Abstract
Background
Next-generation sequencing is widely used for comprehensive molecular profiling for many cancers including lung cancer. However, the complex workflows and long turnaround times limit its access and utility. ChromaCode’s High Definition PCR Non-Small Cell Lung Cancer Panel (HDPCR™ NSCLC Panel) is a low-cost, rapid turnaround, digital polymerase chain reaction assay that is designed to detect variants in nine NSCLC genes listed in National Comprehensive Cancer Network guidelines.
Methods
This assay uses TaqMan® probe limiting chemistry and proprietary analysis software to enable multi-target detection within a single-color channel. We compared the performance of the HDPCR™ NSCLC Panel against an in-house, laboratory-developed, targeted next-generation sequencing panel used in the Molecular Diagnostics Laboratory at the University of Minnesota Medical Center to detect biomarkers for NSCLC.
Results
The overall accuracy of the HDPCR panel was 99.48% (95% confidence interval 99.01–99.76) with a sensitivity of 95.35% (95% confidence interval 88.52–98.72) and a specificity of 99.69% (95% confidence interval 99.29–99.90). The HDPCR wet lab workflow was 4 h, and the time to generate variant calls from raw data using the ChromaCode Cloud was 2 minutes.
Conclusions
We demonstrated that the HDPCR™ NSCLC Panel provides timely, comprehensive, and sensitive mutation detection in NSCLC samples with results in less than 24 h.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
We have shown that the High Definition PCR Non-Small Cell Lung Cancer (HDPCR™ NSCLC) Panel provides timely, comprehensive, and sensitive mutation detection in non-small cell lung cancer samples with results in less than 24 h. |
Not accounting for nucleic acid extraction, the HDPCR NSCLC panel yields results in a single day compared with a minimum of 3 days required for the next-generation sequencing workflows. |
While the HDPCR NSCLC Panel has its limitations with comprehensive coverage of all possible mutations in NSCLC compared with next-generation sequencing, it stands out for its cost effectiveness, fast turnaround times, and user friendliness, making it a promising alternative in various clinical settings. |
1 Introduction
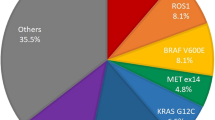
Lung cancer remains a global leading cause of cancer-related deaths, with approximately 1.8 million deaths worldwide each year [1]. The most frequent histopathologic subtype is non-small cell lung cancer (NSCLC), which accounts for 80–85% of new lung cancer cases [2]. Molecular profiling of NSCLC has identified several targets that have therapeutic relevance, notably KRAS and EGFR [3]. KRAS is the most commonly altered gene (25–30% of NSCLC cases), with 13% of lung adenocarcinomas harboring the KRASG12C mutation, which are targeted by the recently US Food and Drug Administration-approved drugs sotorasib and adagrasib [4, 5]. Activating EGFR alterations are heterogeneous, occurring in 10–15% of Caucasian patients and up to 50% in East-Asian patients, as well as in advanced-stage NSCLC [6]. Current and emerging EGFR tyrosine kinase inhibitors are being developed to target the most common EGFR alterations and their resistance mechanisms [7]. However, the presence of other targetable genomic alterations in a smaller percentage of patients necessitates comprehensive molecular profiling for a large number of targets to improve prognostic and predictive accuracy among patients with NSCLC. These rare targetable alterations in NSCLC include BRAFV600E (1.37%) [3], MET exon 14 skipping events (2.6%) [8], ALK rearrangements (5–6%) [9], and oncogenic fusions in ROS1 and RET (1–2%) [10]. This has made next-generation sequencing (NGS) the platform of choice for comprehensive detection of various single base substitutions, insertion/deletions, and fusions that guide targeted therapies for NSCLC [11, 12]. However, the large capital investment, complex technical workflows, and relatively long turnaround times for NGS-based assays limit its access and utility.
