Abstract
riceXIP, a XIP-Type xylanase inhibitor gene identified in rice, was cloned and expressed in Escherichia coli. Recombinant protein riceXIP was active against xylanase from Aspergillus niger, suggesting correct expression. By using transgenic techniques, we achieved the overexpression of the riceXIP gene and gene knock-down plants and elucidated that riceXIP may likely participate in plant defence against herbivores. The defence-related genes were significantly elevated in riceXIP-overexpressing transgenic plants treated with rice brown planthopper infestation. The full-length promoter (2009 bp, RP1) of riceXIP gene and 1534 (RP2), 1179 (RP3), 891 (RP4), 491 (RP5), 400 (RP6), 214 (RP7) and 112 bp (RP8) 5′ deletion constructs were fused with β-glucuronidase (GUS) gene and transgenic rice plants were used to clarify its function. The transgenic lines transformed with promoter fragments of riceXIP differentially responded to methyl jasmonate and wounding stress by quantitative GUS analysis. The 5′ deletion analysis also showed that two repressor elements exist between −1502 and −1147 bp and between −182 and −80 bp, respectively, whereas an enhancer element exists between −1147 and −859 bp. The study strengthens the possibility that riceXIP participates in the resistance against herbivory in rice and provides a helpful insight for understanding the cis-regulation of the riceXIP gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Endo-β-1,4-xylanases (EC 3.2.1.8, xylanases) are one of the important cell wall degrading enzymes secreted by plant-pathogenic microorganisms. It can degrade xylan, which is a major cell wall component of graminaceous plants. Xylanase inhibitors (XIs) are proteins that inhibit the activity of xylanases. Three classes of XIs, namely Triticum aestivum Linnaeus xylanase inhibitor (TAXI)-type (Debyser et al. 1999), xylanase inhibiting protein (XIP)-type (McLauchlan et al. 1999), and thaumatin-like xylanase inhibitor (TLXI)-type XIs (Fierens et al. 2007), have been identified in a variety of cereals. Even a chitinase-like XI has been identified in coffee, Coffea arabica Linnaeus (Vasconcelos et al. 2011). Based on the reason that XIs inhibit only xylanases of microbial origin but not plant origin and were induced following insect herbivores and fungal pathogens infestation, a function in plant defence has been proposed for these proteins. A systematic overview indicating their function in plant defence has been reported (Dornez et al. 2010).
Direct evidences of XIs involved in plant defence against herbivores or pathogens have been reported in recent years. Wheat transgenic plants expressing XIP-I or TAXI-III could limit Fusarium graminearum Schwein. infection (Moscetti et al. 2013; Tundo et al. 2015, 2016). XIP-I could inhibit a xylanase from the coffee berry borer, Hypothenemus hampei, Ferrari the most important pest of coffee plants (Padilla-Hurtado et al. 2012). To date, four XIP-type XIs, namely riceXIP (Goesaert et al. 2005), RIXI (Durand et al. 2005), OsXIP (Tokunaga and Esaka 2007) and OsHI-XIP (Xin et al. 2014), have been reported in rice crops. Transgenic rice expressing OsHI-XIP or RIXI enhanced resistance against herbivores or pathogens Magnaporthe oryzae, T.T. Hebert respectively (Xin et al. 2014; Hou et al. 2015).
XI genes were induced by different abiotic and biotic stress. The transcripts of OsXIP and riceXIP were increased by methyl jasmonate (MeJA) and wounding treatment in root (Tokunaga and Esaka 2007). OsHI-XIP expression was induced not only by jasmonic acid (JA) and mechanical wounding but by herbivory. The transcriptional expression of XIP-I was increased in wheat leaves infected with Erysiphe graminis (DC.) Speer and MeJA (Igawa et al. 2005). The expression of XI genes also showed organ-specific. For instance, the expression of RIXI was constitutive in shoot tissues and weak in stamen (Tokunaga and Esaka 2007; Hou et al. 2015). In general, a gene promoter region can provide significant information on the factors that induce expression or on tissue-specific expression. Moreover, promoter deletion analyses to verify their importance of the XI promoter regions have yet to be reported.
riceXIP is the first XIP-type XI that has been identified as a non-chitinolytic homologue of chitinases in rice plants (Goesaert et al. 2005). However, knowledge of its properties and function in plant defence is limited. In this study, we isolated the gene encoding riceXIP from rice genomic DNA and expressed it in Escherichia coli (Migula) Castellani & Chalmers BL21, and its expression levels were investigated under different treatments.
