Abstract
A host-pathogen surpasses the plant defense machinery and successfully infects the plant to serve its needs. In contrast, a nonhost pathogen is restricted by plant immune responses. In this study, we deciphered the differential responses of Arabidopsis against the host-pathogen (Pseudomonas syringae pv. maculicola, Psm) and the nonhost pathogen (P. syringae pv. tabaci, Pst) infection. The Pst multiplication was restricted in Arabidopsis plant coinciding with the absence of any disease symptoms which was also associated with the increase in defense associated, pathogenesis related gene1 (PR1) expression and callose deposition. Host-pathogen infection, on the other hand, caused chlorotic symptoms with much less activation of the defense marker genes compared to the nonhost infection. Proline content was decreased in plants infected with Psm but not in case of Pst infection. Proline is a crucial plant metabolite, and osmolyte suggested to be involved in plant defense responses. However, the differential regulation of proline pathway under a susceptible or resistance response has not yet been unravelled. The expression profile of proline metabolism genes in a time course post-infection revealed drastic differences after Psm and Pst pathogen infection. The elevated expression of proline catabolic genes, AtProDH and AtP5CDH were noted under Psm infection. In contrast, plants infected with Pst showed upregulation of AtProDH but a decline in AtP5CDH transcripts along with an upregulation of the biosynthesis genes i.e. AtP5CS and AtP5CR. Our study shows that proline metabolism is tightly regulated under host and nonhost pathogen infection and impacts susceptibility and resistance of the plants, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants encounter a plethora of potential pathogens at different stages of their life cycle (Rejeb et al. 2014). However, only a few of these pathogens can cause disease (Lipka et al. 2008). Most plant species depict an active defense mechanism which is not suppressed by a potential plant pathogen. The robust and broad-spectrum resistance mechanism of plant species to defend all the non-adapted pathogen species which are potential threat is called as nonhost resistance (NHR) (Thordal-Christensen 2003; Mysore and Ryu 2004; Nuernberger and Lipka 2005, Senthil-Kumar and Mysore 2013). A plant species which confers tolerance against all races of a potential plant pathogen which are pathogenic to another plant species are called nonhost plant and these pathogens are known as nonhost pathogen (Senthil-Kumar and Mysore 2013). NHR requires preformed and inducible defense responses and has multiple layers of defense which involve many underlying mechanisms and makes it very complex (Nicks and Marcel. 2009; Gill et al. 2015; Ayliffe and Sørensen 2019; Fatima et al. 2019; Fonseca and Mysore. 2019).
Plant immune system is induced by pathogen attack and predominantly relies on basic innate immunity as well as systemic signals which are translocated from the infected cells to the whole plant to restrict the infecting pathogen from spreading, which is referred as systemic acquired resistance (SAR) (Dangl and Jones 2001; Ausubel 2005). Plant innate immunity consists of two layers; first, pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs) are recognized by pattern recognition receptors (PRRs) present on the plant extracellular surface and triggers immunity (called as PTI) (Jones and Dangl 2006; Yu et al. 2017). The second one is elicited in response to a pathogen overriding the PTI and releasing effector molecules inside the plant cells. These effector molecules are recognized inside the cytosol by plant resistance proteins (R-protein). These R-proteins act as surveillance system in the cell, which interrupts effectors and leads to the effector-triggered immunity (ETI) (Jones and Dangl 2006; Cui et al. 2015). A host-pathogen is competent enough to suppress the plant defense machinery and successful in gaining the entry and extracting nutrients from the host plant cell. For example, the foliar pathogens, Pseudomonas syringae pv. tomato (Pto) (causes bacterial speck of tomato) and P. syringae pv. maculicola (Psm) (causes necrosis and water-soaked symptoms in cruciferae plant species) enters the plant by natural openings such as stomata (Melotto et al. 2008; Griffin and Carson 2015; Zhao et al. 2000).
The pathogen which surpasses the preformed defense mechanism and is recognized by the extracellular plant receptors which cause activation of plant immune system are adapted pathogens. While non-adapted pathogens are restricted by preformed or induced defenses upon pathogen entry. Based on the adapted and non-adapted pathogens, plant generally shows two types of resistance termed as host resistance and nonhost resistance (NHR) mechanism respectively (Gill et al. 2015). Host resistance occurs due to specific R protein present in plant and this is accession or cultivar specific and this is often used in breeding programs but less durable while NHR is multi-layered and more durable (Mysore and Ryu 2004; Dangl et al. 2013). NHR is of two types; type I resistance mechanism does not produce any visible symptom on the plant while type II resistance mechanism shows hypersensitive response mediated cell death similar to R-gene mediated cell death (Mysore and Ryu 2004; Senthil-Kumar and Mysore 2013).
