Abstract
Flag leaf is the last leaf to senesce and its life span plays a critical role in determining grain quality and yield in rice. Little is known about the molecular changes that occur and their regulation in time-dependent manner during flag leaf senescence in rice. Several studies have explored different aspects of miRNA functions in plant development; however, a diminutive account is available about their role in flag leaf senescence. With an aim to unravel the role of miRNAs in ageing of flag leaf in rice, four small RNA libraries were prepared from three stages of senescence and sequenced by Illumina deep sequencing technology. Thirty-eight known and 494 novel miRNAs were identified in the senescing flag leaves. Digital expression analysis revealed that 21 known and 116 novel miRNAs were differentially expressed during senescence. Family member(s) of miR156, miR159, miR160, miR164, miR169, miR171, miR393, miR396, miR535, miR827, miR1428, miR1432 and miR1861 were differentially expressed in at least one stage of flag leaf senescence. The present study has generated a repository of senescence-related miRNAs that can be utilized to contemplate molecular approaches for manipulating the timing of flag leaf senescence in rice and other related crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Life cycle of plants involves multiple growth stages, which in turn, are governed by plethora of physiological, biochemical and molecular processes. The last stage in plants’ life cycle is senescence that represents an age-dependent complex phenomenon wherein orchestrated deterioration occurs in cells, tissues, organs or whole organism, eventually leading to death (Lim et al. 2007). Around harvesting time organismal level senescence is observed in mature crop plants such as rice and wheat, whereas falling of mature leaves during autumn exemplify organ level senescence. The onset of senescence is primarily accompanied by greatly reduced efficiency of photosynthetic machinery owing to degeneration of chloroplasts (Guo et al. 2004). Although, senescence is a genetically pre-defined phenomenon, various external components, such as nutrient availability, pathogen infection and sub-optimal growth conditions (temperature fluctuations, drought, high salt and oxidative stress), are known to initiate early senescence (Guo and Gan 2005). At the molecular level, extensive gene reprogramming occurs during senescence which involves change in expression of several senescence-associated genes (SAGs). In Arabidopsis, microarray analysis of leaves undergoing senescence revealed that more than 800 SAGs had significant altered levels of expression (Buchanan-Wollaston et al. 2003). Transcriptome studies in maize and cotton revealed that many transcription factors like members of NAC, WRKY, MYB, C2H2 zinc-finger, bZIP and AP2/EREBP families accumulated during senescence (Zhang et al. 2014; Lin et al. 2015). The gene ontology analysis of transcripts which were altered during senescence in Arabidopsis and maize indicated that most of the up-regulated transcripts were categorized as transporters, lipid metabolism, protein degradation, carboxylic acid catabolic process and hormone metabolism. Concomitant down regulation of photosynthesis-related genes during senescence has also been reported (Breeze et al. 2011; Zhang et al. 2014).
Small RNAs (sRNAs) play a crucial role in controlling plant development processes. However, little is known about the function of small RNAs in regulating senescence in plants. Two small RNAs, miR164 and miR319, have been shown to control leaf senescence in Arabidopsis. miR164, whose expression declines during senescence, is a posttranscriptional regulator of the positive ageing regulator, ORE1/ANAC092 (ORESARA 1 or NAM, ATAF, and CUC). EIN3 (ETHYLENE INSENSITIVE 2) transcriptionally upregulates the transcription factor ORE1 by releasing the repression by miR164. ORE1 is believed to positively regulate the expression of senescence-associated genes (SAGs) during leaf ageing. Therefore, a trifurcate-feed forward pathway finely regulates cell death and leaf senescence in an ethylene-dependent manner (Kim et al. 2009). A recent study by Kim et al. (2018) shows that PSEUDO-RESPONSE REGULATOR (PRR9), a vital component in circadian clock in plants, directly activates the transcription of ORE1 and indirectly suppresses miR164, thereby unravelling the link between ageing and circadian clocks in plants (Kim et al. 2018). Another miRNA, miR319, which targets TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) transcription factor genes controls the senescence program through jasmonic acid biosynthesis, which is a positive regulator of leaf senescence (Schommer et al. 2008).
Rice (Oryza sativa) is one of the major staple food crops and cultivated throughout the world. Pre-mature senescence is a major concern in several crops, including rice, which leads to compromised growth and development, eventually leading to lower productivity. Reduced photosynthetic efficiency arising due to environmentally induced pre-mature senescence at the time of grain filling adversely affects the yield of the plant (Luquez and Guiamét 2001; Rampino et al. 2006). A couple of high throughput sequencing studies have identified gene regulatory networks in leaf senescence in maize and rice (Xu et al. 2014; Wu et al. 2016). Deep sequencing of small RNAs, in combination with degradome sequencing of age-resistant and age-sensitive rice varieties, found involvement of members of 6 miRNA families (miR156, miR160, miR164, miR167, miR172 and miR1848) in flag leaf senescence, primarily by regulating phytohormone signaling pathways (Xu et al. 2014). A similar study in maize also identified six miRNAs (miR159, miR167, miR171, miR172, pre-miR11 and pre-mir131) as key regulators of leaf senescence which potentially target hormone pathways, chlorophyll metabolism and energy metabolism (Wu et al. 2016). In view of limited information available on leaf ageing in cereals, it is worthwhile to understand the molecular mechanisms governing leaf senescence in rice, especially flag leaf which contributes to grain filling and plant productivity. This would provide a platform to modulate associated molecular components/pathways so as to delay the natural senescence process and augment crop yield.