Emerging polymerase chain reaction (PCR) methods, such as digital PCR (dPCR), have enhanced sensitivity and have been increasingly exploited in molecular research and diagnostics [13]. This technology is based on compartmentalizing single nucleic acid molecules into numerous identical partitions or compartments, followed by PCR amplification. Each compartment harbors a micro-PCR reaction, containing at least one or none of the amplified target molecules (DNA or RNA), which are then precisely quantified based on the presence or absence of reaction fluorescence [13, 14]. An example of a clinical application of a dPCR assay is the validation of dPCR to detect the common EML4-ALK gene rearrangement in lung adenocarcinoma samples [15]. One significant drawback of dPCR assays is that they are not easily multiplexed with other assays. For instance, most multiplex dPCR assays have only been validated to target a few genes, such as EGFR and KRAS in both formalin-fixed paraffin-embedded (FFPE) tumor specimens and circulating tumor DNA liquid biopsy samples [16,17,18,19,20,21,22,23,24]. This limited multiplexing capability limits the utility of traditional dPCR assays for comprehensive mutation detection in NSCLC.
ChromaCode’s High Definition PCR (HDPCR™) enables multiplexing within fluorescence color channels on dPCR instrumentation, yielding a 10× target density over traditional dPCR. HDPCR allows for the detection of up to 50 targets in a single dPCR reaction well with a relatively low sample input and familiar PCR workflow. The HDPCR panel uses TaqMan® probe-limiting chemistry and proprietary analysis software to enable multi-target detection within a single color channel on the QIAcuity dPCR system (Qiagen, Hilden, Germany). HDPCR allows targets to have unique endpoint fluorescence intensity levels, resulting in distinct partition populations within the same color channel. The fluorescent signal is deconvoluted through digital signal processing in the ChromaCode Cloud™ analysis software for result interpretation. This multiplexing capability has been demonstrated by Rajagopal et al. [25] using existing reverse transcription-PCR instruments with traditional chemistries. The ChromaCode assay targets somatic cancer variants associated with NSCLC listed in the current National Comprehensive Cancer Network guidelines [26], including variants in EGFR, BRAF, KRAS, ERBB2, ALK, ROS1, RET, MET, and NTRK (Table 1). We evaluated the performance of a research use-only qualitative HDPCR NSCLC assay developed by ChromaCode and compared its performance against a targeted NGS assay.
2 Materials and Methods
2.1 HDPCR NSCLC Assay (ChromaCode, Carlsbad, CA, USA)
ChromaCode’s HDPCR NSCLC assay is designed for multiplexed detection of ten DNA and five RNA alterations split across three subassay wells, including single nucleotide variants, indels, and fusion variants in nine NSCLC genes according to National Comprehensive Cancer Network guidelines, EGFR, BRAF, KRAS, ERBB2, ALK, ROS1, RET, MET, and NTRK on the QIAcuity, 5-plex digital PCR system (Table 1). This assay requires nucleic acid samples to be diluted to a working concentration of 1 ng/μL for a total sample input of 20 ng per reaction (40 ng of DNA and 20 ng of RNA), though a previous study illustrated limit of detection down to 15 ng of DNA and 5 ng of RNA [27]. The assay comprises three separate reaction mixes: a single RNA detection well and two DNA detection wells. Each reaction mix includes calibrator material, which defines the different fluorescence intensity levels for proper target identification. These built-in calibrators, along with genomic DNA primers and probes, allow well-specific quality-control. A built-in coding technology redundantly encodes select targets across multiple channels, allowing for higher multiplexing capabilities. This coding algorithm enables multiple targets to be detected within each channel by replicating some targets across color channels at different signal intensity levels, as well as maximizing the distance between targets in single and multiple presences to ensure proper identification. The data analysis and interpretation of results are facilitated using ChromaCode’s automated software, ChromaCode Cloud. ChromaCode Cloud is a web-based software platform that enables the deployment of HDPCR assays without the need for on-premise installation. It provides full automation of the processing, interpretation, and reporting of assay data. The algorithm determines the positive call by analyzing the combination of each detected partition population across all color channels compared to the internal calibrator and displays the results on ChromaCode Cloud. The ChromaCode Cloud software helps streamline the lab workflow and data processing to accelerate turnaround times for results. Of note, a local data analysis solution is currently unavailable. However, the cloud-based solution is fully compliant with Health Insurance Portability and Accountability Act regulations and facilitates data analysis without requiring additional IT approvals. Data can be stored in the HIPAA-compliant cloud, or if necessary, it can be locally stored and subsequently removed from the cloud following analysis.