In the recent past, a virus-induced gene silencing (VIGS)-based forward genetic screening showed involvement of proline catabolic enzyme, proline dehydrogenase (ProDH1) and ornithine amino transferase (OAT) in nonhost resistance mechanism (Senthil-Kumar and Mysore 2012). In another study, proline degradation enzyme proline dehydrogenase (ProDH) has been shown to act in defense against the avirulent bacterial pathogen, Pto-AvrRpm1 in Arabidopsis (Cecchini et al. 2011). Besides, proline has been well known to accumulate in a plant cell to confer tolerance against most abiotic stresses such as osmotic stress, salinity stress, and waterlogging stress (Verslues and Sharma 2010; Hayat et al. 2012). Proline has also been shown to be accumulated under biotic stress imposed by Pto AvrRpm1 and Pto AvrRpt2 during plant-pathogen incompatible reaction in Arabidopsis as a defense strategy (Fabro et al. 2004). Thus, proline metabolism seems to be an important regulatory mechanism for plant defense against pathogens. However, a clear understanding of the role of proline and its metabolism in defence against host and nonhost bacterial pathogen is still missing. In this study, we have studied the differential response of Arabidopsis plant after infection with host (Psm) and nonhost (Pst) pathogen with regards to basal innate immune responses and the genes involved in proline metabolism.
Materials and methods
Plant growth
Arabidopsis thaliana ecotype Columbia-0 (Col0, Arabidopsis Biological Research Centre, accession number CS70000) was grown in plastic pots with dimensions of 7-inch × 3-inch (height × diameter) filled with a substratum of agro peat and vermiculite (3:1 vol/vol) mix. The top layer of the substratum was covered with mesh. Pots were saturated with water and seeds were sown followed by incubation in a cold room (4 °C) for 2 days. Pots were transferred to growth room under conditions of 23 °C day and 19 °C night temperature, 150–200 μE m−2 s−1 light intensity and photoperiod of 12 h light/12 h dark. Plants were watered at 3 days interval with Hoagland solution [Solution A containing 1 mL/L of 1 M KH2PO4, 1 M KNO3, 1 M Ca(NO3)2, 1 M MgSO4, solution B containing H3BO3, MnCl2, ZnSO4, CuSO4, H2MoO4 and solution C containing Fe-EDTA] till full-grown rosettes were observed. Thirty-days-old well-grown rosette plants were used for pathogen inoculation.
Bacterial strains and inoculum preparation
Pseudomonas syringae pv. maculicola ES4326 (Psm) was used as a host-pathogen and P. syringae pv. tabaci 6605 (Pst) was used as a nonhost pathogen on A. thaliana (Mysore and Ryu 2004). Primary culture was grown from a single colony of Psm and Pst in 10 mL King’s B (KB) medium (Cat# M1544, HiMedia Laboratories, Mumbai, India) with or without kanamycin (50 µg/mL), respectively at 28 °C till the culture optical density at 600 nm (OD600), reaches at 0.2. Secondary culture of Psm and Pst was grown in 500 mL KB medium from primary inoculum till 0.5 OD600 was obtained. Bacterial cultures were centrifuged at 4270 g at room temperature, and the supernatant was discarded. Pellet was washed in sterile distilled water (500 mL) twice and finally the pellet was suspended in 500 mL sterile distilled water. OD600 was checked for the bacterial suspension, and the desired concentration to be used was obtained by diluting with sterile water for the dip inoculation of plants.
Dip inoculation method
Dip inoculation method was adapted from the protocol described by Jacob et al. 2017. Bacterial inoculum was prepared in a plastic tray containing bacterial suspension (final OD600 = 0.05) and 0.02% silwet L-77 (Lehle seeds, Round Rox, Texas, USA). Siwet L-77 acts as a surfactant which helps the bacteria to stick to the leaf surface. A. thaliana plants were dipped in bacterial suspension for 2 min. Plants were covered with tray for 12 h to maintain high humidity for successful infection and incubated in a growth room with the above-mentioned growth conditions.
In-planta bacterial number assessment
To determine bacterial multiplication in A. thaliana leaf, in planta bacterial population was measured at 0, 1, 2, and 3 days post-inoculation (dpi) by following similar method described by Jacob et al. (2017) with some modifications. Leaf disc was cut from 1 cm cork-borer and was used for bacterial number assessment. Leaf discs were surface sterilized with 70% ethanol for 30 s and rinsed thrice with sterile distilled water followed by blot drying on a tissue paper. The sample was homogenized in 1 mL sterile distilled water in a micro-centrifuge tube. The diluted sample was plated onto KB plate containing appropriate antibiotics and incubated at 28 °C. Colonies of Psm and Pst were counted after 2 days and 1-day post-plating respectively. In planta bacterial population was represented as log10 CFU/cm2. Ten biological replicates were used for the assay.
Ion leakage assay
Damage caused by pathogen infection was estimated by measuring ion leakage in the inoculated plants by following the protocol of Tripathy et al. (2000). Ion leakage was measured from 1 cm diameter leaf discs. Leaf discs were thoroughly rinsed with water after punching and gently agitated in 10 mL distilled water at 20 °C. Ion leachates in the bathing medium were determined by measuring ion conductivity (microprocessor-based EC-TDS-SAL meter, model-1602, Esico International, Himachal Pradesh, India) (C1). Subsequent to this, the samples were boiled at 100 °C such as to completely kill plant cells and release the total ions into the bathing medium. The bathing medium was cooled down to room temperature, and ion leachates were measured (C2). Three biological replicates were used for the measurement. Percent ion leakage was measured by using the following formula.