The present study is an attempt to explore the involvement of small RNAs, especially miRNAs, at different stages of flag leaf senescence in rice. To prepare an atlas of miRNAs that are differentially expressed during the progression of senescence, four small RNA libraries were constructed from three stages of flag leaf senescence followed by their deep sequencing using Illumina’s NGS platform. High throughput sequencing data was computationally analyzed for prediction of known and putative novel miRNAs. The target genes of identified miRNAs were computationally predicted, and their functional categorization was performed. The study documents an inventory of miRNAs that exhibit differential expression kinetics at different stages of flag leaf senescence. Our analysis shows that in addition to the previously known miRNAs, several novel miRNAs regulate flag leaf senescence by potentially targeting members of key transcription factor families and components of hormone pathways.
Materials and methods
Collection of tissues
Rice (Oryza sativa cv. Pusa Basmati 1) seeds were procured from National Seed Centre, Indian Agricultural Research Institute (IARI), Delhi, India and were grown in the field during rice-growing season. Fully expanded flag leaves at panicle development stage and different stages of grain filling namely, milky stage (early senescence), dough stage (mid-senescence) and grain maturity stage (late senescence) were collected based on their chlorophyll content. In each case, tissues were pooled from six individual plants, immediately frozen in liquid nitrogen and stored at -80 °C until further use.
Chlorophyll estimation
The chlorophyll content of leaves was estimated by using spectrophotometric analysis (Burman et al. 2018). Briefly, tissues (0.2 g) were incubated in 5 ml of DMSO at 65 °C for 16 h in the dark. Subsequently, absorbance was recorded at 645 nm and 663 nm using a spectrophotometer (Bio-Rad, USA). Amount of chlorophyll was deduced according to the following formulae.
where V = Volume of DMSO (in ml); X = Path length i.e. 1 cm; W = Fresh weight (in grams).
Total chlorophyll content was calculated followed by estimation of percentage chlorophyll content in senescing tissue compared to unsenesced tissues (fully expanded flag leaves).
RNA extraction, construction of small RNA libraries and high-throughput sequencing
Total RNA was isolated from frozen tissues following modified guanidinium isothiocyanate (GITC) method (Chomzynski and Sacchi 1987). The concentration of the isolated RNA was assessed using a spectrophotometer (Bio-Rad, USA) followed by a quality check on MOPS-formaldehyde-agarose gel. For the preparation of small RNA libraries, low molecular weight RNA (LMW) fraction was extracted by resolving total RNA on TBE-urea-PAGE gel and excising 15–30 nt portion. The LMW RNA was ligated with 5′ and 3′ RNA adapters (Additional file 1: Table S1) using T4 RNA ligase. Successively, first strand cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, USA) and 3′ adapter specific reverse transcriptase primer (Additional file 1: Table S1). The cDNA library was enriched using adapter-specific primers and the amplified product was purified. Quality of the cDNA libraries were evaluated using Bioanalyzer (Agilent Technologies, USA). Sequencing of prepared libraries was performed at DBT-funded high-throughput sequencing facility at University of Delhi South Campus, New Delhi, India on Illumina GA IIx sequencing platform as per manufacturer’s instructions.
Computational analysis of sequencing data and prediction of target genes
UEA small RNA Workbench (v2.4; Stocks et al. 2012) was employed for the analysis of purity filtered raw reads. Briefly, non-small RNA sequences were eliminated from sequencing data, and putative small RNAs were mapped on to the rice reference genome (Kawahara et al. 2013). miRBase (version 21; http://www.mirbase.org/) was used as a reference for the identification of known miRNAs. For the identification of differently expressed miRNAs, read counts were normalized by calculating tags per million (TPM) and subsequently used for calculating fold change. The small RNA sequencing data was submitted to Sequence Read Archive (SRA) database available at National Center for Biotechnology Information (NCBI) (Accession number GSE123171; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123171).
Target prediction was carried out using plant small RNA target finder (http://plantgrn.noble.org/psRNATarget/) tool with default settings. Rice Genome Annotation dataset (v7, Kawahara et al. 2013) was used as mapping reference. Only those targets were considered which exhibited maximum expectation value between 0 and 3. Functional categorization of predicted target genes was performed using MapMan annotation tool (http://mapman.gabipd.org/).
Experimental validation of miRNAs and their target genes by qPCR
Total RNA (2 µg) was poly-adenylated using poly (A) tailing kit (Ambion, USA) and the reaction mixture was subsequently purified with RNAeasy Mini Elute Cleanup Kit (Qiagen, Germany). Poly-adenylated RNA was reverse transcribed using 1 µg RTQ primer (Additional file 1: Table S1) and Superscript III reverse transcriptase (Invitrogen, USA) according to manufacturer’s instructions. Quantitative PCR (qPCR) was performed on Mastercycler Realplex 2 (Eppendorf, Germany) using SYBR green chemistry (KAPA Biosystems, USA). Rice 5S ribosomal RNA was used as an internal control. Primers for the amplification of miRNAs and 5S rRNA are listed in Table S1.
