were analyzed. Overall, more than 57% of the tested samples (130 out of 228) were positive for orthotospovirus infection. Tables 2 and 3 and Fig. 1 show the comparison of virus populations in both countries. In Brazil, which can be considered an environment with high orthotospovirus diversity, GRSV was prevalent, infecting approximately 41% of all tested plants, whereas TSWV and TCSV were present in approximately 4% and 9% of the samples analyzed, respectively. In the Dominican Republic, which shows a low orthotospovirus diversity, 55% of the samples analyzed were infected by TSWV and 11% by TCSV. So far, GRSV has not been detected (Fig. 1, Tables 2 and 3).
Comparison of orthotospovirus populations in Brazil (BR) and in the Dominican Republic (DR). The RNA segments of groundnut ringspot virus (GRSV), tomato chlorotic spot virus (TCSV, and tomato spotted wilt virus (TSWV) were detected in symptomatic plants by RT-PCR with universal and/or specific primers. The percentage values indicate the number of single and mixed infections out of all samples tested. The colored areas (orange) indicate the regions where samples were collected in each country
In Brazil, GRSV was detected in the majority of the infected samples collected in the states of São Paulo, Ceará, Goiás and Distrito Federal (Table 2). The mixed infection index was approximately 5% for all tested samples, which represents 10% of the total infected samples. Mixed infections between GRSV, TSWV and TCSV were detected only in samples collected in Brazil, occurring in tomato (Solanum lycopersicum; GRSV + TCSV), sweet pepper (Capsicum annuum; GRSV + TSWV), and Datura stramonium (GRSV + TSWV). Interestingly, the TSWV species was detected in the Brazilian samples only in mixed infections.
In the Dominican Republic, TCSV was identified in tomato, sweet pepper, hot pepper (C. frutescens), bean pod (Vigna unguiculata) and common clover (Trifolium sp.) plants collected in the province of La Vega and in the tobacco (Nicotiana tabacum) samples from the province of Santiago. TSWV was found in tomato and potato (Solanum tuberosum) plants from the province of La Vega, in sweet pepper plants from Monseñor Nouel and in American black nightshade (S. americanum), hot pepper (Capsicum sp.) and tomato plants from San Jose de Ocoa (Tables 2 and 3).
Using the primers designed for specific detection of the L, M and S segments of GRSV, TCSV and TSWV (Table 1), no new (reassortant) genome rearrangement was detected among the orthotospoviruses assessed. However, the recently proposed reassortant TCSV genome (Silva et al. 2019) was present in both countries. It is worth noting that out of 62 samples that were collected in San Jose de Ocoa in the Dominican Republic and further analyzed, 17 from tomato plants and 15 from sweet pepper plants were positive for both PVY and TSWV, as confirmed by ELISA and/or RT-PCR (Table 3). TSWV-infected samples were negative for TMV, CMV, TCSV and GRSV. Hence, no double infections between orthotospovirus species were detected (Table 3).
Phylogenetic analyses based on the M RNA segment
To investigate the presence of reassortant genomes in Brazil and the Dominican Republic, the M RNA segment was used. To perform these analyses, a viral isolate of each orthotospovirus species, found in each plant species analyzed, from the different geographic regions sampled in both countries, was completely sequenced and included in a comparison. New reassortant genomes were detected neither in Brazil nor in the Dominican Republic by analyzing the genome composition of the orthotospovirus isolates. However, based on the recent work performed by Silva et al. (2019), the TCSV found in this study, most likely is a reassortant species.
The sequences of the NSm gene available in the GenBank and the sequences obtained in this work were used to evaluate whether TCSV and GRSV isolates share a highly identical M segment. NSm genes of TSWV isolates were also included for comparison. By using the GRSV and TCSV genome sequences previously made available by Silva et al. (2019), phylogenetic trees were built with protein sequences derived from the M RNA segments. In the trees based on protein sequences of the S and L segments, TCSV and GRSV isolates clearly clustered separately, forming two independent groups (data not shown). In contrast, in the phylogenetic tree based on protein sequences of M segments, TCSV isolates intercalated in a single group together with a previously assumed GRSV reassortant genome (Webster et al. 2011) and a GRSV isolate from peanut recently identified in Brazil (Fig. 2 and Fig. S1). It is worth mentioning that GRSV isolates grouped in two distinct clusters, one containing only GRSV isolates and another in which GRSV isolates intercalate with TCSV isolates. These observations confirm the recent proposal by Silva et al. (2019) that TCSV represents the first reported interspecific reassortant orthotospovirus genome.