2.2 NGS Assays (Comparator Assays)
Genomic nucleic acid was extracted using the Qiagen (Qiagen, Hilden, Germany) QIAamp® FFPE Tissue Kit (Cat. 56404) or the Promega (Promega, Madison, WI, USA) Maxwell® RSC DNA FFPE kit (Cat. AS1450). DNA and RNA quantification was performed by the Qubit™ 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) using the following kits: Qubit™ 1X dsDNA High Sensitivity Kit (Cat. Q33231) and Qubit™ RNA High Sensitivity Kit (Cat. Q10210), respectively.
The DNA-based genomic alterations were detected using two clinically validated, laboratory-developed NGS tests by the Molecular Diagnostics Laboratory at the University of Minnesota. Before December 2022, an amplicon-based NGS assay was used for molecular profiling of NSCLCs. Since December 2022, a hybrid capture-based NGS assay has been employed for genomic testing of all NSCLCs.
Amplicon-based NGS libraries were prepared using an amplicon-based target enrichment method on either the Fluidigm Biomark™ (Standard BioTools, South San Francisco, CA, USA) Access Array System (integrated microfluidics circuit PCR, “IFC”) or on a parallel low-input PCR method (off-chip, “OC”) for a subset of genes. The enriched DNA libraries were then sequenced on an Illumina (Illumina, San Diego, CA, USA) MiSeq system. Amplicons with less than 500× minimum coverage were flagged for limited analytic performance. Variant call files were filtered to remove calls with variant allele fractions (VAF) less than thresholds defined for single nucleotide variants (5–10%) and insertion/deletion variants (1–5%). The estimated analytic sensitivity from validation data was 98.6% (95% confidence interval [CI] 0.9511–0.9962), and the specificity was 99.1% (95% CI 0.9667–0.9974). For more information on the amplicon-based assay methodology, please refer to the detailed description provided by Henzler et al. [28].
Hybrid capture-based NGS libraries were prepared using a custom-designed hybrid capture-based assay (Integrated DNA Technologies, Coralville, IA, USA). The enriched DNA libraries were then sequenced on an Illumina MiSeq or NextSeq 550 instrument. The specificity threshold was empirically set at 5% VAF but the bioinformatic pipeline variant call file output includes all variants with five or more sequencing reads with the variant (1% VAF). The estimated analytic sensitivity from validation data was 99.4% (95% CI 0.968–0.999), and specificity was 99.9% (95% CI 0.996–0.999).
In both methodologies, FASTQ files were processed through a custom-developed bioinformatics pipeline to call sequence variants (single nucleotide variants and insertion-deletion variants). Variant call files were annotated with GenomOncology (GenomOncology, Cleveland, OH, USA) software and reviewed for data quality and clinical utility by genomic analysts and board-certified molecular pathologists.
Similarly, two approaches were employed for detecting RNA-based clinically significant gene fusions and rearrangements. Before 2021, an amplicon-based NGS assay was utilized to identify targeted, recurring, clinically relevant gene fusions with or without fusion partners. Total nucleic acids (TNA) were isolated from the sample, and sequencing libraries were prepared using a laboratory-validated amplicon-based protocol. The extracted TNA were converted to cDNA via initial reverse transcription-PCR, followed by gene-specific PCR for relevant regions of ALK, ROS1, RET, MET, NTRK1, and NTRK3. This was followed by tagging PCR, purification, quantification, and loading on an Illumina MiSeq instrument for sequencing. FASTQ files were processed through a laboratory-validated bioinformatics pipeline provided by Asuragen (Austin, TX, USA). Copy number determination was performed for quality control following reverse transcription-PCR. During the Clinical Laboratory Improvement Amendments validation, the assay’s analytical sensitivity and specificity were determined to be 95.9% (95% CI 0.80–0.99) and 100% (95% CI 0.87–1.0), respectively.