2 Materials and Methods
Plant growth conditions and treatments – Rice seeds of Oryza sativa Linnaeus cv. Nipponbare [wild-type (WT)] and transgenic Oryza sativa cv. Nipponbare expressing riceXIP or its promoter (see below) were sterilised with 1% sodium hypochlorite for 30 min and then thorough washing with sterile water. Rice plants grown by hydroponic method (Wang et al. 2009a) in a greenhouse with a light/dark cycle of 14/10 h at 28/18 °C.
Phytohormone treatment was conducted by submersion method: MeJA were dissolved in 50 μL of dimethylsulphoxide and added to the culture solution. Fourteen-day-old rice seedlings were submerged in 200 µM MeJA, and shoots and roots were harvested at different time intervals (hours). Rice seedlings grown at 28 °C using a 14-h light and 10-h dark cycle. For mechanical wounding, 14-day-old plants were cut into 1–2 cm length and then floated on sterile water for the indicated time. For rice brown planthopper (BPH) treatment, 14-day-old seedlings were infested with adult BPH confined for 1 day in a glass cylinder.
Quantitative real-time polymerase chain reaction (qRT-PCR) – Total RNA was isolated from shoot or root tissues of rice seedlings with different treatments by using a miniBEST universal RNA extraction kit (TaKaRa). cDNA was prepared according to the instruction of the PrimeScript first-strand cDNA synthesis kit (TaKaRa). qRT-PCR was conducted on a LightCycler480 instrument (Roche) using a SYBR® Premix Ex TaqTM kit (TaKaRa). Osactin gene was selected as an endogenous reference gene. The primers for qRT-PCR of all genes are presented in Table 1.
Gene construct and rice transformation – To construct the riceXIP promoter vector, we amplified the full-length riceXIP gene promoter named RP1 (−1977 to +32 bp) by PCR using the genomic DNA of rice (WT) as the template with primers containing Sal I or Kpn I restriction sites (forward: 5′-ACGCGTCGACGGGGTCGTGGATGTTCTGGT-3′; reverse: 5′-GGGGTACCGTTTGCAGCAATGTAAAGC-3′). Then, a series of 5′ deletion of RP1 fragments, RP2 (−1502 to +32 bp), RP3 (−1147 to +32 bp), RP4 (−859 to +32 bp), RP5 (−459 to +32 bp), RP6 (−368 to +32 bp), RP7 (−182 to +32 bp) and RP8 (−80 to +32 bp) were amplified by PCR from pMD19T-RP1 by using the common reverse primer of RP1 and the forward primers RP2-U (5′-ACGCGTCGACCGGCTGGAAGAACACCGCTAA-3′), RP3-U (5′-ACGCGTCGACCCCCTTCCTCTGCCACTTTT-3′), RP4-U (5′-ACGCGTCGACCGTAGAGAAATTTGAAACAG-3′), RP5-U (5′-ACGCGTCGACTTGCACCTGTTTTTCCTGGG-3′), RP6-U (5′-ACGCGTCGACTAGCTTCTCCTGTGTCCACC-3′), RP7-U (5′-ACGCGTCGACGCAGCACCGACGAAGTAGCC-3′) or RP8-U (5′-ACGCGTCGACCACATCACGCACACCACAAG-3′), respectively. The full-length promoter and 5′-deletion derivatives were cloned into the pBI101.3-GUS (Wang et al. 2009b) upstream of GUS. The empty vector pBI101.3-GUS was transformed into rice used as a control (vector control, VC).
To construct the riceXIP-OVER vector, we amplified the complete riceXIP coding sequence from pMD19T-riceXIP plasmid (see below) with a pair of specific primers that contain Kpn I or Sac I restriction sites (forward: 5′-GGGGTACCATGGGCCTCGTGCACGCACT-3′; reverse: 5′-CGAGCTCTTAACCCTCACCAGTGTAGT-3′). Then, the PCR product was inserted into the vector pTCK303, which contains the CaMV35S promoter, the reporter gene GUS and the resistance gene hygromycin.
To yield the riceXIP-RNAi vector, we amplified a 336 bp coding sequence of riceXIP by PCR using the pMD19T-riceXIP plasmid as template with forward primer containing Kpn I and Spe I restriction sites (5′-GGGGTACCACTAGTGTGCTCTTGACGGCGACGA-3′) and reverse primer containing BamH I and Sac I restriction sites (5′-CGGGATCCGAGCTCACCCTCACCAGTGTAGTTG-3′), which were digested with Sac I and Spe I followed by Kpn I and BamH I and subsequently inserted into pTCK303 (Wang et al. 2004).