Cell death assay
Trypan blue staining of the inoculated leaves was performed to assay cell death by following method described by Fernández-Bautista et al. (2016). Infected leaf at 1 dpi was placed in trypan blue solution (0.05%) (Cat#TCL005, HiMedia Laboratories) for 2 h and then washed overnight with ethanol: acetic acid solution (3:1, vol/vol). Stained leaves were observed under Nikon Stereozoom microscope (Nikon Instrument Inc., Melville, NY, USA) and the image was captured with the camera attached to the microscope at 0.6 × magnification and visualized by NIS element software.
Callose deposition assay
Callose deposition was observed by aniline blue staining by following the method described in Schenk and Schikora (2015). Plant leaves infected with the host or nonhost pathogen at 1 dpi were bleached with ethanol and acetic acid solution (3:1, vol/vol) to remove the background chlorophylls. Destained leaves were washed with 150 mM solution of K2HPO4 for 30 min. This was followed by staining with aniline blue (Cat#GRM901, HiMedia) (0.01%) and 150 mM K2HPO4 (Cat#GRM1045, HiMedia) solution for 2 h. Leaves were observed under a Leica TCS SP8 confocal microscope under DAPI filter (excitation filter 390 nm; dichroic mirror 420 nm; emission filter 460 nm).
Proline estimation
Proline estimation was done according to the protocol mentioned by Bates et al. (1973). Leaf tissue (100 mg) was homogenized in 1 mL of 3% sulfosalicylic acid (Cat# A297-500, Fisher Scientific, Mumbai, India) and centrifugation at 7000 rpm for 5 min to get the plant extract. This extract was used for proline estimation by adding acid ninhydrin (Cat# T349-10, Fisher Scientific), glacial acid (Cat# A38S-500, Fisher Scientific) and incubated at 70 °C for 1 h. Toluene (Cat# T324-1, Fisher Scientific) was added to the reaction mix and mixed well on ice. The topmost clear layer was transferred into a fresh tube and OD at 520 nm was measured by FLUOstar Omega plate reader (UV Visible absorbance) (Biotron Helthcare Pvt. Ltd, Mumbai, India). A standard curve was prepared using l-proline (P5607, Sigma Aldrich Inc. Merck KGaA, Darmstadt, Germany). As per the standard curve, OD520 = 1 corresponds to 36.2 μg of l-proline reacted with ninhydrin. This proline value (36.2) was used as a factor in the below-mentioned formula.
RNA isolation and cDNA preparation
Total RNA was isolated from leaf tissue (100 mg) which was harvested from the mock or pathogen-treated plants at 3, 12, and 24-h post-infection (hpi). The sample was homogenized in liquid nitrogen to make a fine powder. Total RNA was extracted by TRIzol™ reagent (Cat# 543 15596026, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA was treated with DNAseI (Cat# M0303, New England Biology, Ipswich, MA, USA) to remove DNA from the RNA sample. The RNA integrity was checked on agarose gel (1%) and quantified using NanoDrop 1000 UV–Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was prepared from 2 µg RNA using verso cDNA synthesis kit (Cat# K1621, Thermo Fisher Scientific) according to the mentioned protocol.
Expression profiling by quantitative real-time PCR
Expression profile of proline metabolic pathway genes was done by quantitative real-time PCR (RT-qPCR) using ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA, USA). The gene-specific primers were designed with the National Centre for Biotechnology Information (NCBI) primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Supplementary Table S1. The reaction mixture contained cDNA template (1 µL), gene-specific primer (10 mM/mL) (1 µL), SYBR Green master mix (5 µL) (Cat# 4309155, Thermo Fisher Scientific) and the reaction volume was made up to 10 µL with sterile water. Four biological replicates were used and the expression values of AtACTIN8 were used as an endogenous control. Comparative cycle threshold (ΔCt) values for each gene were obtained after normalization with the AtACTIN8 expression value. The fold change values for the gene expression were estimated by normalizing the ΔCt value of pathogen inoculated samples with mock-treated sample (Livak and Schmittgen 2001) and values were represented as 2−∆∆CT. Student’s t test was used for evaluating the statistical significance at p value < 0.05.
Statistics or data analysis
The total number of biological replicates are mentioned in each figure legend. One-way ANOVA with HolmSidak correction and student’s t test were used for statistical analyses. Significance differences were measured at p < 0.05 and shown with different letters. Error bars indicate standard error of the mean (SEM). Statistical analysis was done using SigmaPlot 11.0 (Systat Software Inc., Chicago, IL, USA).
Results
Bacterial host or nonhost pathogen infection on Arabidopsis plants and disease development
Arabidopsis plants were infected with host (Psm) or nonhost (Pst) pathogen by dip inoculation. Plants were kept in a growth room and covered with transparent covers to maintain humidity and assist bacterial infection. Host-pathogen (Psm) infected Arabidopsis plants showed chlorotic, water-soaked symptoms on a leaf at 3 dpi. The visible signs of chlorosis started appearing from day two and progressed to prominence at 3 dpi (Fig. 1A). Nonhost (Pst) pathogen was also inoculated in the same manner as that of host (Psm) pathogen by facilitating high humidity to allow bacterial entry. But nonhost pathogen did not show any visible chlorotic symptom and the infected plant was as healthy as a mock-inoculated plant (which was dipped in sterile water for 2 min) (Fig. 1B).