Starting in 2021, the Archer® (ArcherDx, Boulder, CO, USA) FusionPlex® Pan Solid Tumor Panel was implemented in-house as a fusion assay. This is an RNA-based NGS test that has been validated to detect clinically significant gene fusions and oncogenic isoforms across 109 genes targeted by this assay. The assay uses anchored multiplex PCR chemistry to enable the detection of both known and unknown fusion partners. Anchored multiplex PCR was utilized to prepare the library, which is then sequenced on an Illumina instrument. Archer Analysis software is used to process the generated FASTQ files and produce results. During Clinical Laboratory Improvement Amendments validation, the assay exhibited analytical sensitivity and specificity of 100% (95% CI 0.921–1 and 95% CI 0.998–1, respectively). All reference DNA and RNA NGS assays offered comprehensive coverage of all genes and hotspots targeted by ChromaCode’s HDPCR assay.
2.3 Sample Collection
Samples were selected using an internal LIS software search tool using the gene symbol and disease name “lung cancer”. Negative control samples were searched in a similar manner using the disease name and the result “negative”. Total nucleic acids and/or DNA samples extracted from FFPE/cytology/frozen specimens were used in this study. Total nucleic acid samples were stored at −80 °C and DNA samples were stored at 4 °C till analysis. The original FFPE specimens had a minimum tumor cellularity of 10%. A total of 126 sample aliquots were retrieved. Of note, all but seven samples were obtained from lung cancer specimens owing to the rarity of variants available for our validation. These samples included samples from (a) colon cancer; METe14 skipping, (b) papillary thyroid cancer; TPM3-NTRK1, (c) papillary thyroid cancer; NTRK3-ETV6, (d) tumor of unknown origin; EGFR D770delinsGY, (e) papillary thyroid cancer; ANKRD26-RET, (f) papillary thyroid cancer; EML4-RET, and (g) CAP survey 2023A; EGFR S768I.
2.4 Validation Study Design
We evaluated the performance of the HDPCR NSCLC panel against an in-house, laboratory-developed, targeted NGS panel used in the Molecular Diagnostics Laboratory at the University of Minnesota Medical Center to detect biomarkers for NSCLC. The validation study included 126 samples from stored post-extraction aliquots. Six samples did not produce valid results because of insufficient sample material. The samples came from FFPE, frozen, and cytology specimens. The remaining 120 samples consisted of 89 positive samples containing at least one DNA/RNA alteration and 31 negative samples (from specimens without clinically relevant mutations/fusions in the listed genes).
DNA and RNA samples were analyzed using the HDPCR assay per the manufacturer’s instructions. The assay comprises two wells for all DNA targets and one well for all RNA targets. Samples were run on the Qiagen QIAcuity® dPCR instrument, and results were generated with ChromaCode Cloud™, a proprietary cloud-based technology platform and reporting software. Samples that yield discordant results for RNA targets were resolved using the Archer® FusionPlex® Pan Solid Tumor Panel.