Phylogenetic trees based on concatenated protein sequences encoded in M RNAs of tomato chlorotic spot virus (TCSV), groundnut ringspot virus (GRSV) and tomato spotted wilt virus (TSWV) from different geographical regions. Maximum likelihood tree and protein indentity plots of TCSV, GRSV and TSWV. The TCSV from Dominican Republic is shown in yellow
Discussion
In this work, two distinct environmental conditions for orthotospovirus infection were compared. In Brazil, considered an ecological region with high orthotospovirus diversity and presence of virus species for a long period of time, GRSV was detected in the majority of the infected samples. This result shows that GRSV continues to be the most abundant species of the genus in horticultural regions of Midwestern and Northeastern Brazil, and that its population has been expanding to the Southeastern region. In contrast, the incidence of TSWV has been clearly decreasing in all regions. Our results are different from those obtained in previous surveys performed more than 20 years ago, in which the distribution of species varied according to the geographic area (Nagata et al. 1995; Avila et al. 1996).
The reasons for the prevalence of GRSV in the field are probably related to virus-vector interactions. In Brazil, the same species of thrips, namely, Frankliniella occidentalis and F. shultzei, that transmit TSWV also transmit GRSV (Nagata et al. 2004). However, the transmission efficiency of GRSV by F. shultzei, which is the prevalent vector species in the country, is significantly higher when compared to that of TSWV.
In the Dominican Republic, an emergent ecological niche for orthotospovirus showing a low virus diversity, TSWV was found in tomato and potato plants from the province of La Vega, in sweet pepper plants from Monseñor Nouel, and in nightshade, hot pepper and tomato plants from San Jose de Ocoa. These new reports demonstrate TSWV is expanding its incidence to new vegetable crops and new areas in the country. On the other hand, TCSV is reported for the first time in the province of La Vega in this study. It was detected in approximately 11% of the total samples tested, which corresponds to 17% of the orthotospovirus positive samples infecting tomato, sweet pepper, hot pepper and bean pod plants. Furthermore, it was even found in alternative hosts, such as common clover. TCSV was also found infecting tobacco plants for the first time in the province of Santiago.
Taken together, these findings clearly demonstrate that TCSV was introduced in the Dominican Republic, probably around five years ago (Almeida et al. 2014; Martínez et al. 2018) and was able to infect several crops and spread to different vegetable-growing areas in the country. The transmission efficiency of TSWV and TCSV by the main thrips species present on the Island will likely determine which orthotospovirus will prevail. It should be noted that GRSV, the most prevalent orthotospovirus in Brazil and efficiently transmitted by F. shultzei, has not yet been reported in the Dominican Republic. However, this species has already been reported in Central America and the Caribbean basin and will most likely be introduced in the Dominican Republic soon. This fact will certainly make the orthotospovirus population and its evolution in the country much more complex.
As shown here, TSWV has been disseminated for the last few years to various crops and localities in the Dominican Republic, causing yield reductions particularly in tomato and sweet pepper crops. In samples collected in San Jose de Ocoa, TSWV and PVY were detected in mixed infection on tomato plants and to a lesser extent on sweet pepper samples. This finding is important from the epidemiological point of view, in order to design control strategies for tospoviruses. Both viruses have been reported causing mixed infection. PVY was reported in mixed infections with viruses such as Tobacco etch virus (TEV), TMV and CMV in pepper plants (Nuez et al. 2003). However, from the data obtained here, more work needs to be done to address the question of whether PVY could somewhat reduce the presence of TSWV (Gil-Soler et al. 2012) or if, on the contrary, synergism between the two viruses might exist (Chávez-Calvillo et al. 2016). Typical PVY symptoms were not observed in any of the samples collected in the Dominican Republic, which could imply that TSWV infection masked the PVY symptoms.