Disease symptom development on Arabidopsis plants after infection with host and nonhost pathogen. Representative photographs show chlorotic symptoms in a plant infected with host (Pseudomonas syringae pv. maculicola, Psm) pathogen after 3 dpi (A). Red color arrows show chlorotic disease symptoms. No disease symptoms were observed in a plant infected with nonhost pathogen (P. syringae pv. tabaci, Pst) pathogen after 3 dpi (B). dpi represents days post inoculation; mock, water-inoculated control. Scale bar = 1 cm (color figure online)
In planta bacterial number continued to increase in Arabidopsis plants infected with host (Psm) pathogen and corroborated with the appearance of disease symptoms on plant leaves. At 0 dpi bacterial number in log10 CFU was four which significantly increased to 10 at 3 dpi (Fig. 2A). Nonhost (Pst) bacteria also entered in the plant and 0 dpi bacterial number in log10 CFU was ~ 3 and it increased to 4.8 at 2 dpi but did not multiply further at 3 dpi (Fig. 2A). We checked the membrane leakage as a parameter to assess the damage caused by infecting pathogen. The results revealed that membrane leakage was significantly increased due to host (Psm) infection at 3 dpi (Fig. 2B) but there was not a significant difference between nonhost Pst treated plants and mock plant (Fig. 2C). The results thus indicated that most likely the plant defended the nonhost pathogen successfully.
Differential response of Arabidopsis plants inoculated with host and nonhost pathogen. Graphs represent the progression of bacterial multiplication of host pathogen (Psm) and nonhost pathogen (Pst) (A) in inoculated leaves. Graphs show membrane leakage (%) in leaves inoculated with host pathogen (B) and nonhost pathogen (Pst) (C). Values are represented as an average of ten biological replicates and error bars are the standard error of the mean. Different letters indicate significant difference among different dpi in (A) between mock and treatment in (C, D). Significance was determined by one-way ANOVA, Holm-Sidak method (p < 0.05). CFU, colony forming unit
Defense response of Arabidopsis against host and nonhost pathogen infection
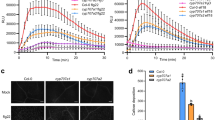
Arabidopsis plants have been demonstrated to deposit callose on cell wall as a strategy to restrict pathogen entry. We checked the callose deposition in plant leaves after infection with host (Psm) or nonhost (Pst) pathogen at 1 dpi. We found nonhost pathogen-infected plant leaf had callose deposits, but these were absent in the mock-inoculated plants (Fig. 3A). This shows that callose is deposited in response to nonhost pathogen infection as a defense strategy. Host-pathogen treated plant did not contain callose deposits, but cell wall was ruptured and stained by aniline blue as patches indicating disease cell death. The bright-field image showed no callose deposition and cell wall degradation (Fig. 3A). Next, we checked cell death with trypan blue staining in Arabidopsis leaves after infection with host (Psm) or nonhost (Pst) pathogen infection at 3 dpi. Psm infected leaves showed the blue staining of the dead region and no staining was observed in the case of Pst infected leaves (Fig. 3B). The results depict that there was cell death due to host-pathogen infection.
Influence of host and nonhost pathogen inoculation on plant resistance. The representative photograph shows callose deposition (A) and cell death (B) in Arabidopsis plants at 1 dpi with Psm and Pst. Callose deposition was assayed in the inoculated plant by aniline blue staining. The samples were observed under DAPI filter and brightfield using Leica TCS SP8 MP confocal microscope at 1 dpi after staining for 2 h with aniline blue solution. Blue arrow indicates the callose deposited on a leaf. Nonhost pathogen (Pst) infected leaves exhibit callose deposition which are shown in navy blue colour dots, indicated with a blue arrow while host (Psm) pathogen did not cause callose deposition. Host-pathogen infection caused cell death but callose deposition was not observed (A). Scale bar = 100 µm. Cell death was visualized by trypan blue staining. Mock leaf did not stain with trypan blue showing no cell death. Psm infected leaf was stained with trypan blue at the cell death region whereas Pst infected leaf did not show any cell death (B). Scale bar = 500 µm (color figure online)
To check the defense response after infection with host or nonhost pathogen, we analyzed the expression of PR1 (Pathogenesis Related protein 1) and nonhost resistance marker gene NHO1 (Nonhost of P. s phasiolicola 1). PR1 gene expression was upregulated in both Psm and Pst infected plant leaf. PR1 expression in Psm infected plant was 3.3-fold at 3 hpi, 72-fold at 12 hpi and 260-fold at 24 dpi while PR1 expression was way higher in Pst (nonhost pathogen) infected plants such that it was 29-fold at 3 hpi, 665-fold at 12 hpi and 1430-fold at 24 hpi (Fig. 4A). Psm infected plant had a basal level of NHO1 relative expression 1-fold at 3 dpi which was 0.8-fold at 12 hpi and around 1.2-fold at 24 hpi. In contrast, under nonhost Pst infection NHO1 relative expression was up by 1.2-fold at 3 dpi which was increased to 4.7-fold at 12 hpi and reached 2.5-fold at 24 hpi when compared with mock-inoculated plants (Fig. 4B).