2.5 Statistics
The sensitivity of the HDPCR assay was calculated using the formula: sensitivity = (true positive)/(true positive + false negative). True positive (TP) refers to samples that tested positive for a genomic alteration by both the reference NGS method and the HDPCR method. False negative (FN) refers to samples that tested positive for a genomic alteration by the reference NGS method but were negative for the alteration by the HDPCR method. The specificity of the HDPCR assay was calculated using the formula: specificity = (true negative)/(true Negative + false positive). True negative (TN) refers to samples that tested negative for a genomic alteration using both the reference NGS and HDPCR methods. False positive (FP) refers to samples that tested positive using the HDPCR method but tested negative using the reference NGS method. The accuracy of the HDPCR assay was calculated using the formula: accuracy = (TP + TN) (TP + TN + FP + FN). The 95% CIs for the sensitivity, specificity, and accuracy were determined using the Clopper-Pearson Exact Method with the MedCalc Software Ltd. diagnostic test evaluation calculator:
(https://www.medcalc.org/calc/diagnostic_test.php, Version 22.026).
2.6 Ethics Approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the University of Minnesota (protocol code STUDY00017413 on 31 October, 2022).
3 Results
Out of 126 retrieved samples, only 120 underwent analysis, as six samples had insufficient amounts of TNA/RNA. There were 86 distinct DNA/RNA alterations out of a total of 1725 alterations that were evaluated using the ChromaCode HDPCR NSCLC Panel. Four samples had two concurrent alterations. Table 2 summarizes the number of positive and negative target calls and the preliminary performance data. Before discordant resolution and assay optimization, the HDPCR panel had a sensitivity of 88.17% (95% CI 79.82–93.95) and a specificity of 99.69% (95% CI 99.29–99.90) with an overall accuracy of 90.07% (95% CI 98.50–99.47) (Table 2).
3.1 Discordant Resolution
Sixteen discordant results were further investigated by re-evaluating the data from the comparator and HDPCR assays or retesting the sample with a comparator assay when possible (Table 3). Six discordant results occurred because the HDPCR assay was not inclusive to specific genomic regions where alterations were detected by the comparator assay (Table 3). In one case (sample ID: 049), a RET fusion was detected through the former amplicon-based fusion assay but could not be confirmed by the newer Archer assay (both assays are detailed in the methods section). Therefore, the negative HDPCR result for this particular sample was deemed to be a true-negative result (Table 3). Among the remaining nine discrepancies, two samples were insufficient to evaluate the cause of the discrepancy between the two assays. One of these two samples that could not be further evaluated had low levels of ALK (sample ID:076) in the HDPCR assay that could not be confirmed by the original fusion assay. Specifically, the ALK alteration was detected at low levels in the presence of a RET fusion, suggesting that there may be some bleed-over of signals between different RNA targets. Of the remaining seven discrepancies, one sample (sample ID: 021) with a MET exon skipping alteration (VAF: 73.78%) was not detected, which should be targeted by the HDPCR assay. A false-positive EGFR L858R alteration was detected by HDPCR (sample ID: 022), while a manual review of the NGS data showed no evidence of any genetic alteration at this position (Table 3). This sample showed inflated internal control counts, indicating the sample may have been overloaded. As such, the observed EGFR L858R false positive may be explained by increased noise because of sample overloading, as the observed count for the variant was just above the threshold. The third sample (sample ID:067) had an EGFR L858R that was correctly called by the HDPCR assay; however, a false-positive ROS1 fusion was found that could not be confirmed by the newer Archer assay. The fourth discrepant sample showed low levels of RET fusions (sample ID: 084) in the HDPCR assay that could not be confirmed by either the original fusion assay or retest by the newer Archer assay, representing a false-positive result for the RET fusion. The fifth sample failed to detect an ALK rearrangement that was detected by the original fusion assay (sample ID: 075). A review of the HDPCR internal control in this sample showed low counts, suggesting sample degradation or underloading of the sample. The sixth sample produced two discordant results (sample ID: 2.29). One result was an ALK fusion with breakpoints known to be inclusive, which was therefore deemed false negative for the ALK fusion. The other result (RET fusion) could not be confirmed by the newer Archer assay, representing a false-positive result for the RET fusion.