All resulting constructs were transformed into WT through Agrobacterium tumefaciens Smith & Townsend-mediated transformation (Hiei et al. 1994; Mei et al. 2006). Then, the independent transgenic lines were selected using GUS staining identification and qRT-PCR assay. The T2 homozygous lines were used for further research.
GUS quantitative analysis – The measurement of quantitative GUS activity was conducted based on the method described by Jefferson et al. (1987) with certain modifications. Briefly, the tissues of 14-day-old rice seedlings were homogenised in extraction buffer (10 mM EDTA, pH 8.0, 50 mM NaPO4 buffer, pH 7.0, 20% methanol, 0.1% sodium lauryl sarcosine, 10 mM β-mercaptoethanol and 0.1% Triton X-100) and centrifuged (14,000 g, 20 min, 4 °C). Then, the supernatant (50 μL) was obtained and added to GUS assay buffer [extraction buffer containing 2 mM 4-methylumbelliferyl-β-D-glucuronide (MUG)] (450 μL) at 37 °C for 45 min. Thereafter, 200 μL of the reaction mixture was added to 800 μL of stop buffer (0.2 M Na2CO3). The 4-methylumbelliferone fluorescence was detected in a spectrofluorophotometer (RF-5301PC, Shimadzu). Protein concentration was determined by methods described by Bradford (1976).
Expression of riceXIP proteins in E. coli – To obtain the recombinant riceXIP protein, we amplified the DNA sequence by PCR by using the genomic DNA of rice (WT) as a template, with primers containing EcoR I or Xho I restriction sites (forward: 5′-CGGAATTCATGGGCCTCGTGCACGCACT-3′; reverse: 5′-CCCTCGAGTTAACCCTCACCAGTGTAGT-3′). Then, the PCR product was inserted into the vector pMD19T (TaKaRa). This vector was named pMD19T-riceXIP. And it was ultimately digested with EcoR I and Xho I and ligated into the pET30a vector (Invitrogen). This expression vector was named pET30a-riceXIP.
pET30a-riceXIP and pET30a were introduced into E. coli BL21, respectively. One millilitre of positive transformant of BL21[pET30a-riceXIP] or BL21[pET30a] was added to 100 mL of liquid Luria–Bertani (LB) with 50 μg mL−1 kanamycin. The mediums were grown until an A600nm of 0.5–1.0 at 37 °C (200 rpm.), and then induced with isopropyl-1-beta-D-galactopyranoside (IPTG) (1 mM final concentration) at 20 °C for 8 h at 200 rpm. The cultures were centrifuged (14,000 g, 15 min, 4 °C) and resuspended with the McIlvaine’s buffer (0.2 M Na2HPO4/0.1 M citric acid, pH 6.0). Then, cells were sonicated and centrifuged (14,000 g, 15 min, 4 °C). Finally, the supernatant and sediment were collected. Electrophoresis was conducted on 10% sodium dodecyl sulphate–polyacrylamide gel (SDS-PAGE) as the method described by Laemmli (1970). The concentration of protein was determined by the method of Bradford (1976).
Assay for xylanase inhibitor activity – Xylanase inhibitor activity was measured as the described by Miller et al. (1959) through comparing the decrease in Aspergillus niger van Tieghem xylanase (ANX) activity in the absence or presence of recombinant riceXIP. ANX was provided by the College of Animal Science, Zhejiang University (Sun 2003). The recombinant riceXIP (80 μg) was pre-incubated with the recombinant GH11 xylanase ANX (60 μg) for 30 min at 50 °C. The prewarmed (30 °C) 1% birchwood xylan (Sigma) (450 μL) was then added to initiate the assay for 5 min at 50 °C. It was terminated with 3, 5-dinitrosalicylic acid (1 mL) after boiling for 10 min. Reaction products in absorbance was measured at 540 nm.
Crude proteins of riceXIP-overexpressing and riceXIP-suppressed rice plants were extracted to determine the xylanase inhibitor activity of transgenic lines (Elliott et al. 2003). In brief, leaves of two-week were ground into a powder with 4 mL of McIlvaine’s buffer and centrifuged (14,000 g, 20 min, 4 °C). Then, the supernatant namely crude protein was obtained. The concentration of protein was determined by the method of Bradford (1976). Xylanase inhibitor activity was measured through comparing the decrease in ANX activity in the absence or presence of crude protein (80 μg) from WT, OX-8 and RNAi-3, respectively.
Statistical analysis – All data were expressed as mean ± standard deviation (SD) of three replicates (n = 3). Statistical analysis was performed using SPSS 16.0 (SPSS Inc., Chicago, IL, USA), and OriginPro 8.0 (OriginLab Co., Northampton, MA, USA) was used for mapping. Differences were considered significant at P < 0.05 or P < 0.01.