For expression analysis of putative target genes, 10 µg of total RNA was treated with DNase I (NEB, USA) and subsequently reverse transcribed using iScript cDNA synthesis kit (Bio-rad, USA). Real-time PCR was performed on Mastercycler Realplex 2 (Eppendorf, Germany) using SYBR green chemistry (KAPA Biosystems, USA). Rice elongation factor 1-alpha was used as internal reference gene for qPCR. Primers for target genes and internal control genes are listed in Table S1. Three independent biological replicates and two technical replicates were used for all qPCR experiments.
Results
Staging of natural flag leaf senescence in rice
Rice plants were grown under field conditions during the growing season, and different stages of progressive natural flag leaf senescence were identified based on total chlorophyll content. Fully expanded unsenesced flag leaf (US) at panicle development stage was harvested as control, and the chlorophyll content was considered as 100%. Flag leaves were continuously monitored for progression of senescence and were harvested at three different stages. When the chlorophyll content of flag leaf was 60–80% to that in unsenesced flag leaf, the stage was referred to as early senescence (ES) stage. The flag leaves harvested when the chlorophyll content was 45–60% to that of US stage were considered in the mid senescence (MS) phase. Finally, late senescence flag leaf (LS) tissue was harvested when the chlorophyll content was 30–45% of US stage. The morphological changes observed during the progression of senescence along with the relative chlorophyll content are illustrated in Fig. 1a, b. At the molecular level the progression was confirmed by performing the expression of two senescence-associated genes (SAGs), RED CHLOROPHYLL CATABOLITE REDUCTASE (RCC reductase) and WRKY6. It was observed that steady state levels of both the SAG genes (RCC reductase and WRKY6) increased in flag leaves with the progression of senescence (Fig. 1c). The inverse correlation between the chlorophyll content and levels of two SAGs strongly endorses the staging of natural flag leaf senescence in the present study.
Flag leaf senescence in rice plants. a Morphological appearance of flag leaf in field grown plants at different stages of senescence. b Percent chlorophyll content in flag leaves harvested at three stages of senescence. c Expression profile of senescence-associated genes (SAGs): red chlorophyll catabolite reductase (RCC reductase) and WRKY6 at different stages of flag leaf senescence. (US: unsenesced flag leaf; ES: early senescence flag leaf; MS: mid senescence flag leaf; LS: late senescence flag leaf)
Deep sequencing and length distribution of small RNA population in rice
To identify small RNAs associated with natural flag leaf senescence in rice, four small RNA libraries were prepared individually from unsenesced flag leaves (US), early senescence flag leaves (ES), mid senescence flag leaves (MS) and late senescence flag leaves (LS). Qualitative and quantitative assessment of small RNA libraries was performed with Bioanalyzer (Additional file 2: Fig. S1), and the libraries were subsequently sequenced using Illumina Genome Analyzer IIx platform. In total, 93,569,768 reads were sequenced out of which 28,304,505 were unique. The reads obtained from sequencing of all the four libraries were pooled for further downstream analysis using UEA sRNA workbench v2.4. Stepwise, the 3′ adapter was first trimmed followed by elimination of reads with length smaller than 16 nt (nucleotides) and longer than 30 nt. Further, reads containing invalid nucleotides (represented as ‘N’ in the sequence) and those with homology to other non-coding RNAs (rRNA, tRNA, snRNA, snoRNA) were also eliminated from the total pool of sequences. Post-filtration, the remaining 19,729,356 unique reads, hereafter referred to as putative small RNA population, were mapped onto rice genome. Summary of filtration criteria and the number of eliminated reads is presented in Table 1. Further size distribution analysis performed with the putative small RNAs revealed that the most abundant sRNA reads were in the size range of 21 to 24 nt, which is a canonical feature of Dicer-like processed sRNAs (Xie et al. 2004). The 24 nt reads were maximally represented followed by 21 nt reads, in the individual libraries as well as in the pooled unique putative small RNA population (Fig. 2). These results are consistent with the earlier reports from Arabidopsis and rice (Rajagopalan et al. 2006; Zhu et al. 2008; Xu et al. 2014; Wu et al. 2016).