Transcript expression profile of defence genes in plants inoculated with host and nonhost pathogen. Graphs show fold change in gene expression of PR1 (pathogenesis related gene 1) (A) and NHO1 (nonhost of phaseolicola 1) (B) in host and nonhost inoculated samples when compared with the expression profile in mock samples at 3, 12 and 24 hpi. Host pathogen Psm and Nonhost pathogen Pst is used. The expression profile was performed by RT-qPCR and the expression value of AtACTIN8 was used to normalize data in 2−∆∆ct method. Bars represent data averaged from four biological replicates and error bars are the standard error of the mean. Significance was calculated by one-way ANOVA and different letters indicate significant differences at p < 0.05
Differential regulation of proline metabolism in Arabidopsis plants inoculated with host or nonhost pathogen
We observed a decrease in free proline levels in Psm but no significant difference in Pst infected sample as compared to mock at 1 dpi (Fig. 5). To decipher further, we investigated the expression profile of proline metabolism genes in Arabidopsis leaves after infection with Psm and Pst at 3-, 12- and 24-hpi by quantitative real-time PCR. Proline biosynthetic gene P5CS1 and P5CS2 were upregulated in Psm-infected samples at 3 hpi with a higher expression of P5CS2 than P5CS1. However, at later time points of 12 hpi and 24 hpi, both these genes were downregulated with P5CS1 being the most downregulated than P5CS2. However, Pst infection led to an increase in the P5CS1 and P5CS2 gene transcript levels at 3, 12 and 24 hpi. P5CS2 expression was more compared to P5CS1 at 12 and 24 hpi. Similarly, proline synthesizing P5CR gene was upregulated at 3 hpi in Psm infected leaves but downregulated at 12 and 24 hpi. In contrast, this gene was upregulated at all the tested time-points in Pst infected leaves. This shows that proline synthesis increases in case of Pst infected plants but decreases in case of Psm infected plants. That is why we found more proline in Pst infected leaves as compared to Psm infected leaves at 1 dpi (Fig. 5). Proline degradation genes were upregulated under both the pathogen stresses. ProDH1 and ProDH2 genes were upregulated upon Psm infection and ProDH1 gene transcript levels were higher compared to ProDH2. Pst infected leaves also showed an increase with ProDH1 being highest compared to ProDH2. Thus, plants exhibit elevated expression of ProDH genes upon host and nonhost pathogen infection, however, the ProDH gene expression was more in the case of Pst infection than Psm infection. The expression level of another gene encoding proline catabolic enzyme P5CDH was higher in Psm infected leaves at 24 hpi but this gene was downregulated in Pst infected leaves at all three time points (Fig. 6). The expression level of gene encoding ornithine amino transferase (OAT, catabolizes ornithine and forms P5C) was slightly upregulated in Psm infected samples at all three time points while OAT gene expression was found higher in Pst-infected samples at 12 and 24 hpi as compared to Psm-infected samples. Proline degradation occurs in both cases (Psm and Pst infected plants) while proline synthesis is increased in case of Pst but not in Psm infected plants. That is why we found decrease in proline levels in Psm infected plants while no significant decrease in case of Pst infected plants compared to mock (Fig. 5). SRO5 (similar to RCD One 5) (SRO5) gene is present in tandem to the P5CDH gene in the Arabidopsis genome and both genes overlap (Borsani et al. 2005). This junction produces natural cis acting siRNAs which degrade the P5CDH transcripts. The SRO5 gene expression was downregulated in host (Psm) infected samples at 12 and 24 dpi while its expression was upregulated in nonhost (Pst) infected samples at 3, 12 and 24 hpi. Pst infected samples had downregulation of P5CDH transcripts. While SRO5 was downregulated in Psm infected sample which causes no degradation of P5CDH transcripts by siRNA and P5CDH transcripts were upregulated in Psm infected sample at 24 dpi (Fig. 6). Our results thus suggest that P5C is increased more in mitochondria upon Pst infection due to increase in ProDH, OAT and downregulation of P5CDH transcripts.