3 Results
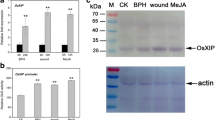
Transcript expression of riceXIP gene responding to MeJA and wounding – The transcript level of the riceXIP was studied by qRT-PCR under MeJA and wounding treatments at different time (2 h to 24 h) (Fig. 1). The transcript of riceXIP in shoot was induced with both two treatments concomitantly with time in comparison with the control. At maximum, about 14- and 160-fold transcript expressions were observed at 6 h under MeJA condition and 12 h under condition of wounding, respectively (Fig. 1a). The expression of riceXIP in root tissues increased rapidly and reached a maximum level at 2 h under both treatments (Fig. 1b). The expression level was significantly higher under MeJA treatment than that under wounding treatment, and this finding was contrary to the expression of riceXIP in shoot tissues.
Expression of riceXIP in wild-type rice seedlings after different treatments by qRT-PCR. Relative expression of riceXIP gene in shoots (a) and roots (b) of 14-day-old WT seedlings. Total RNA for expression analysis was isolated from shoots or roots of 14-day-old seedlings after 200 μM MeJA or wounding treatment for 0, 2, 6, 12 and 24 h. Experiments were conducted in triplicate, and data were averaged from three independent replicates with standard deviation. Bars with different letters are significantly different at P < 0.05 by using Duncan’s multiple range
Specificity expression of riceXIP gene – As mentioned above, the transcript level of riceXIP was markedly induced by MeJA and wounding treatments by transcriptional analysis (Fig. 1). We analysed the promoter sequence of riceXIP using PlantCARE and PLACE software. As expected, a number of biotic and abiotic stress-responsive cis-acting elements, such as TGACG-motif, MYB core, GATA box and ABRE, were identified in the promoter region. The full-length promoter (2009 bp, RP1) of riceXIP and 1534 (RP2), 1179 (RP3), 891 (RP4), 491 (RP5), 400 (RP6), 214 (RP7) and 112 bp (RP8) 5′ deletion constructs were amplified from rice genomic DNA. Then, the transgenic rice lines that carried RP1 construct or the 5′ deletion promoter::GUS construct, RP2, RP3, RP4, RP5, RP6, RP7 or RP8 were obtained and applied to investigate the expression pattern of the riceXIP gene.
Quantitative GUS activity of shoots of 14-day-old transgenic rice with promoter fragments was measured by a MUG assay (Fig. 2). The expression of GUS was detected to be about sixfold and sevenfold higher in RP1 and RP2, respectively, compared with VC, whereas an unexpected increase of GUS expression was detected in RP3. Thereafter, expression decreased in RP4 and was almost similar to those of the subsequent deletion constructs RP5, RP6 and RP7 in comparison with RP3. However, the expression of GUS of RP8 reached the maximum level, which was 66 times higher in comparison with VC.
GUS expression of the 5′ deletion promoter of riceXIP. Schematic of 5′ deletion promoter constructs of riceXIP and their β-glucuronidase (GUS) expression relative to vector control in shoot tissues. Three independent T2 transgenic lines were used for every construct, and similar results were obtained. The data were averaged from three independent replicates with standard deviation
Then, the response of the riceXIP promoter to MeJA and wounding stress was studied by measuring the expression level of GUS gene in shoot of MeJA or wounding stress-treated 14-day-old transgenic rice plants (Fig. 3). Under MeJA stress (200 μM), GUS expression in the shoots of PR1 and PR8 transgenic rice increased markedly and was approximately 3- and 1.3-fold higher in comparison with the control, respectively. Transgenic line PR6 showed increases in GUS expression, whereas the GUS expression of RP3, RP4 and RP7 was decreased compared with the control. However, RP2 and RP5 only showed minimal changes in expression level (Fig. 3a). Similar to MeJA stress, GUS expression increased in the shoots of PR1, RP2, RP4, RP5, RP6 and PR8 transgenic lines after wounding stress, except in RP3 and PR7, which showed decreases in expression compared with the control (Fig. 3b).