3.2 Turn-Around Time Comparison
Post-sample extraction, the HDPCR wet lab workflow was 4 hours, and the time to generate variant calls from raw data using the ChromaCode Cloud was 2 minutes. In comparison, the wet lab workflow for the DNA NGS assay averaged 30 non-consecutive hours and required approximately 4 h for bioinformatics interpretation spanning a minimum of 3 days. The wet lab workflow for the RNA NGS assay averaged 40 non-consecutive hours and required about 4 h for bioinformatics interpretation, also spanning a minimum of 3 days. Figure 1 summarizes a workflow comparison between the HDPCR NSCLC Panel and the comparator NGS assay.
Comparison of High Definition PCR (HDPCR) workflow against next-generation sequencing (NGS) comparator methods. This graph compares the turnaround times (in hours) between the HDPCR non-small cell lung cancer (NSCLC) assay and the DNA and RNA NGS panels, which are used as comparators. The breakdown of steps is also included in the graph and color coded for clarity. Please refer to the methods section for further details. Not accounting for nucleic acid extraction, the HDPCR NSCLC assay yields results in a single day compared to a minimum of 3 days required for the NGS workflows. LDT laboratory developed test, Prep preparation, QC quality control, RUO research use only
3.3 Default Detection Threshold Adjustments to Improve Assay Performance
The ChromaCode Cloud software enables the user to adjust target-specific detection thresholds to maximize assay performance with consideration to system variability. Furthermore, it is recommended that detection thresholds be determined based on limit of blank and limit of detection studies to ensure confidence in distinguishing a positive signal from noise while maintaining detection sensitivity. In this study, four false-positive calls for fusion variants were observed near (within two-fold of) the default detection thresholds that could be mitigated with detection threshold adjustments. Figure 2 illustrates the observed target-specific counts and detection threshold.
Observed target-specific counts and detection thresholds. Observed target-specific counts color coded by false negative (FN, blue), false positive (FP, red), true negative (TN, purple), and true positive (TP, green). Red dotted lines indicate the default target-specific detection thresholds. Each target’s detection threshold could be customized to mitigate FP calls. Please note that the data presented above may not directly match the specific numbers in the tables. The data are preliminary and may include overlapping colored dots or replicate testing. This is intended to demonstrate the customizable feature of this assay
To illustrate the utility of customizable thresholds, the data from this study were reprocessed through the ChromaCode Cloud analysis software using updated thresholds that aimed to mitigate false positives and increase sample quality-control passing rates. Table 4 details the default and updated thresholds for each target across the three subassay wells. The thresholds for EGFR L858R, ALK, RET, and ROS1 were increased to mitigate false-positive detection while the internal control threshold was decreased for the well 1 and well 2 assays to increase sample passing rates, leading to an increase in true-positive detections. There were four samples that yielded passing results that had previously failed because of internal control failures. There were five false-positive calls including one ALK, two RET, one ROS1, and one EGFR L858R that were converted to true-negative calls. Thus, customizing the default thresholds can help improve the assay performance in individual laboratories.
3.4 Enhanced HDPCR Assay Performance Data
After discordant resolution and assay optimization, the HDPCR panel had an improved sensitivity of 95.35% (95% CI 88.52–98.72) and a specificity of 99.69% (95% CI 99.29–99.90) with an overall accuracy of 99.48% (95% CI 99.01–99.76). The performance characteristics after discordant resolution and assay optimization for individual alterations are shown in Figs. 3 and 4 and in Table 2 of the Electronic Supplementary Material (ESM).
