Abstract
MicroRNAs are known to regulate almost all developmental processes in plants. These are ubiquitously expressed and most importantly their abundances vary by several orders of magnitude in different tissues and developmental stages. MicroRNA profiles in seedlings and leaves of switchgrass have been reported but not from the inflorescence or tillers. The overall small RNA population in inflorescence differs from the other vegetative organs, and, moreover, miRNAs are important regulators of flowering and flower development, thus inflorescence was chosen. Likewise, emerging tillers were chosen to identify miRNAs that might play a role in tillering, which is an important trait contributing to higher biomass production. The sequencing followed by computational analyses of small RNA libraries generated from inflorescence and emerging tillers revealed the identification of 28 conserved and 4 novel miRNA families. The expression levels of most miRNA families and miRNA variants within a family displayed greater differences between the tissues. Importantly, this study has offered insights into differences in abundances of miRNA families in the inflorescence and tillers. Specifically, the reported miRNA profiles from tillers are a valuable resource to examine which of these miRNAs play important roles in tillering, an important agronomic trait in this bioenergy crop.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the need for biofuel production from plants that are not being used in producing the human diet, several species such as switchgrass, Sorghum, Brachypodium and Miscanthus have emerged as promising feedstock species. Of these, switchgrass has received much attention, however, to advance the utility of switchgrass as a major biofuel feedstock, unlocking the molecular switches that controls biomass production (increased foliar number and size, plant height, tiller numbers etc.,) is critically important.
In plants, the importance of miRNA-guided target gene regulation is indispensable for various physio-biochemical processes/pathways associated with the development and maintaining cellular homeostasis under stress conditions (Sunkar and Zhu 2007; Sunkar et al. 2012). Earlier studies have reported the identification miRNAs in switchgrass including their response to PEG-induced osmotic stress and salinity (Matts et al. 2010; Sun et al. 2012; Xie et al. 2014). Additionally, we reported the expression profiles of miRNAs in switchgrass exposed to long-term drought or heat (Hivrale et al. 2016). These studies have explored the miRNA profiles either in leaves or seedlings with or without the stress treatments. It is known that the plant small RNA population especially the small interfering RNA (siRNA) population varies greatly in inflorescence (reproductive tissues) compared to the vegetative tissues. Further, the miRNA profiles and their abundances in inflorescence of switchgrass is unknown. Keeping this in view, we analyzed small RNAs in the inflorescence or flowers of switchgrass.
Biomass production in the case of herbaceous bioenergy plant species is dependent on more tillers, thus tillering is an important trait in switchgrass (Fu et al. 2012; Wang et al. 2012, 2013; Xu et al. 2015). Tillering involves initiation of tiller buds and subsequently their outgrowth (Xu et al. 2015). At the molecular level genes such as the MONOCULM 1, TEOSINTE BRANCHED 1, PIN FORMED FAMILY OF TRANSPORTER 1 (PIN1), HIGH TILLERING DWARF/DWARF17(D17), DWARF 10 (D10), DWARF 14 (D14), DWARF 27 (D27), RICEFLORICULA/LEAFY (RFL) and MOC3 (MONOCULM 3)/TILLERS ABSENT1(TBA1) are known to regulate the process of tillering in rice and other monocots (Xu et al. 2015). MicroRNAs largely fine-tune the abundances of target gene expression in a tissue- or cell-specific manner. Tillering is controlled by multiple transcription factors and pathways, therefore fine-tuning their expression is critically important, which in turn suggests an important role for miRNAs in the process of tillering. More recently, a role for miR156 and consequently their target genes (Squamosa promoter-binding transcription factors) in the tillering of switchgrass has been revealed (Fu et al. 2012; Chuck et al. 2011). However, a role for the other miRNAs in tillering cannot be excluded and this requires a comprehensive miRNA profiling in emerging tillers, which was undertaken in this study.
