Abstract
Background
The risk of various types of kidney disease is significantly increased in the presence of APOL1 high-risk genotype (carriage of two risk alleles), particularly HIV-associated nephropathy (HIVAN). However, there are discrepancies in the existing evidence about the level of association between APOL1 high-risk genotype and the risk of kidney diseases in people living with HIV (PLWHIV).
Methods
This systematic review and meta-analysis was conducted to assess the relationship between the APOL1 genotypes and kidney disease in the HIV population. An a priori protocol registered on PROSPERO (ID: CRD42021253877), was followed by a systematic search of five electronic databases. Database-specific search terms were used to identify observational studies that evaluated the outcomes chosen in the review, based on a set of prespecified eligibility criteria. Using a random effect model, the odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were pooled for the meta-analysis.
Results
After screening 4418 citations, 14 articles comprising 11,069 participants were included in this review. The risk of chronic kidney disease (CKD) in the HIV positive population was significantly increased in the presence of two APOL1 risk alleles (OR 4.65 [95% CI 3.51–6.15]). Also, a significant association was observed between the carriage of two risk APOL1 variants and proteinuria (OR 2.58 [95% CI 2.05–3.25]), HIVAN (OR 16.67 [95% CI 10.22–27.19]), and progression to end-stage kidney disease (ESKD) hazard ratio: 1.79 (95% CI 1.20–2.66).
Conclusion
This review highlights a strong association between the presence of two risk APOL1 variants and an increased risk of kidney disease in PLWHIV, and provides a more precise estimate of the effect size, with smaller 95% CIs for CKD, HIVAN, and progression to ESKD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The global impact of chronic kidney disease (CKD) is enormous and continues to increase, leading to a quest to unravel novel risk factors that may be targeted to alleviate this burden. Additionally, a major ethnic disparity in the development of CKD is well documented, with the Black population being at disproportionately higher risk for CKD and end-stage kidney disease (ESKD) [1, 2]. Therefore, in an attempt to understand this ethnic disproportion for kidney disease risk, Kopp et al. [3] identified single-nucleotide polymorphisms in non-coding regions of MYH9 on chromosome 22q12 locus. This single-nucleotide polymorphism is strongly associated with focal segmental glomerulosclerosis (FSGS) which predominates in Black populations. Two years later, another landmark study by Genovese et al. [4] discovered apolipoprotein L1 (APOL1) risk genetic variants, namely G1 (a haplotype consisting of two missense variants, S342G and I3484M) and G2 (consisting of two–amino acid deletion, 388NY) as novel genetic renal risk factors that are also strongly linked to FSGS. Subsequently, studies have linked the carriage of two APOL1 risk alleles (APOL1 high-risk genotype) to hypertension-attributed kidney disease, human immunodeficiency virus-associated nephropathy (HIVAN), and as a progressor to ESKD [5,6,7]. Most of these studies attributed the increased kidney disease risk to the presence of APOL1 G1 and G2 in the gene encoding APOL1. For example, Genovese et al., reported that individuals of African descent carrying two copies of the APOL1 G1 and G2 (G1/G1, G1/G2, or G2/G2) risk alleles have five–sevenfold higher odds of nondiabetic kidney disease and non-HIV-associated FSGS [4]. This increased risk was found to be substantially higher in HIV settings, with some studies reporting odds ratios (ORs) of 29 (95% CI 13–68) from the US [8], and as high as 89 (95% CI 18–912) from a South African study [9]. These strikingly high ORs in the setting of HIV further support the second hit hypothesis which states that the presence of APOL1 high-risk variants alone is not enough to cause APOL1-associated nephropathy, and thus the need for a second hit or presence of other modifying risk factors [10]. HIV has remained one of the best-studied examples of the second hit or modifying factor. However, despite this strong association, a few studies have reported minimal to moderate association between APOL1 and kidney diseases in the HIV positive population [11]. This non-uniformity in findings from these studies in addition to the fact that a subset of HIV positive patients with high-risk alleles still did not develop kidney disease suggested the need for a meta-analysis to better quantify the strength of this association. Therefore, this systematic review and meta-analysis was conducted to determine the association between APOL1 genetic variants and kidney diseases in HIV positive patients.