DNA targets and performance of the High-Definition PCR non-small cell lung cancer assay. This graph illustrates the performance metrics (sensitivity, specificity, and accuracy) for the detection of each DNA target by the High Definition PCR non-small cell lung cancer assay. The x-axis displays the name of the specific DNA alteration, while the y-axis represents the performance metric range from 0.8 to 1, where 1 is considered maximum performance. Three separate-colored bars correspond to the sensitivity, specificity, and accuracy for each alteration. Note that this figure represents assay performance following discordant resolution and assay optimization. *EGFR T790M was consistently found with other EGFR driver mutations
RNA targets and performance of the High-Definition PCR non-small cell lung cancer assay. This graph illustrates the performance metrics (sensitivity, specificity, and accuracy) for the detection of each RNA target by the High Definition PCR non-small cell lung cancer assay. The x-axis displays the name of the specific RNA alteration, while the y-axis represents the performance metric range from 0.5 to 1, where 1 is considered maximum performance. Three separate-colored bars correspond to the sensitivity, specificity, and accuracy for each alteration. Note that this figure represents assay performance following discordant resolution and assay optimization
4 Discussion
This study demonstrates the robust and accurate performance of the ChromaCode HDPCR NSCLC assay in detecting 15 alterations in nine genes targeted by this assay. Thus, the information obtained provides a comprehensive overview of the patient’s disease state and supports informed decisions for precision medicine-targeted therapies, with results possible in 24–48 h. Accompanied by the proprietary bioinformatics suite, the ChromaCode Cloud™, data processing, and storage are simplified into reports with quality-control results, variant calls, and summary statistics. We have demonstrated that this assay is sensitive enough to detect DNA variants down to below a 10% allele fraction (the lowest detectable target was at 6.4% VAF) with high concordance with sequencing-based assays. In addition, the HDPCR NSCLC assay requires a minimum input (15 ng of DNA and 5 ng of RNA), which makes it a suitable alternative for testing low-input samples that do not meet the quality requirements for NGS assays [29]. Furthermore, we utilized the built-in customization tools to further enhance our assay performance. The internal control reads were within the manufacturer’s recommended cutoff values. However, they were close to the manufacturer recommended cutoff for the discrepant samples and substantially lower than the average internal control counts for the remaining samples. Customizing the assay by modifying the internal control counts and detection limits is a strength of the HDPCR NSCLC panel, and each laboratory should determine the optimal internal control cutoffs based on assay validation prior to clinical implementation.
A previous study by Cabrera et al. [27] has demonstrated the limit of detection at 0.8% for mutant allele fractions of DNA targets with 20 ng per well of DNA input and 23 positive partition counts for RNA targets. In contrast to our study, which primarily concentrated on analyzing samples from post-extraction stored aliquots, Cabrera et al. tested pre-extraction samples from FFPE blocks. Moreover, only clinical samples that were known to be positive were included, and no negative samples were tested in their cohort. Among the tested unique DNA alterations, all were inclusive to the assay, and only five out of nine false-negative RNA results were attributed to novel fusions that were outside the scope of the HDPCR panel. Similar to our findings, this study achieved an accuracy greater than 99% compared with results from the reference method after discordant resolution. This lower limit of detection of the HDPCR NSCLSC assay compared to the comparator NGS assays could be a possible reason for detecting the EGFR L858R mutation that NGS could not confirm. However, we acknowledge that a manual review of NGS data did not support the presence of low levels of the EGFR L858R mutation in this sample. Furthermore, because of logistical constraints, our sample cohort consisted entirely of stored TNA and DNA samples. Additional freeze-thaw of the TNA and DNA aliquots and storage of these nucleic acid samples at − 80 °C/− 20 °C and 4 °C, respectively, could have contributed to some of the discrepancies, and we are unable to rule out the effect of these parameters on the observed discordance in our analysis.
As with all targeted PCR assays, the HDPCR assay does not have comprehensive coverage for all mutations, especially for highly variable regions such as EGFR exon 20 insertions [30]. The HDPCR NSCLC Panel was specifically designed to minimize this issue by ensuring high inclusivity of highly variable targets such as EGFR exon 20 insertions (89%), EGFR exon 19 deletions (99%), and RNA fusions (95–100%) [27]. However, the initial sensitivity observed in our study for all EGFR exon 20 insertion variants is 60%, without considering the assay coverage. Hence, a significant percentage of EGFR exon 20 insertion mutations could go undetected owing to the limited coverage of targeted regions in EGFR exon 20 of the HDPCR NSCLC assay. This underscores the limitations of targeted assays, such as our assay, as they are unable to offer comprehensive coverage for all actionable mutations. Therefore, our preliminary performance data (Table 2), before addressing any discrepancies and performing assay optimization, may mirror real-world clinical performance. For the genomic regions targeted by the HDPCR NSCLC assay, this panel has an aggregate population prevalence-based coverage of 99.6% based on data from the COSMIC database [27, 31]. However, the HDPCR NSCLC assay covers only 56.6% (Table 1 of the ESM) of all known mutations in NSCLC (data from the COSMIC database) [31].