Tomato plants appear to be more susceptible to mixed infection than sweet pepper plants. For example, TSWV was reported causing mixed and synergistic infections with Tomato chlorosis virus (ToCV) in tomato plants (García-Cano et al. 2006). Perhaps, it could also be the case for the mixed infections found in tomato and sweet pepper plants in the Dominican Republic. Furthermore, mixed infections between different orthotospoviruses, namely TSWV and INSV, have been documented in other crops (Mavrič 2001).
Comparing the orthotospovirus populations in the two distinct environmental conditions, the occurrence of a mixed infection between species was relatively low, reaching approximately 5% of total tested samples, which represents 10% of orthotospovirus infected samples in Brazil, but no mixed infection was detected in the Dominican Republic. This fact may prevent the appearance of new reassortant genomes, especially in Brazil, where an overlapping of orthotospovirus and vector species is expected. Indeed, comparing the viral isolates of each orthotospovirus species found in each plant species analyzed, from the different geographic regions in both countries, no new reassortant genomes were detected. However, based on recent work by Silva et al. (2019), TCSV isolates present in both countries should be considered as reassortant orthotospovirus genomes.
Our analyses comparing orthotospovirus species revealed that the TCSV and GRSV M segments exhibit lower diversity accumulation in comparison with the S and L segments. The M segment of TCSV isolates presented less nucleotide diversity than GRSV isolates, suggesting that the TCSV isolates may represent reassortant genomes. Based on the phylogenetic tree organization, it could posited that reassortment may have occurred between TCSV and GRSV, since TCSV sequences are scattered throughout the TCSV and GRSV cluster for the NSm gene (data from this work and Silva et al. 2019). The acquisition by TCSV of the M RNA region from GRSV could have influenced the recent spread of TCSV to North and Central America and to the Caribbean basin, including the Dominican Republic (Almeida et al. 2014; Webster et al. 2015; Londoño et al. 2012; Martínez et al. 2018), but that hypothesis remains to be proven.
References
Adkins S, Quadt R, Choi TJ, Ahlquist P, German T (1995) An RNA-dependent RNA polymerase activity associated with virions of Tomato spotted wilt virus, a plant- and insect-infecting bunyavirus. Virology 207:308–311
Almeida MMS, Orílio AF, Melo FL, Rodriguez R, Feliz A, Cayetano X, Martínez RT, Resende RO (2014) The first report of Tomato chlorotic spot virus (TCSV) infecting long beans and chili peppers in the Dominican Republic. Plant Disease 98:1285. https://doi.org/10.1094/PDIS-04-14-0348-PDN
Avila, AC, Lima, MF, Resende, RO, Pozzer, L, Ferraz, E, Maranhao, EA, Candeia, JÁ, Costa, ND (1996) Identificacao de tospovirus em hortalicas no submedio São Francisco utilizando Das-Elisa e Dot-Elisa. Fitopatologia Brasileira, 21:503–508
Chávez-Calvillo G, Contreras-Paredes CA, Mora-Macias J, Noa-Carrazana JC, Serrano-Rubio AA, Dinkova TD, Carrillo-Tripp M, Silva-Rosales L (2016). Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 489:179–91
De Oliveira AS, Melo FL, Inoue-Nagata AK, Nagata T, Kitajima EW, Resende RO (2012) Characterization of Bean necrotic mosaic virus: a member of a novel evolutionary lineage within the genus Tospovirus. PLoS One 7:e38634
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–1797
García-Cano E, Resende OR, Fernández-Muñoz R, Moriones E (2006) Synergistic interaction between Tomato chlorosis virus and Tomato spotted wilt virus results in breakdown of resistance in tomato. Phytopathology 96:1263–1269
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59:307–321
Kormelink R, Storms M, Vanlent J, Peters D, Goldbach R (1994) Expression and subcellular location of the NSM protein of Tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200:56–65
Londoño A, Capobianco H, Zhang S, Polston JE (2012) First record of Tomato chlorotic spot virus in the USA. Tropical Plant Pathology 37:333–338