Length distribution of putative small RNA reads obtained by high-throughput sequencing of small RNAs in rice flag leaf senescence libraries. a Reads were pooled from all the four libraries and size distribution analysis was performed with redundant and unique reads. b Length distribution of unique reads obtained in stage-specific libraries. US: unsenesced rice flag leaf; ES: early senescence flag leaf; MS: mid senescence flag leaf; LS: late senescence flag leaf. Read counts are shown in millions (M); nt: nucleotides
Identification of known and novel miRNAs, their chromosomal distribution, and digital expression profile at different stages of flag leaf senescence in rice
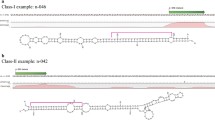
The unique putative small RNA population was mapped onto rice reference genome (http://rice.plantbiology.msu.edu/), and the mapped reads were aligned to known miRNA sequences available in miRBase v21.0 by using miRCAT tool of UEA sRNA workbench. Putative sRNAs having an exact identity (0 mismatch) and up to 3 mismatches with miRBase entries were categorized as ‘known miRNAs’. The remaining mapped reads (21–24 nt in length) were scanned using miRCAT tool to fetch hairpin foldback structure (Meyers et al. 2008). The precursor structures were screened manually according to the criteria proposed by Meyers et al. (2008) and the miRNAs present within the authentic foldback structures were referred to as ‘novel miRNAs’. Among the identified known and novel miRNAs only those miRNAs were selected for further analysis which exhibited one or greater than one TPM in at least one library. Our analysis identified 38 known miRNAs belonging to 28 miRNA families. Star sequences of 26 known miRNAs was found in the sequenced small RNA population (Additional file 3: Table S2). Furthermore, 244 and 250 novel miRNAs with and without star sequences were identified in the small RNA dataset, respectively. Precursors of both the known and novel miRNAs were mapped onto rice chromosomes and the predicted precursors of known miRNAs were found to be randomly distributed on all, except chromosome 10 (Fig. 3), whereas precursors of the novel miRNAs were present on all the 12 chromosomes. Overall, the highest number (61) of precursors of known and novel miRNAs mapped onto chromosome 2, followed by those on chromosome 3 and chromosome 1, which exhibited equal number (56) of miRNA precursors. Least number of miRNA precursors (20) mapped onto chromosome 10 (Fig. 3).
Chromosomal distribution of predicted known and novel miRNA precursors in rice. Precursors of known and novel miRNAs were mapped onto rice chromosomes using the map tool of Oryzabase Integrated Rice Science Database (https://shigen.nig.ac.jp/rice/oryzabase/). Known miRNA precursor loci are shown as unshaded text and loci for novel miRNAs are highlighted as black shaded text. The number of novel miRNA precursor loci is mentioned in brackets
In order to predict the possible function of miRNAs in regulating flag leaf senescence in rice, their digital expression levels were analyzed at different stages of senescence and the miRNAs exhibiting ≥ 1.5-fold change with respect to unsenesced leaves (US), were considered as differentially regulated. Digital expression analysis revealed that 21 known and 116 novel miRNAs were differentially expressed during senescence (Fig. 4, 5). Four known miRNAs (osa_miR159b, osa_miR159f, osa_miR827 and osa_miR1432-5p) were up-regulated in at least two stages of flag leaf senescence. osa_miR1432-5p was highly induced at ES stage (4.5-fold), whereas osa_miR160a-5p and osa_miR827 showed 1.98 and 4.1-fold change in the LS stage, respectively. Six known miRNAs (osa_156f-3p, osa_miR160e-5p, osa_miR164a, osa_miR169r-5p, osa_miR535-3p and osa_miR1428e-3p) were found to be down-regulated in at least two stages of senescence (Additional file 3). Among novel miRNAs, 80 and 21 miRNAs were up-regulated and down-regulated in at least two stages of senescence in flag leaves, respectively (Additional file 3).
Heat map illustrating the expression profiles of novel miRNAs at different stages of flag leaf senescence in rice. a Expression profile of upregulated novel miRNAs, b Expression profile of downregulated novel miRNAs. ES: early senescence flag leaf; MS: mid senescence flag leaf; LS: late senescence flag leaf
Validation of expression profile of known and novel miRNAs at different stages of flag leaf senescence in rice
In order to validate the sequencing results and digital expression profiles of predicted known and novel miRNAs, we utilized quantitative RT-PCR (qPCR) based approach. For this purpose, four known (osa_miR164a, osa_miR827, osa_miR319a-3p and osa_miR396c-5p) and four novel (N_osa_221, N_osa_297, N_osa_445 and N_osa_524) candidate miRNAs were randomly selected. We found that the expression pattern of all miRNAs tested by qPCR was largely consistent with the digital expression profile (Fig. 6). On the basis of these results it is concluded that osa_miR827, N_osa_524 and N_osa_297 were significantly upregulated, whereas miR164a, miR396c-5p and N_osa_445 were substantially downregulated.
Computational prediction of target genes of rice miRNAs, their functional categorization and expression profiling during flag leaf senescence in rice
To decipher the biological function of identified miRNAs, putative targets of these miRNAs were predicted using Plant small RNA Target Finder (http://plantgrn.noble.org/psRNATarget/) tool. MSU Rice Genome Annotation v7 dataset (http://rice.plantbiology.msu.edu/) was utilized as a reference. Computational prediction of putative targets of known and novel miRNAs identified 386 and 3642 target genes, respectively. To understand the miRNA-gene regulatory network in flag leaf senescence, functional categorization of target genes was performed using MapMan annotation tool. Several of the target genes were involved in development, signaling, transport, stress and hormone metabolism (Fig. 7a, b). Among the transcription factors identified as putative miRNA targets, MYB family members were maximally represented followed by bHLH and C2H2 zinc finger proteins (Fig. 7c, Additional file 4). Two predicted target genes each for eight qPCR-validated miRNAs were shortlisted for validation by qPCR, and their expression profile was compared with the corresponding miRNAs during the progression of senescence (Fig. 8). Both the targets tested for osa_miR164a, N_osa_221, N_osa_297 and N_osa_524 and one target tested for osa_miR827 and N_osa_445 exhibited inverse correlation in expression profile to that of miRNAs. All these results provide us an insight into the possible involvement of miRNAs and their target genes in regulating flag leaf senescence in rice.