Methods
Study design
An a priori protocol (S1 File) was developed for this SR&MA according to the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocol (PRISMA-P) guidelines (S2 File) [12]. The protocol was then registered on the National Institute for Health Research International Prospective Register of Systematic Reviews (PROSPERO 2021 CRD42021253877 available at: https://www.crd.york.ac.uk/prospero/displayrecord.php?ID=CRD42021253877). This systematic review was conducted according to the PRISMA (S3 File) guideline [13].
Eligibility criteria
The eligibility criteria were defined as follows:
Inclusion criteria: any study that satisfies the following criteria was included:
-
Context: all observational (cohort, case–control, and cross-sectional) studies that evaluated the association between kidney disease and APOL1 genotypes in the HIV positive population.
-
Study location: all accessible published full articles from any country across the globe.
-
Time period: no time limit was placed on the year of publication.
-
Language of Publication: studies that used English as the language of publication.
-
Participants: any study that recruits patients that meet the diagnostic criteria of HIV according to the standard guideline stipulated or adopted in the country of the research.
-
Exposure: the measurement of APOL1 genetic variant genotypes in the study.
-
Age: studies with participants of any age range.
Exclusion criteria: studies with any of the following criteria were excluded:
-
Animal studies, Case reports, case series, letters to editor, editorials, books, dissertations, review articles, unpublished reports, and conference papers.
-
Studies without a clear study design.
-
Articles published in languages other than English.
Outcomes:
Primary outcome:
-
Association of APOL1 and CKD
Secondary outcomes:
-
Association of APOL1 with proteinuria.
-
Association of APOL1 with eGFR decline.
-
Association of APOL1 with progression to ESKD.
-
Association of APOL1 with HIVAN
-
Association of APOL1 with FSGS.
Search and selection strategy
Prespecified search strategies were developed and used to search five online databases. The strategy also included a literature search via hand searching of references of selected review articles and conference proceedings. Additionally, an internet search was carried out on Google Scholar and Google search.
Databases
The selected databases were PUBMED, MEDLINE, Web of Science, Scopus, and Embase. The specific search terms used, the dates the searches were conducted, and the results for each of the databases searched are detailed in the study protocol (S1 File), and S4 File. The search terms used in the PubMed database are given as follows; (apolipoprotein l1 OR Apolipoprotein L1 OR Apolipoprotein-L1 OR APOL 1) AND (Kidney disease OR Chronic kidney disease OR CKD OR Renal disease OR End stage renal disease OR End stage kidney disease OR ESRD OR ESKD) AND (Human immunodeficiency virus OR human immunodeficiency virus-associated nephropathy OR HIV OR HIVAN).
Data management
The citations obtained from the online database search (search results) were compiled and de-duplicated in an MS Excel spreadsheet (S5 File). All steps of the systematic review from screening to data extraction were carried out on an Excel spreadsheet.
Selection process
The search and screening process of the study was conducted by two independent reviewers (YR & BW) and a third reviewer who decided about uncertainties (UE).
Data collection process
Extraction of data was conducted after the full-text screening. Relevant information was extracted from each eligible article included and recorded immediately in the data extraction form. The process of the extraction was carried out by two independent reviewers (BW & YR) and two others checked the information (UE & SN).
Study quality assessment
After evaluation for the inclusion and exclusion criteria, all included articles were subjected to quality assessment using the Newcastle Ottawa Scale (NOS) for observational cohort studies [14]. For the observational case–control and cross-sectional studies, a modified Newcastle Ottawa Scale appraisal tool was used (S6 File). The critical appraisal was carried out by three independent reviewers (YR, UE & BW) and cross-checked by one other reviewer (SN).
Meta-analysis
A meta-analysis was used to summarize association data between APOL1 and prevalent CKD; APOL1 and prevalent persistent proteinuria; APOL1 and HIVAN; and APOL1 and progression to ESKD. We pooled the study-specific estimates using a random-effects meta-analysis model (DerSimonian-Laird) to obtain an overall summary estimate for each of the analyses. Heterogeneity was assessed using the χ2 test on Cochrane’s Q statistic [15] and quantified by calculating the I2 (with values of 25%, 50%, and 75% representing low, medium, and high heterogeneity, respectively [16]). We assessed the presence of publication bias using Begg’s test [17]. Data were analyzed using Stata 17.0 for Windows (Stata Corp. 2021. Stata Statistical Software: Release 17. College Station, Tx: Stata Corp USA).