Materials and methods
Small RNA library construction and sequence analysis
Switchgrass cultivar Alamo was grown to maturity under greenhouse conditions. Emerging tillers from the young plants and inflorescence from the adult plants were harvested, snap frozen in liquid nitrogen, and stored at − 80°C until total RNA was isolated. Total RNA from the frozen samples was extracted using the Trizol reagent (Invitrogen, USA) following the manufacturer’s instructions. Construction of small RNA libraries and their sequence analyses were accomplished as described earlier (Matts et al. 2010; Jagadeeswaran et al. 2012).
Small RNA blot analysis
Small RNA blot analysis was performed as described previously (Matts et al. 2010). In brief, low-molecular weight (LMW) RNA was precipitated using NaCl and PEG from the total RNA. Twenty micrograms of LMW RNA was resolved on a denaturing 15% acrylamide/8 M urea gel, transferred to a hybond-N + (Amersham) membrane, and probed with the labeled 32P oligonucleotides complementary to the miRNA sequence. The membranes were pre-hybridized for at least 1 h, and hybridized overnight with PerfectHYB + buffer (Sigma) at 38 °C and then washed, and exposed to a phosphoscreen. Images were acquired by scanning the phosphoscreen using a Typhoon scanner.
Results and discussion
Sequencing of small RNA libraries provided a glimpse of miRNAs expressed in switchgrass seedlings or leaves (Matts et al. 2010; Sun et al. 2012; Xie et al. 2014; Hivrale et al. 2016). To identify miRNAs and to determine their abundances in inflorescence and emerging tillers of switchgrass, two small RNA libraries were generated from these tissues and sequenced using Illumina sequencing platform. This yielded approximately 10 and 11 million total reads as well as 3,757,827 and 5,175,081 unique reads from emerging tillers and inflorescence libraries, respectively. The degradation products from the rRNAs, tRNAs and from the protein-coding mRNAs were removed and the remaining unique small RNAs were mapped to miRBase to identify known miRNAs.
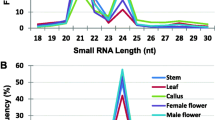
The small RNAs ranging in size between 18 and 27 nt were analyzed for their abundances, which revealed two peaks (Fig. 1); one at the 21 nt size that largely includes miRNAs and the other at 24 nt size class which mostly comprises heterochromatic siRNAs, and these findings were consistent with small RNA populations from diverse plant species (Jagadeeswaran et al. 2012; Zhang et al. 2011; Li et al. 2013). Although the total reads of 24 nt small RNAs were approximately similar in numbers both in tillers and inflorescence, but the unique read numbers differed greatly between these tissues. The count of unique small RNAs of 24 nt size class was far greater in inflorescence than in tillers (Fig. 1), which suggests a greater diversity of 24-nt long small RNAs in inflorescence but less diverse (greater redundancy) in tillers (Fig. 1). Unlike 24 nt long total reads, the 21 nt long total read count differed greatly between inflorescence and tillers, i.e., inflorescence has much greater frequencies than in tillers. A similar trend was observed for 21 nt long unique small RNA population (Fig. 1). This observation suggests that the overall miRNA abundances were much greater in inflorescence relative to emerging tillers. Consistent with this suggestion, the expression levels of the most miRNA families were several folds greater in inflorescence compared to tillers (Table 1).
In plants, most conserved miRNAs exist as families (multiple loci) with or without sequence variation. Our analysis has revealed the identification of 88 distinct conserved miRNAs belonging to 28 miRNA families (miR156, miR159, miR160, miR162, miR164, miR165/166, miR167, miR168, miR169, mIR170/171, miR172, miR319, miR390, miR393, miR394, miR395; miR396, miR397, miR398, miR399, miR408, miR444, miR528, miR529, miR535, miR827, miR2118, and miR5025) in both tissues of switchgrass. The identified distinct miRNAs and their frequencies in inflorescence and emerging tillers was shown in Table 1. On the basis of number of miRNA variants that differ at least by one or more nucleotides for each family, miR169 was found to be the largest family represented by 12 members in these tissues of switchgrass (Table 1). This was followed by miR170/171 and miR399 families with 6 members each; four miRNA families (miR156, miR165/166, miR393 and miR2118) were represented by 5 members each and another four miRNA families (miR164, miR167, miR172 and miR395) were represented by 4 members each in these tissues of switchgrass. The miRNA families such as miR160, miR319, miR393, miR397, miR398, miR444, miR529, miR1432 were only represented by 2 members each whereas miR162, miR168, miR390, miR394, miR408, miR528, miR535, miR827 and miR5025 were represented by only one member in these two tissues of switchgrass (Table 1).