Expression profiling of putative target genes of identified miRNAs during flag leaf senescence in rice. Quantitative PCR-based expression profiles of computationally predicted targets of known (a) and novel miRNAs (b). ES: early senescence flag leaf; MS: mid senescence flag leaf; LS: late senescence flag leaf
Discussion
High throughput sequencing approaches have aided functional characterization of miRNAs in diverse developmental and physiological processes in several plant systems (Zeng et al. 2015; Shriram et al. 2016; Djami-Tchatchou et al. 2017; Liu et al. 2017; Xiong et al. 2018). However, few sequencing-based studies have been used to associate miRNAs with regulation of leaf senescence. In cereal crops, flag leaf and its senescence are crucial in deciding the grain quality and/or quantity. Therefore, understanding the molecular basis of flag leaf senescence could possibly be used to alter grain quality and quantity in rice. In the present study deep sequencing was carried out to identify the miRNAs that are differentially expressed at different stages of flag leaf senescence in rice.
Out study reports identification of 38 known and 494 novel miRNAs in flag leaf senescence of rice. Digital expression profiles were generated and fold change in expression was calculated at three stages of senescence relative to unsenesced stage. One of the known miRNAs, osa_miR160a-5p, predicted to target ARF18 (auxin response factor), was highly upregulated specifically at late stage of senescence (LS). ARFs are known to play an important role in regulating auxin-mediated leaf senescence (Lim et al. 2010). Further, the expression of two members of miR159 family, osa_miR159b and osa_miR159f, increased during two consecutive stages, ES-MS and MS-LS, of senescence, respectively. These miRNAs are predicted to target specific members of MYB family transcription factors, which in turn are known to act as transcriptional regulators of plant leaf senescence (Zhang et al. 2011; Huang et al. 2015). Wu et al. (2016) reported induction of miR159 with simultaneous downregulation of the MYB target gene in maize inbred line, ELS-1 (early leaf senescence-1) (Wu et al. 2016). Altered senescence phenotypes were observed when levels of MYBH gene were modulated in Arabidopsis (Huang et al. 2015). Expression of another known miRNA, osa_miR827, was high in MS and LS stages. The levels of two putative targets of osa_miR827 were determined by qPCR and it was found that WAK receptor like protein kinase (WAK20) levels were elevated at all three stages but expression of F-box containing protein (FBX285) declined in MS and LS, thereby indicating that F-box containing protein could possibly a target of osa_miR827 during mid and late phases of flag leaf senescence in rice. F-box proteins have been demonstrated to be involved in diverse biological processes of plant development, including senescence (Kanneganti and Gupta 2008; Stefanowicz et al. 2015). Remarkably, osa_miR1432-5p exhibited drastic upregulation (4.5-fold) at ES and although its level declined slightly in MS (2.9-fold) and LS (2.5-fold), its level was significantly higher as compared to UN stage. Multiple target genes were predicted for osa_miR1432-5p, such as EF-hand family protein, alpha-amylase precursor and serine/threonine protein phosphatase. Although no direct evidence is available for the role of EF-hand family proteins and alpha-amylase precursor in plant senescence, genes encoding certain serine/threonine protein phosphatases such as SSPP (SENESCENCE SUPPRESSED PROTEIN PHOSPHATASE) are known to be suppressed during senescence, thereby negatively regulating leaf senescence in Arabidopsis (Xiao et al. 2015).
Survey of digital gene expression analysis revealed that three members of miR171 family (osa_miR171b, osa_miR171 h and osa_miR171i-3p) and osa_miR396c-5p were particularly downregulated in LS stage, while the levels of osa_miR393a and osa_miR1861 declined only in ES and MS stage, respectively. Expression levels of osa_miR160e-5p and osa_miR164a declined in MS as well as LS stages of flag leaf senescence. Interestingly, expression of 4 miRNAs (osa_miR156f-3p, osa_miR169r-5p, osa_miR535-3p and osa_miR1428e-3p) consistently declined at all the three stages of senescence. miR171 and miR164 are already known to be downregulated during senescence in maize and Arabidopsis leaves, respectively. It was predicted that members of miR171 family target scarecrow-like transcription factor genes, which are involved in leaf development (Dhondt et al. 2010). However, whether SCARECROW proteins directly regulate leaf senescence is still not clear. Members of miR164 family target NAC transcription factors. Our results showed that two genes, NAM2 and NAM3, predicted as targets for osa_miR164a were upregulated at all the three stages of senescence, which is precisely opposite to the expression trend of osa_miR164a. NAM2 and NAM3 belong to NAC transcription factor family, whose members form a network and act as central regulators (both negatively and positively) of leaf senescence in plants (Dhondt et al. 2010). Overexpression of NAM2 or ONAC011 in rice resulted in early heading and precocious flag leaf senescence, while its repression caused a delay in both heading and natural senescence (Mannai et al. 2017).