GUS expression of 5′ deletion promoter constructs treated by MeJA or wounding in T2 transgenic plants. Relative (to VC) GUS activity in shoots of different transgenic rice plant (RP1–RP8) after 200 μM MeJA (a) and wounding treatment (b) for 12 h, respectively. Three independent 14-day-old T2 transgenic lines were used for every construct, and similar results were obtained. Experiments were conducted for three times, and data were averaged from three independent replicates with standard deviation. Asterisks indicate statistically significant differences in comparison with CK (before MeJA or wounding treatment) (** P < 0.01; Student’s t test)
Expression of recombinant riceXIP protein in E. coli – We cloned the riceXIP gene from rice genomic DNA because the gene contains no introns. The coding sequence of riceXIP was connected to an expression vector pET30a. This expression vector contains 71 amino acids in the N-terminus, which can be fused with the expressed protein. The expression of protein in E. coli BL21 was induced by IPTG after 8 h. SDS-PAGE showed that after IPTG induction, the crude proteins from pET30a-riceXIP strain displayed an obvious thick band with a relative molecular mass of between 35 and 40 kDa compared with the control (Fig. 4a, lane 2), in line with the predicted molecular mass, including the additional 71 amino acids in the N-terminal of the pET30a vector.
SDS-PAGE and xylanase inhibitor activity of the recombinant riceXIP expressed in Escherichia coli BL21. a SDS-PAGE of the protein fraction obtained from strains of E. coli BL21. The protein was stained with Coomassie brilliant blue. M, marker of proteins; Lane 1, crude protein extracts from control strain of E. coli containing pET30a vector induced by IPTG after 8 h; Lane 2, crude protein extracts from E. coli containing pET30a–riceXIP vector induced by IPTG after 8 h; Lane 3, supernatant of E. coli containing pET30a–riceXIP vector induced by IPTG after 8 h; lane 4, sediment of E. coli containing pET30a–riceXIP vector induced by IPTG after 8 h. The sizes of the markers are indicated at the left of each gel. b Xylanase inhibitor activity of recombinant inhibitor protein riceXIP. Inhibition of riceXIP to xylanases, using birchwood (1,4)-β-xylan as substrate and a GH11 xylanase from Aspergillus niger (ANX) as the target enzyme. The residual xylanase activity is shown. ‘riceXIP+’ and ‘riceXIP−’ represent the presence and absence of riceXIP, respectively. Data were averaged from three independent replicates with standard deviation. Asterisks indicate statistically significant differences between ‘riceXIP+’ and ‘riceXIP−’ (** P < 0.01; Student’s t test)
To detect whether the supernatant contained recombinant riceXIP after sonication, we measured xylanase inhibition activity using xylanase from A. niger (ANX) as the target enzyme. When incubating with the supernatant, the xylanase ANX activity remained at 25.97% (Fig. 4b), suggesting that ANX was markedly inhibited by riceXIP. In addition, this finding suggested that the supernatant contained recombinant riceXIP and that its expression was correct as wild-type.
Generation and characterisation of riceXIP-overexpressing and riceXIP-suppressed transgenic rice plants – The riceXIP-overexpressing and riceXIP-suppressed transgenic plants were obtained to elucidate its physiological function. A minimum of four independent lines of riceXIP overexpression showed that the transcript of riceXIP was 50 times higher than that in WT (Fig. 5a). Similarly, the obtained riceXIP-suppressed lines were analysed. We confirmed that riceXIP mRNA of more than four lines was actually decreased in comparison with the WT plant (Fig. 5b). Two independent T2 generation lines (OX-8 and RNAi-3) of riceXIP-overexpressing and riceXIP-suppressed transgenic rice plants were selected for further analysis.