Results
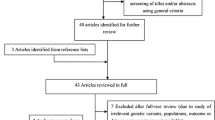
Study selection process and characteristics of included studies
The initial literature search retrieved 4418 articles, of which 24 were selected after the title and abstract screening for full-text review (Fig. 1, PRISMA flow chart). Out of the 24 articles screened for full-text review, 14 were included for the review and for the meta-analysis (S5 File). There were 11,069 participants across all included studies, five studies [9, 18,19,20,21] comprising 5075 participants from Africa and eight studies [5, 8, 11, 22,23,24,25,26] from North America with a cumulative sample size of 3130 participants, and one study [27] from Europe with a sample size of 2864. The countries of origin for the component studies included the United States of America (eight studies [8, 11, 22,23,24,25,26, 28]), the Democratic Republic of Congo (one study [21]), Nigeria (two studies [18, 19]), South Africa (one study [9]), United Kingdom (UK; one study [27]), and one study [20] was done across Burkina Faso, Senegal, and Cameroon (Table 1). The year of publication of the studies spanned from 2011 to 2022 (Table 1). Participants were black in most (85.7%) of the studies. Two of the studies [11, 21] included pediatric patients, while the remaining 12 were undertaken in adult populations. The participants in the pediatric studies had an average age of 9.0–15.3 years, while the mean age for the adult studies ranged from 37.1 to 48.1 years. Two of the studies [25, 26] were composed of only females, and the rest had a female proportion in the range of 33.0–69.9%. Seven of the studies [18,19,20,21, 24,25,26] were not based on renal histopathology findings; two [11, 27] had a mixture of patients with or without renal histopathology, while five studies [8, 9, 22, 23, 28] were histopathology-based. The HIV positive CKD patients were heterogeneous, ranging from HIVAN, FSGS-only, to HIV-associated immune complex kidney disease (HIVICK) across the component studies. The outcomes of the component studies were also heterogeneous. CKD, as defined by the Kidney Disease Improving Goals Outcome (KDIGO) as either kidney damage or a reduced eGFR of less than 60 mL/min/1.73 m2 for at least 3 months, was the outcome of interest in five studies [9, 11, 18, 19, 27]; proteinuria in five studies [19, 21, 25,26,27]; progression to ESKD in three [8, 22, 23]; eGFR decline in two [20, 24]; association with having FSGS or HIVAN in three studies [8, 27, 28]. One study [9] disaggregated the effect estimates of APOL1 renal-risk variants for HIVAN, FSGS, HIVICK, other types of glomerular nephtitis, and other kidney diseases. A pooled estimate of the findings in this study was used for further computations of the association of APOL1 and CKD in the HIV positive population.
Quality (risk of bias) assessment
For all the included studies, the quality assessment was conducted using the Newcastle Ottawa Scale appraisal tool [14]. All studies included were found to be of high quality and the result of the appraisal is presented in the S8 File.
Meta-analysis
Primary outcome: APOL1 association with CKD
Five studies (total sample size of 7164; 3 from Africa, 1 from the UK, and 1 from the USA) examined the association of APOL1 high-risk variants with the occurrence of CKD in the HIV positive population [9, 11, 18, 19, 27]. With the exception of the South African study, the reported odds ratios for CKD are fairly comparable between the African and European studies. The carriage of two APOL1 risk alleles significantly increased the risk of CKD occurrence in the HIV positive population [pooled odds ratio (OR) 4.65 (95% CI 3.51 – 6.15); n = 7164, 5 studies, I2 = 52.4%, p-value for heterogeneity = 0.08) (Fig. 2). There was no evidence of publication bias (the p-value of Begg’s test was 0.81).
Secondary outcomes
APOL1 association with proteinuria
Five studies [19, 21, 25,26,27] examined the association between renal-risk APOL1 variants with proteinuria in the HIV positive population. The presence of two APOL1 risk alleles significantly increased the risk of proteinuria in the HIV positive population [pooled odds ratio (OR) 2.58 (95% CI 2.05–3.25); n = 7450, 5 studies, I2 = 85.3%, p < 0.001) (Fig. 3). There was no evidence of publication bias (the p-value of Begg’s test was 0.09).
APOL1 association with HIVAN
Four studies [8, 9, 27, 28] examined the association between renal-risk APOL1 variants with HIVAN. The reported odds ratios for HIVAN by the South African study far exceeded those reported by the European studies. The presence of two APOL1 risk alleles significantly increased the risk of HIVAN [pooled odds ratio (OR) 16.67 (95% CI 10.22–27.19); n = 769, 4 studies, I2 = 84.6%, p < 0.001) (Fig. 4). There was no evidence of publication bias (p-value of Begg’s test was 0.99).