Auxin negatively regulates leaf senescence by downregulating the expression of SAGs or senescence-associated genes (Kim et al. 2011; Xu et al. 2014). The expression of auxin response genes is controlled by auxin response transcription factors (ARFs). It would be interesting to check the expression pattern of ARFs targeted by osa_miR160e-5p, whose expression was consistently low and osa_miR160a-5p whose expression levels were elevated during late stages of flag leaf senescence, respectively. The putative target genes predicted for osa_miR393a are OsFBLs which exhibited high homology with auxin receptor OsAFBs. Whether auxin homeostasis and signaling mediated by ARFs and AFBs are involved in regulating senescence needs to be addressed. Target prediction analysis showed that osa_miR169r-5r may target NF-Y (nuclear factor Y) genes which are a class of transcription factors involved in regulation of diverse developmental and stress responses in plants. Upregulation of NF-Y gene during leaf senescence is reported in Arabidopsis (Breeze et al. 2011).
Prediction of targets for identified known and novel miRNAs revealed more than 4000 target genes. Transcription factors are key regulators of differential gene expression as they bind to specific cis-regulatory elements in the promoter regions of numerous genes leading to their activation or suppression (Zou et al. 2011). Subcategorization of the transcription factors targeted by miRNAs revealed maximum representation of MYB family members followed by bHLH and C2H2 zinc proteins. Several transcription factors such as NAC, WRKY, AP2, zinc finger proteins, MYB and bZIP have previously been implicated in plant senescence (Guo et al. 2004; Zhou et al. 2011; Koyama et al. 2013; Mao et al. 2017). Our study identified miR156, miR159, miR160, miR164, miR171 and miR172 as prominent miRNA family members, which target zinc finger proteins, MYB proteins, auxin responsive factors, NAC proteins and APETALA2 (AP2), respectively. These results correlate with the previous studies on rice and Arabidopsis and further confirms a crucial role of miRNAs-mediated regulation of transcription factors during natural plant senescence (Liu et al. 2008; Koyama 2014; Leng et al. 2017).
Peroxidases are antioxidative enzymes, which are involved in the scavenging of reactive oxygen species (ROS). An enhanced level of peroxidases was previously reported during senescence (Veljovic-Jovanovic et al. 2006). In our study, we found that the putative target of N_osa_445, a peroxidase precursor was up-regulated during senescence. In addition, novel miRNA, N_osa_297 was found to target zinc ring finger genes. In wheat, two zinc ring finger genes were reported to be down-regulated during leaf senescence (Kam et al. 2007). The expression analysis of zinc ring finger transcripts in flag leaf senescence is in agreement with this report (Fig. 8b). Moreover, zinc finger proteins and bHLH transcription factors, which are known to be involved in leaf senescence were targeted by novel miRNAs. For instance, N_osa_221 targets both zinc finger protein and bHLH family members in leaf senescence.
Plant senescence is a highly coordinated process which involves an intricate interplay between phytohormones for e.g., ethylene, jasmonic acid, abscisic acid, salicylic acid and strigolactones are positive regulators, whereas cytokinins and auxins are inhibitors of senescence (Pourtau et al. 2004; Schippers et al. 2007; Jibran et al. 2013; Kim et al. 2015). The Arabidopsis mutants, ethylene-receptor1-1 and ethylene-insensitive2, display delayed senescence phenotype, while overexpression of EIN3 accelerated leaf senescence (Grbic and Bleecker 1995; Alonso et al. 1999). The conserved nature of ethylene-mediated senescence process in rice and Arabidopsis is demonstrated by high degree of homology in the targets of Arabidopsis EIN3 (EIN3-SAGs or EIN3-bound SAGs) with that in rice and presence of EIN3 core binding sites in these genes (Goodstein et al. 2012). In the present study, the putative targets of miRNAs included 4 AP2/ERF transcription factors and 24 genes of ethylene pathway. Higher levels of abscisic acid (ABA), salicylic acid (SA) and gibberellic acid (GA) were reported in the senescing leaves of Arabidopsis (Breeze et al. 2011). Signaling or biosynthetic mutants of these phytohormones exhibited delayed leaf senescence phenotype (Morris et al. 2000; Moore et al. 2003; Chen et al. 2014). ABA promotes leaf senescence by regulating sugar signaling in plants (Pourtau et al. 2004). Recent studies showed that ABA-INSENSITIVE5 (ABI5) directly regulates the expression of ORESARA1, (a NAC transcription factor which in turn regulates developmental leaf senescence; Sakuraba et al. 2014) and STAYGREEN1 (codes for chlorophyll degrading enzyme; Park et al. 2007; Sakuraba et al. 2014). Consistent with these reports an inverse correlation was observed in the levels of miR164a and its targets NAC TFs (NAM2 and NAM3) during senescence. In rice, miR159 controls GA signaling through GAMYB protein (Aya et al. 2009). The upregulated miR159 family members in our study gave a clue about the role of GA and indirectly MYB proteins in leaf senescence. Auxin is a negative regulator of senescence, and its exogenous application could delay leaf senescence by down-regulating the SAGs expression. In senescing flag leaves alteration in expression levels of osa_miR160a-5p and osa_miR160e-5p may modulate the levels of AUXIN RESPONSE FACTORS (ARFs), which would regulate auxin signaling and in turn affect plant senescence. All these results indicate the active involvement of complex miRNA-phytohormone gene regulatory network in controlling flag leaf senescence in rice.