Verification, xylanase inhibitor activity and expression patterns of defence-related genes of riceXIP transgenic rice. Generation and verification of riceXIP-overexpressing (a) and riceXIP-suppressed (b) lines. Relative quantity of riceXIP mRNA in leaves of wild-type (WT) and T2 transgenic lines. Asterisks indicate statistically significant differences between WT and transgenic lines (** P < 0.01; Student’s t test). Relative expression of defence-related genes in shoots of riceXIP-overexpressing and riceXIP-suppressed plants before brown planthopper treatment (c) and at 1-day post-infestation by BPH (d). The expression level of each gene was calculated relative to that in non-infested WT plants. Experiments were conducted for three times, and the data were averaged from three independent replicates with standard deviation. Bars with the different letters are significantly different at P < 0.05 by using Duncan’s multiple range. e Xylanase inhibitor activity in transgenic rice, with ANX as the target enzyme. The residual xylanase activity is shown. ‘riceXIP+’ and ‘riceXIP−’ represent the presence and absence of crude protein from WT, OX-8 and RNAi-3, respectively. Asterisks indicate statistically significant differences between ‘riceXIP+’ and ‘riceXIP−’ (** P < 0.01; Student’s t test)
Function of riceXIP in the resistance against herbivory – To further explore the physiological function of riceXIP, we examined two PR genes and three genes that are known to function in defence signalling pathways in a riceXIP-overexpressing (OX-8) line and a riceXIP-suppressed (RNAi-3) line. Before BPH infestation, the overexpression and suppression of the riceXIP slightly affected the gene expression of the defence-related genes in rice plants (Fig. 5c). The transcript levels of PR protein1 (OsPR1), WRKY transcription factor 24 (OsWRKY24) and allene oxide synthase 2 (OsAOS2) were down-regulated, while Arabidopsis NPR1 homologue 1 (OsNH1) was up-regulated. The transcript level of OsSci2 (a PR protein) was increased in riceXIP-overexpressing plants and decreased riceXIP-suppressed plants. Three of the five genes showed differential expression upon BPH stress in WT rice plants. BPH infestation significantly induced the expression of OsPR1 and OsNH1, and suppressed the expression of OsAOS2 in WT (Fig. 5d). The expression of OsSci2 and OsWRKY24 was not affected by BPH infestation in WT. After infestation, except OsNH1, the transcriptional levels of other genes were notably up-regulated in riceXIP-overexpressing plants in comparison with the WT (Fig. 5d). Among these genes, OsPR1 expression reached a maximum and was induced by about 28 times. The expression levels of OsAOS2 and OsWRKY24 were induced approximately 10- and 18-fold, respectively. Meanwhile, the expression of OsPR1, OsSci2, OsNH1 and OsWRKY24 was induced, while OsAOS2 exhibited no changes in expression levels in riceXIP-suppressed plants (Fig. 5d). These results suggested that overexpression of riceXIP enhanced the expression levels of defence genes upon BPH stress, which may imply higher resistance against herbivores.
Measurement of xylanase inhibitors activity in rice plants – To investigate whether the overexpression of riceXIP enhanced the xylanase inhibitor activity in rice, the relative activity of XIs in transgenic rice and WT was measured. The riceXIP-overexpressing lines showed a significantly higher level of relative activity of total XIs in comparison with WT and riceXIP-suppressed lines. Under normal conditions, xylanase activities of ANX incubated with crude protein from WT, OX-8 and RNAi-3 remained at 42.94, 27.54 and 52.33%, respectively (Fig. 5e), indicating that the overexpression of riceXIP significantly inhibited ANX. The results were consistent with those of recombinant riceXIP mentioned above, indicating that the overexpression of riceXIP heightened the relative xylanase inhibitor activity in rice. This enhanced activity probably subsequently increased the resistance of rice to herbivores.
4 Discussion
It has been reported that the transcript levels of riceXIP and OsXIP genes were slightly enhanced in shoots and drastically in roots after MeJA and wounding treatments (Tokunaga and Esaka 2007). In our experiment, MeJA and wounding treatments induced a higher accumulation of riceXIP transcripts in shoots and root tissues of WT, and the riceXIP promoter also responded to MeJA and wounding stress (Fig. 3). These results were similar to a report on OsHI-XIP gene (Xin et al. 2014). These findings indicated that riceXIP, OsXIP and OsHI-XIP are genes that respond to stress and possibly perform similar biological functions.
Through qRT-PCR experiment, we also found that the mRNA levels of riceXIP under MeJA stress are strongly induced in root tissues and mildly induced in shoot tissues. These results are antithetical to those under wounding stress. These results suggest that rice plants perceive MeJA stress prior to wounding stress, thus reinforcing the speculation that the elevated expression of riceXIP upon wounding stress may generate by way of a JA-mediated signalling pathway (Tokunaga and Esaka 2007). The plant hormone JA performs key functions in defence signalling network (Pieterse et al. 2012). In addition, necrotrophic pathogens and herbivorous insects are generally more sensitive to JA-induced defences (Glazebrook 2005; Howe and Jander 2008).
Then, riceXIP-overexpressing and riceXIP-suppressed rice plants were obtained using a reverse-genetics approach (Fig. 5a, b). The transcriptional expression of defence-related genes was consistently enhanced by the overexpression of riceXIP upon BPH infestation (Fig. 5d). Among these genes, OsPR1 and OsSci2 are PR protein genes, that is, markers for plant defence responses, and XIP-type XIs which is homologous with chitinases belong to class PR-8 proteins. OsNH1 is SA-dependent; OsAOS2 is involved in JA synthesis (Qiu et al. 2007); OsWRKY24 is a herbivore resistance-related gene. In our study, the transcript levels of OsPR1, OsAOS2, OsWRKY24 were significantly higher in riceXIP-overexpressing transgenic line than that in WT and riceXIP-suppressed line upon BPH infestation. Furthermore, compared to the WT and riceXIP-suppressed lines, the riceXIP-overexpressing lines had a higher level of xylanase inhibitor activity (Fig. 5e). These findings suggest that the xylanase inhibitor riceXIP may function as a member of defensive proteins to take part in resistance to herbivory for adaptation to the surrounding environment.