Proline content in Arabidopsis plants after inoculation with host and nonhost pathogen. Proline content was measured in leaves inoculated with host (Psm) and nonhost (Pst) pathogen at 1 dpi. The different letters indicate significant values in treated samples when compared with mock control by one-way ANOVA at p < 0.05. Data are the average of four biological replicates and error bars designate standard error of means
Transcript expression profile of proline metabolism genes in plants inoculated with host and nonhost pathogen. Proline biosynthesis occurs in cytosol and plastid in two steps; which initiates with the conversion of glutamate to P5C by P5CS (P5CS1 and P5CS2) and P5C is eventually converted to proline by P5CR. Proline catabolism occurs exclusively in mitochondria where proline is catabolized to P5C by ProDH (ProDH1 and ProDH2). P5C is subsequently catabolized to glutamate by P5CDH. OAT forms P5C from ornithine and SRO5 act as an inhibitor of P5CDH transcript by natural si-RNA mediated silencing. Differential transcript expression of the genes encoding listed enzymes was determined by RT-qPCR in plants inoculated with host (Psm) and Nonhost (Pst) pathogen at 3, 12 and 24 hpi. The gene expression data were normalized with the expression values of AtACTIN8 gene. Four biological replicates were considered for expression profiling. Expression values are represented as heatmap. P5CS, pyrroline 5-carboxylate synthase; P5CR, pyrroline 5-carboxylate reductase; ProDH, proline dehydrogenase; OAT, ornithine amino transferase; SRO5, Similar to RCD One 5. Colorbar scale in red and blue shows upregulation and downregulation in gene expression, respectively
Discussion
Plants are constantly exposed to a plethora of pathogens in nature where only a few can cause disease. The pathogen which adapts and infects the host plants have evolved various mechanisms to overcome the plant defense barrier and manipulate the host machinery (Lipka et al. 2008; Lee et al. 2017). Pathogens inject a range of effector molecules inside a plant cell to target various plant mechanisms (Speth et al. 2007). Some plants have resistance proteins (R-protein) which can recognize the specific effector protein released by host-pathogen (gene for gene hypothesis). This recognition leads to the R -protein mediated hypersensitive response by the plant which causes cell death to stop further spread of pathogen to another plant cell (Jones and Dangl 2006). But most of plants exhibit nonhost mechanism against the non-adapted pathogen which not only acts at the level of ETI but also at PTI (Senthil-Kumar and Mysore 2013). The nonhost resistance mechanism is a multi-layered robust mechanism which however is complex to understand. We have compared the plant resistance mechanism using both host (Psm) and nonhost pathogen (Pst) to find out a differential response in Arabidopsis plants. Host (Psm) bacterial pathogen showed successful infection in Arabidopsis and results from in planta bacterial multiplication showed an exponentially growing bacterial population causing chlorotic disease symptoms on its host plant. In our study, we found that infact nonhost bacteria (Pst) also infected the plants but was restricted by active plant defense mechanisms as inferred from data on in planta bacteria multiplication assay. There was no disease symptom observed after infection with nonhost pathogen, and visibly the plant was no different than the mock-inoculated plant. Furthermore, we checked the membrane leakage after infection with host-pathogen which increased significantly compared to mock-inoculated plants, while nonhost pathogen did not cause significant damage to the membrane and there was no significant change in electrolyte leakage compared to mock.
The NHR is a multi-layered defense strategy involving pre- and post-invasive machinery stopping the entry of bacteria inside the plant and suppression of bacterial growth and multiplication respectively (Supplementary Table 1). When plant recognizes the PAMPs by the PRRs and triggers PTI, the plant closes the stomata which are the entry point for the pathogen. The plant also makes changes in the cell wall to make it resistant by depositing the callose (a polysaccharide in the form of β-1,3-glucan with some β-1,6-branches) (Chen and Kim. 2009). We, therefore, checked the callose deposition in the cell wall of the plant and found that plants which were infected with nonhost pathogen, deposited the callose on cell wall but a plant infected with host-pathogen failed to do so. Corroborating with a successful infection and disease progression in a host pathogen-infected plant, cell death was observed while nonhost pathogen infection did not lead to any disease symptom causing cell death. We did not observe the hypersensitive cell death in case of Pst infected sample while earlier report by Choudhary et al. 2017 showed HR cell death. The difference is due to the method of inoculation where Choudhary et al. (2017) have used the syringe infiltration of the pathogen inside the apoplast surpassing the plant preinvasive defense mechanism but we have used the dip inoculation mimicking the natural mode of infection.
It is possible that the host-pathogen surpasses the PTI and can release effector molecules inside the plant cell to manipulate the plant machinery and hence successfully establishes and multiplies. The NHR mechanism involves many underlying molecular mechanisms such as robust SA-mediated defense signaling and other hormone signaling. NHO1 is shown as nonhost marker gene which is a glycerol kinase which is required for gene for gene resistance mechanism (Lu et al. 2001, Kang et al. 2003, Li et al. 2005). SA signaling leads to activation of TGA transcription factors which activates the pathogenesis-related protein (PR) genes expression. We found that NHO1 is indeed a nonhost specific marker gene which expressed highly in case of nonhost infection while no significant increase was noted in case of host-pathogen infection. On the other hand, PR1 gene expression was observed under both host and nonhost pathogen infection which suggests SA signaling was activated in plants in both cases. PR1 gene expression was increased thousand-fold in case nonhost pathogen infection but only hundred-fold in case of host-pathogen infection compared to mock.