In the present study, several known and novel miRNAs were found to be differentially expressed at different stages of flag leaf senescence. Functional categorization of the putative target genes revealed their possible involvement in plant development and stress. It would be worthwhile to perform functional characterization of miRNAs that are differentially expressed during senescence and their putative targets to gain an insight into the regulation of flag leaf senescence in rice.
Conclusions
Leaf senescence is a complex but highly orchestrated degradation process which involves massive gene reprogramming and other changes at molecular level. The current study was designed to broaden our knowledge on the involvement of miRNAs and their putative targets during ageing of flag leaf in rice. Several miRNAs, both known as well as novel, were found to be differentially expressed in senescing flag leaves of rice. Computational prediction followed by functional categorization of target genes of identified miRNAs revealed their involvement in signaling and hormone pathways. These results provide valuable information for understanding the molecular basis of miRNA-mediated flag leaf senescence and provides a platform for manipulating ageing of plants for augmented crop yield.
References
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S., & Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science, 284, 2148–2152.
Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K., Nishimura, M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell, 21, 1453–1472.
Breeze, E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell, 23, 873–894.
Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., et al. (2003). The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnology Journal, 1, 3–22.
Burman, N., Bhatnagar, A., & Khurana, J. P. (2018). OsbZIP48, a HY5 transcription factor ortholog, exerts pleiotropic effects in light-regulated development. Plant Physiology, 176, 1262–1285.
Chen, M., Maodzeka, A., Zhou, L., Ali, E., Wang, Z., & Jiang, L. (2014). Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Science, 219–220, 26–34.
Chomzynski, P., & Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Analytical Biochemistry, 162, 156–159.
Dhondt, S., Coppens, F., De Winter, F., Swarup, K., Merks, R. M. H., Inze, D., et al. (2010). SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiology, 154, 1183–1195.
Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., & Dubery, I. A. (2017). Functional roles of microRNAs in Agronomically important plants—potential as targets for crop improvement and protection. Frontiers in Plant Science, 8, 378.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 40, 1178–1186.
Grbic, V., & Bleecker, A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J., 8, 595–602.
Guo, Y., Cai, Z., & Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant, Cell and Environment, 27, 521–549.
Guo, Y., & Gan, S. (2005). Leaf senescence: Signals, execution, and regulation. Current Topics in Developmental Biology, 71, 83–112.
Huang, C. K., Lo, P. C., Huang, L. F., Wu, S. J., Yeh, C. H., & Lu, C. A. (2015). A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Molecular Biology, 88, 269–286.
Jibran, R., Hunter, D. A., & Dijkwel, P. P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Molecular Biology, 82, 547–561.
Kam, J., Gresshoff, P., Shorter, R., & Xue, G. P. (2007). Expression analysis of RING zinc finger genes from Triticum aestivum and identification of TaRZF70 that contains four RING-H2 domains and differentially responds to water deficit between leaf and root. Plant Science, 173, 650–659.
Kanneganti, V., & Gupta, A. K. (2008). Wall associated kinases from plants—an overview. Physiology and Molecular Biology of Plants, 14, 109–118.
Kawahara, Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice, 6, 1–10.
Kim, J., Chang, C., & Tucker, M. L. (2015). To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Frontiers in Plant Science, 6, 1–7.
Kim, H., Kim, H. J., Vu, Q. T., Jung, S., McClung, C. R., Hong, S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proceedings of the National Academy of Sciences, 115, 8448–8453.
Kim, J. I., Murphy, A. S., Baek, D., Lee, S. W., Yun, D. J., Bressan, R. A., et al. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany, 62, 3981–3992.
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science, 323, 1053–1057.
Koyama, T. (2014). The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Frontiers in Plant Science, 5, 650.
Koyama, T., Nii, H., Mitsuda, N., Ohta, M., Kitajima, S., Ohme-Takagi, M., et al. (2013). A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiology, 162, 991–1005.
Leng, Y., Ye, G., & Zeng, D. (2017). Genetic dissection of leaf senescence in rice. International Journal of Molecular Sciences, 18, 2686.
Lim, P. O., Kim, H. J., & Gil Nam, H. (2007). Leaf senescence. Annual Review of Plant Biology, 58, 115–136.
Lim, P. O., Lee, I. C., Kim, J., Kim, H. J., Ryu, J. S., Woo, H. R., et al. (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. Journal of Experimental Botany, 61, 1419–1430.
Lin, M., Pang, C., Fan, S., Song, M., Wei, H., & Yu, S. (2015). Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biology, 15, 43.
Liu, M., Yu, H., Zhao, G., Huang, Q., Lu, Y., & Ouyang, B. (2017). Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics, 18, 1–18.
Liu, L., Zhou, E. Y., Zhou, E. G., Ye, R., Zhao, E. L., Li, X., et al. (2008). Identification of early senescence-associated genes in rice flag leaves. Plant Molecular Biology, 67, 37–55.
Luquez, V. M., & Guiamét, J. J. (2001). Effects of the “stay green” genotype GGd1d1d2d2 on leaf gas exchange, dry matter accumulation and seed yield in soybean (Glycine max L. Merr.). Annals of Botany, 87, 313–318.