The overall abundances of almost all miRNA families were differed between inflorescence and emerging tillers. In general, most miRNA families exhibited relatively much higher levels in the inflorescence than in tillers. For instance, miR165/166 family was most abundantly expressed, followed by miR168, miR156 and miR167 in the flowers (Table 1). Although these miRNA families expressed at much lower levels in emerging tillers, a similar trend was observed with respect to ranking of their abundances (Table 1). Likewise, the abundances of miR172 family that regulates flower development, were at least five-fold higher in inflorescence compared to tillers. Similarly, miR159, miR164, miR170/172, miR827 and miR396 exhibited approximately similar abundances (about 7000 RPTM) in inflorescence, while their levels were far low (ranged between half to one-fourth levels) in emerging tillers (Table 1). By contrast, miR528 levels were almost four-fold greater in emerging tillers compared to inflorescence (Table 1). Additionally, few miRNA families such as miR169, miR397, miR408 and miR529 were also displayed slightly higher abundances in tillers than in inflorescence. Only miR393 family displayed similar abundances between inflorescence and tillers. Thus, the majority of miRNA families were more abundantly expressed in inflorescence and only a small number of families were expressed at higher levels in tillers.
Within the two small RNA libraries (inflorescence and emerging tillers), miR165/miR166 family had the highest abundances. The miR165/166 family targets HD-ZIP transcription factors that are essential for specification of adaxial/abaxial (dorso-ventral) polarity. This polarity is established through the polarized expression of HD-ZIPIII transcription factors that specify adaxial/upper cell fate (Emery et al. 2003; Juarez et al. 2004; Nogueira et al. 2007). In Arabidopsis and maize, the adaxial-specific expression of HD-ZIPIII family members is delineated by the expression pattern of miR166 (Juarez et al. 2004; Kidner and Martienssen 2004; Nogueira et al. 2007). HD-ZIP factors are also implicated in regulating floral development (Jung and Park 2007). Given the fact that HD-ZIPIII family members play diverse roles in leaf and flower development, it was not surprising that their abundance was the highest in inflorescence library. However, the highest abundance in emerging tillers was noteworthy and this requires further attention to scrutinize if miR165/166-HD-ZIP circuitry plays a role in tillering.
miR444, miR528 and miR1432 are well-characterized monocot-specific miRNA families (Sunkar and Jagadeeswaran 2008). Of these, miR528 had the highest abundances in the emerging tillers and deserves further attention. The targets for miR528 include Cu/Zn SODs, which suggests that suppressing transcripts of ROS scavenging Cu/Zn SODs is important in tillers. This in turn could maintain a threshold level of ROS, which might be important for ROS signaling in tillers. Further studies are needed to examine if this is indeed true or not. miR444 family was more abundantly expressed in the inflorescence while miR1432 expressed at very low levels in both tissues (Table 1). Other partially conserved miRNA families identified in these libraries include miR827 and miR2118 that were reported from some of the monocots and dicots (Jagadeeswaran et al. 2009; Arenas-Huertero et al. 2009; Johnson et al. 2009). The miR827 was expressed at much higher levels (at least four-fold greater) in inflorescence compared to tillers.
Within each miRNA family, the abundances of different members appear to vary greatly, which could occur in a tissue- or cell-specific manner. The abundances of different members can be assessed from their normalized frequencies. Interestingly, a few variants/members of the same miRNA family were more abundantly expressed than the other variants. For instance, miR159 that was represented by 3 members in both tissues but only one of them was abundantly expressed (7907 and 3616 RPTM in inflorescence and tillers, respectively) whereas the other two members of miR159 family were barely expressed in these tissues. Similarly, some of the loci belonging to miR165/166 were expressed as abundant as 575,000 RPTM in inflorescence and 137,000 RPTM in tillers whereas the other members of this family were expressed at varying levels (moderate to low) in both tissues (Table 1). Interestingly, the abundances of specific members of miR170/171 family were greater in tillers compared to inflorescence.