We can obtain pivotal information on the transcription factors that regulate gene expression by analysing the promoter of a gene. A number of cis-acting elements exist in the promoter region of riceXIP, including Myb-binding sites, W-box sequences, GCC-box and TGACG-motif sequences, which are related to wound- and pathogen-inducible gene expression. W or W-like box cis-acting elements, the DNA-binding sites of WRKY transcription factors (Eulgem et al. 2000), exist in the numerous defence-related genes promoter regions, such as the gene PR-1 in maize (Raventos et al. 1995). MYB transcription factors that respond to salicylic acid specifically bind to Myb-binding sites and thereby participate in the transcriptional activation of PR genes. The TGACG motif (TGACG) is involved in MeJA responsiveness. The GCC-box (GCCGCC) is known to the binding site of AP2/ERF transcription factors, which is required for transcriptional activation by MeJA (Rushton and Somssich 1998; Van der Does et al. 2013). Furthermore, wounding that caused by insect feeding or mechanical injury induces biosynthesis of JA, which in turn activate the JA defence pathway. Our result showed the promoter activities of RP1 and RP8 were significantly increased in MeJA and wounding treatment, compared to CK (Fig. 3). The data suggested some specific cis-elements related to responding to MeJA and wounding treatment, exist in −1977 to −1502 bp and −182 to −80 bp. We analysed the corresponding sequence and found that three TGACG-motifs and one GCC-box exist in −1977 to −1502 bp, and one GCC-box exists in −182 to −80 bp.
In this study, through the analysis of different 5′-deleted mutants of the promoter, in contrast with RP2, GUS activity of RP3 abruptly increased, decreased subsequently in RP4 and increased to a maximum in RP8 in the shoots (Fig. 2). Thus, at the minimum, a repressor (r1), an enhancer (e) and a repressor (r2) motifs were predicted between RP2 and RP3 (−1502 to −1147 bp), RP3 and RP4 (−1147 to −859 bp) and RP7 and RP8 (−182 to −80 bp), respectively. Therefore, we proposed a model for the cis-regulation of the riceXIP gene. The upstream promoter regions presumably contain a minimum of two repressor sites (r1, r2) positioned at −1502 to −1147 bp and −182 to −80 bp, respectively, and an enhancer site (e) that existed between −1147 and −859 bp. A low level of GUS expression was obtained in RP2 because the repressor (R1) was bound to the r1 site, thus preventing the binding of the enhancer (E) to the enhancer site (e). Deletion of r1 resulted in the up-regulation of GUS expression in RP3 because of the binding of E to the e site. Then that the repressor (R2) was bound to r2 site and the e site was deleted resulted in a decrease in GUS expression in RP4, RP5, RP6 and RP7. Owing to the deletion of the r2 site, RP8 showed a maximum level of GUS expression. We consulted literatures and found core sequence of ATAA and CCTCAA function as enhancer and repressor binding sites, respectively (Tiwari et al. 2014). Analysis of the promoter, three ATAA sequences exist in −1147 and −859 bp; one CCTC sequence exists in −1502 to −1147 bp, and one CCTCA sequence exists in −182 to −80 bp. These speculations need to be validated by construction of a serial deletion promoter and using electrophoretic mobility shift assay.