APOL1 association with progression to ESKD
The APOL1 association with progression to ESKD was assessed in three of the included studies [8, 22, 23]. The summary estimate indicated an increased rate of progression in individuals carrying two APOL1 renal risk alleles (pooled hazard ratio 1.79 (95% CI 1.20–2.66); n = 449, 3 studies, I2 = 74.8%, p = 0.004 (Fig. 5). There was no evidence of publication bias (p-value of Begg’s test was 0.60).
APOL1 association with eGFR decline
The association of APOL1 with eGFR decline was reported in two of the included studies [20, 24]. The two studies used the CKD-EPI equation to determine the eGFR, without the correction for ethnicity in one of the studies [20]. In a longitudinal cohort study among unsuppressed HIV-infected African Americans, Estrella et al., 2015 showed that the APOL1 high-risk group (carriage of two APOL1 risk alleles) experienced a faster annual eGFR decline than the low-risk individuals (carrying one or no risk allele). The study found a significant downward eGFR trajectory in the high-risk group compared with the low-risk group in both unadjusted (− 2.48 mL/min/1.73 m2 [CI − 3.60 to − 1.36] p < 0.001) and adjusted (− 2.42 mL/min/1.73 m2 [CI − 3.52 to − 1.32] p < 0.001) analyses. However, in the second study [20] conducted among people living with HIV of Black African origin (across Burkina Faso, Senegal, and Cameroon), the obtained result is discordant. The researchers determined the APOL1 association with eGFR decline in two cohorts: the day care unit (antiretroviral therapy [ART]-naïve) and the 2LADY trial (on long-term ART) cohorts. The study revealed that there was no direct association between APOL1 high-risk status and eGFR decline over time, showing an average decrease of 0.8 mL/min/1.73 m2 [− 1.0 to − 0.6]. On the other hand, in the 2LADY cohort, the study showed a difference in the eGFR over time (2.42 mL/min/1.73 m2 [− 3.52 to − 1.32]) in the APOL1 low-risk group without any associated change in the APOL1 high-risk group.
APOL1 association with FSGS
Three studies [8, 9, 28] reported APOL1 association with FSGS. APOL1 high-risk genotype (carriage of two risk alleles) was associated with FSGS with almost threefold greater odds (OR 2.95 [1.48–5.84] p = 0.002) than the low-risk in a univariable model as reported by Atta et al. [28]. In an adjusted analysis, the high-risk APOL1 status shows fivefold higher odds (OR 5.25 [2.37–11.62] p < 0.001) for FSGS than the low-risk status [28]. For the Kopp et al., study [8] also, APOL1 high-risk was found to have significantly (p = 1.3 × 10–48) higher odds (OR 16.9 [11–26.5]) for FSGS than the APOL1 low-risk alleles. While the association reported in the Kasembeli et al. study was not significant (p = 0.48), nevertheless, the high-risk APOL1 status showed greater odds (OR 2.1 [0.03–44]) of developing FSGS than the low-risk status [9].
Discussion
In this meta-analysis, HIV positive individuals carrying two copies of the APOL1 risk alleles were found to have an almost three-fold higher risk of developing CKD, a two-fold higher risk of developing proteinuria, and a 16-fold increased risk of developing HIVAN. Although the traditional CKD risk factors and HIV-related factors contribute to the development of CKD, carrying two APOL1 risk alleles appears to further potentiate the risk of kidney disease.
The mechanisms by which the APOL1 variants modulate the risk of kidney diseases is complex and still obscure. However, some of the proposed mechanisms largely supported by animal and in vitro studies include podocyte injury by the increased APOL1 expression through interferon, APOL1 inflammatory-mediated apoptosis leading to proteinuria and glomerular scarring, inhibition of protein synthesis by the APOL1 G1 and G2 variants through activation of protein kinase R, APOL1-triggered mitochondrial dysfunction, and dysregulation of ubiquitin D (UBD)—a ubiquitin-like modifier protein [29,30,31]. An additional mechanistic may be the triggering of cell lysis and intracellular loss of potassium by G1 and G2 APOL1 risk alleles [32].
Surprisingly, since the discovery of the genetic association of APOL1 and kidney disease and the description of its strong link with HIVAN about a decade ago, only few studies have examined this relationship in HIV positive patients.