Identification of novel miRNAs in inflorescence and tillers of switchgrass
Novel miRNAs have been reported from diverse plant species including switchgrass. Here we used two tissues (emerging tillers and inflorescence) that have not been used in previous studies, therefore, we surveyed the small RNA libraries for identifying novel miRNAs as suggested by Meyers et al. (2008). Our analysis revealed at least 4 small RNAs that could be annotated as novel miRNAs based on sequencing of miRNA* reads (Table 2). Fold-back structures for the novel miRNA precursors could be predicted using their precursor sequences (Fig. 2). Interestingly, the frequency of a novel miRNA#1 was substantially higher and even greater than the frequency of several highly conserved miRNAs such as miR396, miR393 and miR398. Additionally, we also assessed for their conservation using BLAST searches against the NCBI EST database. This could identify homologs of some of these novel miRNAs in other monocots such as maize, sorghum, sugarcane, rice and Cenchrus ciliaris.
Temporal expression analyses of miR529 and miR535 as well as novel miRNAs
The abundances of most conserved miRNAs greatly differed between different tissues and developmental stages of switchgrass (lower and upper leaves from seedlings and mature adult plants, stems from seedlings and adult plants as well as roots and inflorescence) as analyzed by small RNA blot analyses (Matts et al. 2010). In this study, we have sequenced two semi-conserved miR529 and miR535 families from both tissues of switchgrass (Table 1). To examine their expression patterns in the above mentioned eight different tissues, small RNA blot analyses were performed (Fig. 3). The analyses revealed that the miR529 was abundantly expressed in the inflorescence and stems but barely detectable in leaves of both seedlings and adult plants (Fig. 3). On the other hand, miR535 was expressed at moderate levels in leaves but at much lower levels in stem (Fig. 3). We also analyzed the expression pattern of three novel miRNAs in these tissues of switchgrass. Novel miRNA#1 was abundantly expressed in leaves, stem and root tissues of switchgrass (Fig. 3). Novel miRNA#2could be detected in inflorescence and stems of seedlings as well as in leaves. Similarly, the novel miRNA#4 was barely detectable in stem of adult plants as well as upper leaves of both seedlings and adult plants but it could not be detected in other tissues (Fig. 3). The small RNA blot analyses revealed that the novel miRNA#1 levels were much higher compared to the novel miRNAs #2 and #4. Indeed, the high abundances of novel miRNA#1 was consistent between both the sequencing profiles (Table 1) and small RNA blot assay (Fig. 3).
The analyses of two independent small RNA libraries sequenced from inflorescence and emerging tillers revealed the identification of 28 conserved and 4 novel miRNA families in switchgrass. Targets for most conserved miRNAs have been predicted and some of them have been confirmed (Matts et al. 2010; Hivrale et al. 2016). Identification of most conserved miRNA homologs in these tissues of switchgrass suggests the possibility that the functions described to these miRNAs from other plant species likely to be true for switchgrass also. In general, this study offered insights into which miRNA families and which variants within a miRNA family were more abundantly expressed in inflorescence and tillers. It was shown earlier that miR156 overexpression significantly improves biomass production of switchgrass (Fu et al. 2012; Chuck et al. 2011). Identification of various conserved and novel miRNAs in tillers of switchgrass forms a valuable resource to examine which of these miRNAs have roles in tillering.
References
Arenas-Huertero, C., Pérez, B., Rabanal, F., Blanco-Melo, D., De la Rosa, C., Estrada-Navarrete, G., et al. (2009). Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Molecular Biology, 70(4), 385–401.
Chuck, G. S., Tobias, C., Sun, L., Kraemer, F., Li, C., et al. (2011). Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proceedings of the National Academy of Sciences USA, 108, 17550–17555.
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD ZIP and KANADI genes. Current Biology, 13, 1768–1774.
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J.-Y., Matts, J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnology Journal, 10, 443–45210.