References
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Debyser W, Peumans WJ, Van Damme EJM, Delcour JA (1999) Triticum aestivum xylanase inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance. J Cereal Sci 30:39–43
Dornez E, Croes E, Gebruers K, De Coninck B, Cammue BPA, Delcour JA, Courtin CM (2010) Accumulated evidence substantiates a role for three classes of wheat xylanase inhibitors in plant defense. Crit Rev Plant Sci 29:244–264
Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, Juge N (2005) Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J 272:3227
Elliott GO, McLauchlan WR, Williamson G, Kroon PA (2003) A wheat xylanase inhibitor protein (XIP-I) accumulates in the grain and has homologues in other cereals. J Cereal Sci 37:187–194
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Fierens E, Rombouts S, Gebruers K, Goesaert H, Brijs K, Beaugrand J, Volckaert G, Van Campenhout S, Proost P, Courtin CM, Delcour JA (2007) TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem J 403:583–591
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Goesaert H, Gebruers K, Courtin CM, Delcour JA (2005) Purification and characterization of a XIP-type endoxylanase inhibitor from rice (Oryza sativa). J Enzym Inhib Med Ch 20:95–101
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hou CX, Lv T, Zhan YH, Peng YY, Huang YY, Jiang DA, Weng XY (2015) Overexpression of the RIXI xylanase inhibitor improves disease resistance to the fungal pathogen, Magnaporthe oryzae, in rice. Plant Cell, Tissue Organ Cult 120:167–177
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66
Igawa T, Tokai T, Kudo T, Yamaguchi I, Kimura M (2005) A wheat xylanase inhibitor gene, Xip-I, but not Taxi-I, is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci Biotechnol Biochem 69:1058–1063
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
McLauchlan WR, Garcia-Conesa MT, Williamson G, Roza M, Ravestein P, Maat J (1999) A novel class of protein from wheat which inhibits xylanases. Biochem J 338:441–446
Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19:1127–1137
Miller GL, Blum R, Glennom WE, Burton AL (1959) Measurement of methods for assay of xylanase activity. Anal Biochem 2:127–132
Moscetti I, Tundo S, Janni M, Sella L, Gazzetti K, Tauzin A, Giardina T, Masci S, Favaron F, D’Ovidio R (2013) Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in durum wheat transgenic plants. Mol Plant Microbe Interact 26:1464–1472
Padilla-Hurtado B, Florez-Ramos C, Aguilera-Galvez C, Medina-Olaya J, Ramirez-Sanjuan A, Rubio-Gomez J, Acuna-Zornosa R (2012) Cloning and expression of an endo-1,4-beta-xylanase from the coffee berry borer, Hypothenemus hampei. BMC Res Notes 5:23
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Qiu DY, Xiao J, Ding XH, Xiong M, Cai M, Cao CL, Li XH, Xu CG, Wang SP (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20:492–499
Raventos D, Jensen AB, Rask MB, Casacuberta JM, Mundy J, Sansegundo B (1995) A 20-bp cis-acting element is both necessary and sufficient to mediate elicitor response of a maize prms gene. Plant J 7:147–155
Rushton PJ, Somssich IE (1998) Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol 1:311–315
Sun JY (2003) Construction and expression of hybrid gene encoding thermostable xylanase and property of hybrid enzyme. PhD Dissertation, Zhejiang University, China (English abstract)
Tiwari V, Chaturvedi AK, Mishra A, Jha B (2014) The transcriptional regulatory mechanism of the peroxisomal ascorbate peroxidase (pAPX) gene cloned from an extreme halophyte, Salicornia brachiata. Plant Cell Physiol 55:201–217
Tokunaga T, Esaka M (2007) Induction of a novel XIP-type xylanase inhibitor by external ascorbic acid treatment and differential expression of XIP-family genes in rice. Plant Cell Physiol 48:700–714
Tundo S, Moscettia I, Faoro F, Lafond M, Giardinac T, Favaron F, Sella L, D’Ovidio R (2015) Fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci 240:161–169
Tundo S, Kalunke R, Janni M, Volpi C, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2016) Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol Plant Microbe Interact 29:629–639
Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Korbes AP, Memelink J, Ritsema T, Van Wees SCM, Pieterse CMJ (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25:744–761
Vasconcelos EAR, Santana CG, Godoy CV, Seixas CDS, Silva MS, Moreira LRS, Neto OBO, Price D, Fitches E, Filho EXF, Mehta A, Gatehouse JA, Grossi-De-Sa MF (2011) A new chitinase-like xylanase inhibitor protein (XIP) from coffee (Coffea arabica) affects Soybean Asian rust (Phakopsora pachyrhizi) spore germination. BMC Biotechnol 11:14
Wang M, Chen C, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22:409–417
Wang C, Ying S, Huang H, Li K, Wu P, Shou H (2009a) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57:895–904
Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P (2009b) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2:823–831
Xin ZJ, Wang Q, Yu ZN, Hu LC, Li JC, Xiang CY, Wang BH, Lou YG (2014) Overexpression of a xylanase inhibitor gene, OsHI-XIP, enhances resistance in rice to herbivores. Plant Mol Biol Rep 32:465–475
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Grant Nos. 30971702, 31271632 and 31672462) and by research grants from the Science and Technology Department of Zhejiang Province, China (2016C32086).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhan, Y., Sun, R., Sun, X. et al. Expression regulation of a xylanase inhibitor gene riceXIP in rice (Oryza sativa L.). Braz. J. Bot 40, 983–991 (2017). https://doi.org/10.1007/s40415-017-0400-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-017-0400-5