In this meta-analysis that included four published articles dealing with patients with biopsy-proven HIVAN, having two risk APOL1 alleles was shown to significantly increase the risk for HIVAN [pooled odds ratio (OR) 16.67 (95% CI 10.22–27.19) compared with being carriers of low-risk APOL1 genotype. All four studies reported a significant increase in susceptibility to HIVAN in patients carrying two APOL1 risk alleles with a strikingly high odds ratio of 89 (95% CI 18, 912) in the South African study [9]. The relatively small sample size of the South African study may have accounted for the huge confidence intervals that overlapped with the study from the USA [8].
The mechanism accounting for the increased susceptibility to HIVAN in individuals with two APOL1 risk variants is likely due to a synergic interaction between HIV-1 protein and APOL1 variant-driven gene expression leading to podocyte injury and glomerular scarring [29, 33]. For example, Mikulak et al. showed that kidney risk variant APOL1 protein enhances the accumulation and persistence of HIV-1 in podocytes, which is facilitated through the priming of a pro-inflammatory cytokine IL-1β [34], while the non-risk variant of APOL1 attenuates accumulation and boarding of HIV-1 within the podocytes [34]. Although most of the studies reported the requirement of two APOL1 risk variants to induce renal cell toxicity, a weak association between a single copy of APOL1 G1 risk allele and HIVAN was reported by the South African study. The reason attributed for this finding by the authors is that an effect from one risk allele is in line with a gain of injury and/or toxicity of these variants in renal cells that manifests in the presence of a potent interactor like HIV, which is in contrast to a loss of gene function of APOL1 risk variants that follow a recessive model of inheritance [9]. In addition, it has also been shown that HIV viremia facilitates the detrimental renal effects of the APOL1 risk alleles and that achieving HIV viral suppression with ART may attenuate these detrimental effects [25]. Therefore, the unexpected effect of the G1 risk allele may have been driven by viremia, since the South African cohort was ART- naïve.
Although sufficient studies were not available to allow for a meta-analysis for APOL1 and other HIV-associated kidney diseases, APOL1 renal risk variants have been strongly associated with non–HIVAN FSGS [5]. For example, carrying two copies of the APOL1 risk alleles has a fivefold higher risk of developing non-HIV-associated FSGS with respect to carrying one or no risk allele [5]. Conversely, HIVICK is not associated with two risk alleles. Fine et al. reported that among 25 HIV-positive study participants who had kidney biopsy with no APOL1 risk alleles, only 3 (12%) had FSGS, while more than 40% (10) had HIVICK [23].
Since the identification of the association of APOL1 renal risk variants with increased progression of CKD by earlier case–control studies [4, 6], prospective studies have provided evidence that the APOL1 high-risk variants are associated with increased CKD progression over a long duration in a non-HIV population [7, 35]. Thus, APOL1 is now termed a disease progressor gene. This association is further supported by this meta-analysis which shows that the high-risk APOL1 variants carry a 79% increased risk of CKD progression to ESKD as compared with low-risk APOL1 genotype. A similar 70% increased risk of progression to ESKD was reported by a previously published meta-analysis in non-HIV patients [36].
Although this is the first meta-analysis conducted to determine the association between APOL1 and kidney diseases in people living with HIV, this study has some limitations. First, substantial heterogeneity was observed across some of the considered outcomes, which is largely due to the heterogeneous population included in the studies, variation in the adjustment for confounding variables across the studies, and non-uniformity of the methodologies used in the ascertainment of CKD progression. For example, some of the included studies did not explicitly state the confounding variables that were adjusted for.
Second, the insufficient number of studies precluded further subgroup analyses and meta regressions to account for the significant heterogeneity across the studies.
Third, the included studies were largely from the USA and Sub-Saharan Africa, thus limiting the global generalization of our findings.
Fourth, despite the fact that the strongest association between APOL1 renal risk variants and kidney disease that has been identified to date is with HIVAN, only four studies included kidney biopsies.
In conclusion, this meta-analysis confirms that APOL1 renal risk variants are significantly associated with proteinuria, CKD, and HIVAN and confer an increased risk of CKD progression to ESKD. It has also provided a more precise estimate of the effect size with smaller 95% CIs for CKD, HIVAN, and progression to ESKD, that can be useful for counseling and risk identification.