Proline acts as an osmolyte and gets accumulated under different abiotic stress condition (Verslues and Sharma 2010; Hayat et al. 2012) to scavenge the reactive oxygen species. Oxidative burst also occurs during defense against pathogen stress and it was observed that proline biosynthetic gene P5CS2 was upregulated during an incompatible host–plant interaction and proline levels were upregulated but not in case of compatible host–plant interaction (Fabro et al. 2004). We observed that proline was decreased in host pathogen-infected sample as compared to mock plants. But there was no significant difference in proline levels between mock and in nonhost pathogen-infected plants. Similarly, Cecchini et al. (2011) showed that proline degradation enzyme ProDH contributes to plant defense in case of avirulent pathogen and host plant interaction. A study based on VIGS (virus-induced gene silencing), a forward genetic approach had identified the proline catabolic and metabolic genes ProDH and OAT (both localized to mitochondria) as a nonhost resistance factors in Nicotiana benthamiana (Senthil-Kumar and Mysore 2013). To delineate the response of the host and nonhost pathogen in proline metabolism we checked the expression of pathway genes and observed the differential response in proline anabolism and catabolism. Our results revealed that generally the proline synthesis genes were upregulated in nonhost pathogen-infected plants but were downregulated in host pathogen-infected plants. While proline catabolism gene AtProDH was upregulated in both cases, AtP5CDH gene was upregulated in the host but not in nonhost where P5CDH was downregulated. This shows that AtProDH and AtP5CDH genes are not coupled and creates a difference in P5C which is catabolized product of proline in mitochondria. AtOAT gene is upregulated in both cases, but the relative expression was higher in nonhost than host pathogen-infected plants. This shows that proline metabolism is differentially regulated leading to differential P5C levels in mitochondria under host and nonhost pathogen infection. P5C has been shown to produce ROS and hypersensitive response (Hellmann et al. 2000; Maxwell and Davis 2000; Deuschle et al. 2004; Nishimura et al. 2012; Qamar et al. 2015) and can be responsible for the avirulent R-gene-mediated defense and nonhost resistance. Host-pathogen manipulate the distinct plant mechanism for their benefit. We observed that proline metabolism can be one of the distinct mechanisms which host-pathogen has manipulated for its successful growth and proliferation inside the plant while the plant has used this metabolism for defense against the nonhost pathogen.
Conclusion
Nonhost resistance (NHR) mechanism is a multi-layered and broadspectrum defense mechanism and is complex due to which it is a more durable resistance mechanism for plant species. Understanding of NHR is much needed to apply this knowledge for developing durable disease resistance crops. We have shown the differential response of Arabidopsis plants to the host and nonhost pathogen. Proline metabolism also plays an important role in defense against pathogen and differential response has been observed against host and nonhost pathogen infection. Differential regulation of proline metabolism acts as a defense mechanism against the nonhost pathogen in nonhost plant while it can also lead to susceptibility in host plant against host-pathogen.
References
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nature Immunology,6(10), 973.
Ayliffe, M., & Sørensen, C. K. (2019). Plant nonhost resistance: Paradigms and new environments. Current Opinion in Plant Biology,50, 104–113.
Bates, L. S., Waldren, R. P., & Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant and Soil,39(1), 205–207.
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., & Zhu, J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell,123(7), 1279–1291.
Cecchini, N. M., Monteoliva, M. I., & Alvarez, M. E. (2011). Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiology,155(4), 1947–1959.
Chen, X. Y., & Kim, J. Y. (2009). Callose synthesis in higher plants. Plant Signaling & Behavior,4(6), 489–492.
Choudhary, A., Gupta, A., Ramegowda, V., & Senthil-Kumar, M. (2017). Transcriptomic changes under combined drought and nonhost bacteria reveal novel and robust defenses in Arabidopsis thaliana. Environmental and Experimental Botany,139, 152–164.
Cui, H., Tsuda, K., & Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology,66, 487–511.
Dangl, J. L., Horvath, D. M., & Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science,341(6147), 746–751.
Dangl, J. L., & Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature,411(6839), 826.
Deuschle, K., Funck, D., Forlani, G., Stransky, H., Biehl, A., Leister, D., et al. (2004). The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell,16(12), 3413–3425.
Fabro, G., Kovács, I., Pavet, V., Szabados, L., & Alvarez, M. E. (2004). Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Molecular Plant-Microbe Interactions,17(4), 343–350.
Fatima, U., Bhorali, P., & Senthil-Kumar, M. (2019). Morpho-pathological and global transcriptomic analysis reveals the robust nonhost resistance responses in chickpea interaction with Alternaria brassicae. Molecular Plant-Microbe Interactions,32(12), 1598–1613.
Fernández-Bautista, N., Domínguez-Núñez, J. A., Moreno, M. C., & Berrocal-Lobo, M. (2016). Plant tissue trypan blue staining during phytopathogen infection. Bio-protocol,6(24), e2078.
Fonseca, J. P., & Mysore, K. S. (2019). Genes involved in nonhost disease resistance as a key to engineer durable resistance in crops. Plant Science,279, 108–116.
Gill, U. S., Lee, S., & Mysore, K. S. (2015). Host versus nonhost resistance: Distinct wars with similar arsenals. Phytopathology,105(5), 580–587.