Mannai, Y., Akabane, K., Hiratsu, K., Satoh-Nagasawa, N., & Wabiko, H. (2017). The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. International Journal of Molecular Sciences, 18, 2165.
Mao, C., Lu, S., Lv, B., Zhang, B., Shen, J., He, J., et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiology, 174, 1747–1763.
Meyers, B. C., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell, 20, 3186–3190.
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.-H., Liu, Y.-X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science, 300, 332–336.
Morris, K., MacKerness, S. A., Page, T., John, C. F., Murphy, A. M., Carr, J. P., et al. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant Journal, 23, 677–685.
Park, S.-Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell, 19, 1649–1664.
Pourtau, N., Mares, M., Purdy, S., Quentin, N., Ruel, A., & Wingler, A. (2004). Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta, 219, 765–772.
Rajagopalan, R., Vaucheret, H., Trejo, J., & Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Development, 20, 3407–3425.
Rampino, P., Pataleo, S., Gerardi, C., Mita, G., & Perrotta, C. (2006). Drought stress response in wheat: Physiological and molecular analysis of resistant and sensitive genotypes. Plant, Cell and Environment, 29, 2143–2152.
Sakuraba, Y., Jeong, J., Kang, M.-Y., Kim, J., Paek, N.-C., & Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature Communications, 5, 4636.
Schippers, J. H. M., Jing, H.-C., Hille, J., & Dijkwel, P. P. (2007). Developmental and hormonal control of leaf senescence. In S. Gan (Ed.), Senescence processes in plants (pp. 145–170). Oxford: Blackwell Publishing Ltd.
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology, 6, e230.
Shriram, V., Kumar, V., Devarumath, R. M., Khare, T. S., & Wani, S. H. (2016). MicroRNAs as potential targets for abiotic stress tolerance in plants. Frontiers in Plant Science, 7, 1–18.
Stefanowicz, K., Lannoo, N., & Van Damme, E. J. M. (2015). Plant F-box proteins—judges between life and death. Critical Review Plant Sciences, 34, 523–552.
Stocks, M. B., Moxon, S., Mapleson, D., Woolfenden, H. C., Mohorianu, I., Folkes, L., et al. (2012). The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics, 28, 2059–2061.
Veljović-Jovanović, S., Kukavica, B., Stevanović, B., & Navari-Izzo, F. (2006). Senescence- and drought-related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. Journal of Experimental Botany, 57, 1759–1768.
Wu, X., Ding, D., Shi, C., Xue, Y., Zhang, Z., Tang, G., et al. (2016). microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biology, 16, 73.
Xiao, D., Cui, Y., Xu, F., Xu, X., Gao, G., Wang, Y., et al. (2015). SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE directly interacts with the cytoplasmic domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and negatively regulates leaf senescence in Arabidopsis. Plant Physiology, 169, 1275–1291.
Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biology, 2, e104.
Xiong, Y. Y., Ma, J., He, Y. H., Lin, Z., Li, X., Yu, S. M., et al. (2018). High-throughput sequencing analysis revealed the regulation patterns of small RNAs on the development of A. comosus var. bracteatus leaves. Scientific Reports, 8, 1–11.
Xu, X., Bai, H., Liu, C., Chen, E., Chen, Q., Zhuang, J., et al. (2014). Genome-wide analysis of microRNAs and their target genes related to leaf senescence of rice. PLoS ONE, 9, e114313.
Zeng, S., Liu, Y., Pan, L., Hayward, A., & Wang, Y. (2015). Identification and characterization of miRNAs in ripening fruit of Lycium barbarum L. using high-throughput sequencing. Frontiers in Plant Science, 6, 1–15.
Zhang, X., Ju, H. W., Chung, M. S., Huang, P., Ahn, S. J., & Kim, C. S. (2011). The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant and Cell Physiology, 52, 138–148.
Zhang, W. Y., Xu, Y. C., Li, W. L., Yang, L., Yue, X., Zhang, X. S., et al. (2014). Transcriptional analyses of natural leaf senescence in maize. PLoS ONE, 9, e115617.
Zhou, X., Jiang, Y., & Yu, D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecular Cell, 31, 303–313.
Zhu, Q.-H., Spriggs, A., Matthew, L., Fan, L., Kennedy, G., Gubler, F., et al. (2008). A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research, 18, 1456–1465.
Zou, C., Sun, K., Mackaluso, J. D., Seddon, A. E., Jin, R., Thomashow, M. F., et al. (2011). Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proceedings of National Academy of Sciences, 108, 14992–14997.
Acknowledgements
JMS and CVK are thankful for research fellowships from University Grants Commission (UGC), India. R&D grants from DBT, DST-PURSE and University of Delhi are acknowledged. We thank Gopal Joshi for helping with submission of small RNA sequencing data to SRA database. Deep sequencing was carried out by DBT-funded High-Throughput Sequencing Facility at University of Delhi South Campus, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sasi, J.M., Kumar, C.V., Mani, B. et al. Identification and characterization of miRNAs during flag leaf senescence in rice by high-throughput sequencing. Plant Physiol. Rep. 24, 1–14 (2019). https://doi.org/10.1007/s40502-019-0436-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40502-019-0436-6