Data availability statement
Data used to support the findings of this study are included within the supplementary information file(s). The study protocol was registered on the National Institute for Health Research International Prospective Register of Systematic Reviews (PROSPERO 2021 CRD42021253877 available at: https://www.crd.york.ac.uk/prospero/displayrecord.php?ID=CRD42021253877).
References
Schwartz EJ, Szczech LA, Ross MJ, Klotman ME, Winston JA, Klotman PE (2005) Highly active antiretroviral therapy and the epidemic of HIV+ end-stage renal disease. J Am Soc Nephrol 16(8):2412–2420
Muntner P, Newsome B, Kramer H, Peralta CA, Kim Y, Jacobs DR Jr, Kiefe CI, Lewis CE (2012) Racial differences in the incidence of chronic kidney disease. Clin J Am Soc Nephrol 7(1):101–107
Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS (2008) MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet 40(10):1175–1184
Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR (2010) Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York, NY) 329(5993):841–845
Atta MG, Estrella MM, Skorecki KL, Kopp JB, Winkler CA, Wasser WG, Shemer R, Racusen LC, Kuperman M, Foy MC, Lucas GM, Fine DM (2016) Association of APOL1 genotype with renal histology among black HIV-positive patients undergoing kidney biopsy. Clin J Am Soc Nephrol 11(2):262–270
Lipkowitz MS, Freedman BI, Langefeld CD, Comeau ME, Bowden DW, Kao WH, Astor BC, Bottinger EP, Iyengar SK, Klotman PE, Freedman RG, Zhang W, Parekh RS, Choi MJ, Nelson GW, Winkler CA, Kopp JB (2013) Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int 83(1):114–120
Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH (2013) APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 24(9):1484–1491
Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S (2011) APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22(11):2129–2137
Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T, Limou S, Sezgin E, Nelson GW, Fogo AB, Goetsch S, Kopp JB, Winkler CA, Naicker S (2015) APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol 26(11):2882–2890
Freedman BI, Skorecki K (2014) Gene-gene and gene-environment interactions in apolipoprotein L1 gene-associated nephropathy. Clin J Am Soc Nephrol 9(11):2006–2013
Purswani MU, Patel K, Winkler CA, Spector SA, Hazra R, Seage GR 3rd, Mofenson L, Karalius B, Scott GB, Van Dyke RB, Kopp JB (2016) Brief report: APOL1 renal risk variants are associated with chronic kidney disease in children and youth with perinatal HIV infection. J Acquired Immune Deficiency Syndromes (1999) 73(1):63–68
Moher D, Liberati A, Tetzlaff J, Altman DG (2010) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 8(5):336–341
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021:372
Wells GA, Shea B, O’Connell Da, Peterson J, Welch V, Losos M, Tugwell P (2000) The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Oxford
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10(1):101–129
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
van Enst WA, Ochodo E, Scholten RJPM, Hooft L, Leeflang MM (2014) Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Med Res Methodol 14(1):70
Ekrikpo UE, Mnika K, Effa EE, Ajayi SO, Okwuonu C, Waziri B, Bello A, Dandara C, Kengne AP, Wonkam A (2020) Association of genetic polymorphisms of TGF-β1, HMOX1, and APOL1 with CKD in Nigerian patients with and without HIV. Am J Kidney Dis 76(1):100–108
Wudil UJ, Aliyu MH, Prigmore HL, Ingles DJ, Ahonkhai AA, Musa BM, Muhammad H, Sani MU, Nalado AM, Abdu A (2021) Apolipoprotein-1 risk variants and associated kidney phenotypes in an adult HIV cohort in Nigeria. Kidney Int. https://doi.org/10.2139/ssrn.3711452
Kabore NF, Cournil A, Poda A, Ciaffi L, Binns-Roemer E, David V, Eymard-Duvernay S, Zoungrana J, Semde A, Sawadogo AB (2021) APOL1 renal risk variants and kidney function in HIV-1–infected people from sub-Saharan Africa. Kidney Int Reports 7(3):483–493