Hivrale, V., Zheng, Y., Puli, C. O. R., Jagadeeswaran, G., Gowdu, K., Kakani, V. G., et al. (2016). Characterization of drought-and heat-responsive microRNAs in switchgrass. Plant Science, 242, 214–223.
Jagadeeswaran, G., Nimmakayala, P., Zheng, Y., Gowdu, K., Reddy, U., & Sunkar, R. (2012). Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genomics, 13, 329.
Jagadeeswaran, G., Zheng, Y., Li, Y. F., Shukla, L. I., Matts, J., et al. (2009). Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytologist, 184, 85–98.
Johnson, C., Kasprzewska, A., Tennessen, K., Fernandes, J., Nan, G. L., Walbot, V., et al. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Research, 19(8), 1429–1440.
Jones-Rhoades, M. W., Bartel, D. P., & Bartel, B. (2006). Micro-RNAs and their regulatory roles in plants. Annual Review of Plant Biology, 57, 19–53.
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A., & Timmermans, M. C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature, 428(6978), 84–88.
Jung, J. H., & Park, C. M. (2007). MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta, 225(6), 1327–1338.
Kidner, C. A., & Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through Argonaute1. Nature, 428, 81–84.
Li, Y. F., Zheng, Y., Jagadeeswaran, G., & Sunkar, R. (2013). Characterization of small RNAs and their target genes in wheat seedlings using sequencing-based approaches. Plant Science, 203–204, 17–24.
Matts, J., Jagadeeswaran, G., Roe, B. A., & Sunkar, R. (2010). Identification of microRNAs and their targets in switchgrasss, a model biofuel plant species. Journal of Plant Physiology, 167(11), 896–904.
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell, 20, 3186–3190.
Nogueira, F. T. S., Madi, S., Chitwood, D. H., Juarez, M. T., & Timmermans, M. C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Development, 21, 750–755.
Sun, G., Stewart, C. N., Jr., Xiao, P., & Zhang, B. (2012). MicroRNA expression analysis in the cellulosic biofuel crop switchgrass (Panicum virgatum) under abiotic stress. PLoS ONE, 7, e32017.
Sunkar, R., & Jagadeeswaran, G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology, 8, 37.
Sunkar, R., Li, Y. F., & Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends in Plant Sciences, 17, 196–203.
Sunkar, R., & Zhu, J. K. (2007). Micro RNAs and short-interfering RNAs in plants. Journal of Integrative Plant Biology, 49, 817–826.
Wang, Y. X., Zeng, X., Iyer, N. J., Bryant, D. W., Mockler, T. C., & Mahalingam, R. (2012). Exploring the switchgrass transcriptome using second-generation technology. PLoS ONE, 7, e34255.
Wang, Y., Zeng, X., Peal, L., Tang, Y., Wu, Y., & Mahalingam, R. (2013). Transcriptome analysis of nodes and buds from high and low tillering switchgrass inbred lines. PLoS ONE, 8(12), e83772.
Xie, F., Stewart, C. N., Jr., Taki, F. A., He, Q., Liu, H., & Zhang, B. (2014). High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnology Journal, 12, 354–366.
Xu, K., Sun, F., Chai, G., Wang, Y., Shi, L., Liu, S., et al. (2015). De novo assembly and transcriptome analysis of two contrary tillering mutants to learn the mechanisms of tillers outgrowth in switchgrass (Panicum virgatum L.). Frontiers. Plant Science, 6, 749.
Zhang, L., Zheng, Y., Jagadeeswaran, G., Li, Y., Gowdu, K., & Sunkar, R. (2011). Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics, 98, 460–468.
Acknowledgements
This work has been supported, in part, by the NSF EPSCoR award EPS0814361 and the Oklahoma Agricultural Experiment Station, to R. Sunkar.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matts, J., Zheng, Y., Jagadeeswaran, G. et al. MicroRNA expression profiles in the emerging tillers and inflorescence of switchgrass, a major feedstock for biofuel production. Ind J Plant Physiol. 22, 558–565 (2017). https://doi.org/10.1007/s40502-017-0343-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40502-017-0343-7