Griffin, Eric A., & Carson, Walter P. (2015). The ecology and natural history of foliar bacteria with a focus on tropical forests and agroecosystems. Botanical Review,81(2), 105–149.
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., & Ahmad, A. (2012). Role of proline under changing environments: A review. Plant Signaling & Behavior,7(11), 1456–1466.
Hellmann, H., Funck, D., Rentsch, D., & Frommer, W. B. (2000). Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiology,122(2), 357–368.
Jacob, C., Panchal, S., & Melotto, M. (2017). Surface inoculation and quantification of Pseudomonas syringae population in the Arabidopsis leaf apoplast. Bio-protocol,7(5), e2167.
Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature,444(7117), 323.
Kang, L., Li, J., Zhao, T., Xiao, F., Tang, X., Thilmony, R., et al. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proceedings of the National Academy of Sciences USA,100(6), 3519–3524.
Lee, H. A., Lee, H. Y., Seo, E., Lee, J., Kim, S. B., Oh, S., et al. (2017). Current understandings of plant nonhost resistance. Molecular Plant-Microbe Interactions,30(1), 5–15.
Li, X., Lin, H., Zhang, W., Zou, Y., Zhang, J., Tang, X., et al. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proceedings of the National Academy of Sciences USA,102(36), 12990–12995.
Lipka, U., Fuchs, R., & Lipka, V. (2008). Arabidopsis non-host resistance to powdery mildews. Current Opinion in Plant Biology,11(4), 404–411.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods,25(4), 402–408.
Lu, M., Tang, X., & Zhou, J. M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. The Plant Cell,13(2), 437–447.
Maxwell, S. A., & Davis, G. E. (2000). Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proceedings of the National Academy of Sciences USA,97(24), 13009–13014.
Melotto, M., Underwood, W., & He, S. Y. (2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Annual review of Phytopathology,46, 101–122.
Mysore, K. S., & Ryu, C. M. (2004). Nonhost resistance: How much do we know? Trends in Plant Science,9(2), 97–104.
Nicks, R. E., & Marcel, T. C. (2009). Nonhost and basal resistance: How to explain specifity. New Phytologist,182(4), 817–828.
Nishimura, A., Nasuno, R., & Takagi, H. (2012). The proline metabolism intermediate Δ1-pyrroline-5-carboxylate directly inhibits the mitochondrial respiration in budding yeast. FEBS Letters,586(16), 2411–2416.
Nuernberger, T., & Lipka, V. (2005). Non-host resistance in plants: New insights into an old phenomenon. Molecular Plant Pathology,6(3), 335–345.
Qamar, A., Mysore, K., & Senthil-Kumar, M. (2015). Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Frontiers in Plant Science,6, 503.
Rejeb, I., Pastor, V., & Mauch-Mani, B. (2014). Plant responses to simultaneous biotic and abiotic stress: Molecular mechanisms. Plants,3(4), 458–475.
Schenk, S. T., & Schikora, A. (2015). Staining of callose depositions in root and leaf tissues. Bio-protocol,5(6), e1429.
Senthil-Kumar, M., & Mysore, K. S. (2012). Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant, Cell and Environment,35(7), 1329–1343.
Senthil-Kumar, M., & Mysore, K. S. (2013). Nonhost resistance against bacterial pathogens: Retrospectives and prospects. Annual review of Phytopathology,51, 407–427.
Speth, E. B., Lee, Y. N., & He, S. Y. (2007). Pathogen virulence factors as molecular probes of basic plant cellular functions. Current Opinion in Plant Biology,10(6), 580–586.
Thordal-Christensen, H. (2003). Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology,6(4), 351–357.
Tripathy, J. N., Zhang, J., Robin, S., Nguyen, T. T., & Nguyen, H. T. (2000). QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theoretical and Applied Genetics,100(8), 1197–1202.
Verslues, P. E., & Sharma, S. (2010). Proline metabolism and its implications for plant–environment interaction. The Arabidopsis Book/American Society of Plant Biologists,8, e0140.
Yu, X., Feng, B., He, P., & Shan, L. (2017). From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annual review of Phytopathology,55, 109–137.
Zhao, Y., Damicone, J. P., Demezas, D. H., Rangaswamy, V., & Bender, C. L. (2000). Bacterial leaf spot of leafy crucifers in Oklahoma caused by Pseudomonas syringae pv. maculicola. Plant Disease,84(9), 1015–1020.
Acknowledgements
Aarzoo Qamar acknowledges UGC (23/12/2012(ii) EU-V, 17/7/2013) and NIPGR for providing funding. Authors thank Dr. Aarti Gupta for critical reading and editing of the manuscript and Dr. Mahesh Patil for internal reviewing of the manuscript. Authors acknowledge Mr. Rahim Tarafdar for plant care and Mr. Sunder Solanki for technical help in the laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qamar, A., Senthil-Kumar, M. Arabidopsis exhibits differential response in basal immunity and proline metabolism during defense against host and nonhost pathogen infection. Plant Physiol. Rep. 24, 496–506 (2019). https://doi.org/10.1007/s40502-019-00480-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40502-019-00480-w