Ekulu PM, Nkoy AB, Betukumesu DK, Aloni MN, Makulo JRR, Sumaili EK, Mafuta EM, Elmonem MA, Arcolino FO, Kitetele FN (2019) APOL1 risk genotypes are associated with early kidney damage in children in sub-Saharan Africa. Kidney Int Reports 4(7):930–938
Atta MG, Estrella MM, Kuperman M, Foy MC, Fine DM, Racusen LC, Lucas GM, Nelson GW, Warner AC, Winkler CA (2012) HIV-associated nephropathy patients with and without apolipoprotein L1 gene variants have similar clinical and pathological characteristics. Kidney Int 82(3):338–343
Fine DM, Wasser WG, Estrella MM, Atta MG, Kuperman M, Shemer R, Rajasekaran A, Tzur S, Racusen LC, Skorecki K (2012) APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol 23(2):343–350
Estrella MM, Li M, Tin A, Abraham AG, Shlipak MG, Penugonda S, Hussain SK, Palella FJ Jr, Wolinsky SM, Martinson JJ (2015) The association between APOL1 risk alleles and longitudinal kidney function differs by HIV viral suppression status. Clin Infect Dis 60(4):646–652
Estrella MM, Wyatt CM, Pearce CL, Li M, Shlipak MG, Aouizerat BE, Gustafson D, Cohen MH, Gange SJ, Kao WL (2013) Host APOL1 genotype is independently associated with proteinuria in HIV infection. Kidney Int 84(4):834–840
Jotwani V, Shlipak MG, Scherzer R, Parekh RS, Kao WL, Bennett M, Cohen MH, Nowicki M, Sharma A, Young M (2015) APOL1 genotype and glomerular and tubular kidney injury in women with HIV. Am J Kidney Dis 65(6):889–898
Hung RK, Binns-Roemer E, Booth JW, Hilton R, Harber M, Santana-Suarez B, Campbell L, Fox J, Ustianowski A, Cosgrove C (2022) Genetic variants of APOL1 are major determinants of kidney failure in people of African ancestry with HIV. Kidney Int Reports
Atta MG, Estrella MM, Skorecki KL, Kopp JB, Winkler CA, Wasser WG, Shemer R, Racusen LC, Kuperman M, Foy MC (2016) Association of APOL1 genotype with renal histology among black HIV-positive patients undergoing kidney biopsy. Clin J Am Soc Nephrol 11(2):262–270
Beckerman P, Bi-Karchin J, Park AS, Qiu C, Dummer PD, Soomro I, Boustany-Kari CM, Pullen SS, Miner JH, Hu CA, Rohacs T, Inoue K, Ishibe S, Saleem MA, Palmer MB, Cuervo AM, Kopp JB, Susztak K (2017) Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med 23(4):429–438
Daneshpajouhnejad P, Kopp JB, Winkler CA, Rosenberg AZ (2022) The evolving story of apolipoprotein L1 nephropathy: the end of the beginning. Nat Rev Nephrol 18(5):307–320
Zhang JY, Wang M, Tian L, Genovese G, Yan P, Wilson JG, Thadhani R, Mottl AK, Appel GB, Bick AG, Sampson MG, Alper SL, Friedman DJ, Pollak MR (2018) UBD modifies APOL1-induced kidney disease risk. Proc Natl Acad Sci USA 115(13):3446–3451
Olabisi OA, Zhang JY, VerPlank L, Zahler N, DiBartolo S 3rd, Heneghan JF, Schlöndorff JS, Suh JH, Yan P, Alper SL, Friedman DJ, Pollak MR (2016) APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci USA 113(4):830–837
Kopp JB, Heymann J, Winkler CA (2017) APOL1 renal risk variants: fertile soil for HIV-associated nephropathy. Semin Nephrol 37(6):514–519
Mikulak J, Oriolo F, Portale F, Tentorio P, Lan X, Saleem MA, Skorecki K, Singhal PC, Mavilio D (2016) Impact of APOL1 polymorphism and IL-1β priming in the entry and persistence of HIV-1 in human podocytes. Retrovirology 13(1):63
Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT Jr, Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ (2013) APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369(23):2183–2196
Jagannathan R, Rajagopalan K, Hogan J, Hart A, Newell KA, Pastan SO, Patzer RE (2021) Association between APOL1 genotype and kidney diseases and annual kidney function change: a systematic review and meta-analysis of the prospective studies. Int J Nephrol Renov Dis 14:97–104
Funding
This review had no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declared no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Waziri, B., Raji, Y.E., Ekrikpo, U.E. et al. Apolipoprotein L1 gene variants and kidney disease in patients with HIV: a systematic review and meta-analysis. J Nephrol 36, 1119–1134 (2023). https://doi.org/10.1007/s40620-022-01512-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40620-022-01512-9