Margaria P, Ciuffo M, Pacifico D, Turina M (2007) Evidence that nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Molecular Plant-Microbe Interactions 20:547–558
Martínez RT, Poojari S, Tolin SA, Cayetano X, Naidu RA (2013) First report of Tomato spotted wilt virus in peppers and tomato in the Dominican Republic. Plant Disease 98:163. https://doi.org/10.1094/PDIS-06-13-0617-PDN
Martínez RT, de Almeida MMS, Rodriguez R, de Oliveira AS, Melo FL, Resende RO (2018) Identification and genome analysis of Tomato chlorotic spot virus and dsRNA viruses from coinfected vegetables in the Dominican Republic by high-throughput sequencing. Virology Journal 15:24–30
Mavrič I (2001) First report of Tomato spotted wilt virus and Impatiens necrotic spot virus in Slovenia. Plant Disease 12:1288–1288
Nagata T, Avila AC, Tavares PC, Barbosa C, Juliatti FC, Kitajima EW (1995) Occurrence of different tospoviruses in six states of Brazil. Tropical Plant Pathology 20:90–95
Nagata T, Almeida ACL, Resende RO, Avila AC (2004) The competence of four thrips species to transmit and replicate four tospoviruses. Plant Pathology, 53: 136-140
Nuez F, Gil OR, Costa J (2003) El cultivo de pimientos, chiles y ajíes. Ediciones Mund-Prensa. Reimpresión. España. 607 p
Pappu HR, Jones RA, Jain RK (2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Research 141:219–236
Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist KA, Schmaljohn CS, Tesh RB (2012) Family – Bunyaviridae. Virus taxonomy: classification and nomenclature of viruses. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (Eds) Ninth Report of the International Committee on Taxonomy of Viruses. San Diego. pp. 725–741
Resende RO, Pozzer L, Nagata T, Bezerra IC, Kitajima EW, Avila AC (1996) New tospoviruses found in Brazil. Acta Horticulturae (431):78–89
Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD (2011) Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology 12:938–954
Silva J, de Oliveira AS, de Almeida MMS, Kormelink R, Nagata T, Resende RO (2019) Tomato chlorotic spot virus (TCSV) putatively incorporated a genomic segment of Groundnut ringspot virus (GRSV) upon a reassortment event. Viruses 11:187–194
Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Letters 532:75–79
Tentchev D, Verdin E, Marchal C, Jacquet M, Aguilar JM, Moury B (2011). Evolution and structure of Tomato spotted wilt virus populations: evidence of extensive reassortment and insights into emergence processes. Journal of General Virology 92:961–973
Webster CG, Reitz SR, Perry KL, Adkins SA (2011) A natural M RNA reassortant arising from two species of plant- and insect-infecting bunyaviruses and comparison of its sequence and biological properties to parental species. Virology 413:216–225
Webster CG, Frantz G, Reitz SR, Funderburk JE, Mellinger HC, McAvoy E, Turechek WW, Marshall SH, Tantiwanich Y, McGrath MT, Daughtrey ML, Adkins S (2015) Emergence of Groundnut ringspot virus and Tomato chlorotic spot virus in vegetables in Florida and the southeastern United States. Phytopathology 105:388–398
Acknowledgements
This work was supported by a grant from the Fondo Nacional de Innovación y Desarrollo Científico y Tecnológico (FONDOCYT) from the Dominican Republic and grants from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Capes (Conselho de Aperfeiçoamento de Pessoal de Nível Superior) and FAP-DF (Fundação de Apoio à Pesquisa do Distrito Federal) from Brazil.
Author information
Authors and Affiliations
Contributions
R.T.M and R.O.R. designed and supervised the study. R. T. M., M.M.S.d.A., R.R., X.C. and A.S.d.O. performed sample preparation and executed the experimental work. M.M.S.d.A., R.T.M. and F.L.M. performed data analyses. R.T.M and R.O.R wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Section Editor: Juliana Freitas-Astua
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 259 kb)
Rights and permissions
About this article
Cite this article
Martínez, R.T., de Almeida, M.M.S., Rodriguez, R. et al. Analyses of orthotospovirus populations and dispersion under different environmental conditions in Brazil and in the Dominican Republic. Trop. plant pathol. 44, 511–518 (2019). https://doi.org/10.1007/s40858-019-00307-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-019-00307-x