An additional limitation is the low number of positive samples, and the large number of negative samples for individual targets may overestimate the sensitivity of the HDPCR assay. In silico and empirical (bench testing) results for inclusivity by target (DNA/RNA) are shown in Table 1 of the ESM. Unique variants listed in the COSMIC Mutation Database for each target were tested via an in silico analysis. Any variants of high prevalence or those with indeterminant binding kinetics during an in silico analysis were also evaluated empirically. The limited coverage of mutations in targeted PCR assays is not unique to the HDPCR NSCLC assay. Despite this inherent limitation, the ability to identify a large proportion of therapeutically actionable alterations in NSCLC makes this assay useful in settings without access to NGS infrastructure. In addition, the rapid 24–48 h turnaround of this assay and the relatively lower cost of this assay compared with NGS may make this assay a cost-effective alternative to NGS in many clinical settings.
Finally, though the HDPCR assay does not offer comprehensive detection of RNA fusion using other assays, such as the Archer fusion assay, we have successfully improved assay performance for multiple targets by utilizing the customizable detection threshold feature of the HDPCR assay (refer to Sect. 3.3). This process was particularly effective in reducing false-positive calls for RNA targets such as ALK, ROS1, and RET (Table 4). However, we recognize that this assay has lower sensitivity in detecting ALK fusions and MET exon skipping events. Though the lower sensitivity for detecting ALK fusions and MET exon skipping events in this study was likely due to sample quality (Table 3), individual laboratories can consider improving detection rates by integrating ancillary studies into their workflow, such as incorporating ALK immunohistochemistry if deemed necessary. Nevertheless, these results do not suggest that it is mandatory to include ALK immunohistochemistry for every case when using the HDPCR NSCLC panel.
5 Conclusions
We have demonstrated that the HDPCR™ NSCLC Panel allows for sensitive and reliable mutation detection in NSCLC samples with a turnaround time of only 1 day compared with at least 3 days required for NGS workflows. Despite certain limitations in comparison to NGS, the rapid turnaround time and the ability to tailor assays to institutional preferences provide significant value for clinical decision making and choosing the appropriate targeted therapy, ultimately contributing to improved outcomes for patients with newly diagnosed NSCLC.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Araujo LH, Horn L, Merritt RE, Shilo K, Xu-Welliver M, Carbone DP. 69: Cancer of the lung: non-small cell lung cancer and small cell lung cancer. In: Niederhuber JE, Armitage JO, Kastan MB, Doroshow JH, Tepper JE, editors. Abeloff’s clinical oncology. 6th ed. Philadelphia (PA): Elsevier; 2020: p. 1108–1158.e16. Available from: https://www.sciencedirect.com/science/article/pii/B9780323476744000694. Accessed 20 Jul 2024.
AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7:818–31.
Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384:2371–81.
Jänne PA, Riely GJ, Gadgeel SM, Heist RS, Ou S-HI, Pacheco JM, et al. Adagrasib in non-small-cell lung cancer harboring a KRASG12C mutation. N Engl J Med. 2022;387:120–31.
Passaro A, Malapelle U, Del Re M, Attili I, Russo A, Guerini-Rocco E, et al. Understanding EGFR heterogeneity in lung cancer. ESMO Open. 2020;5: e000919.
Johnson M, Garassino MC, Mok T, Mitsudomi T. Treatment strategies and outcomes for patients with EGFR-mutant non-small cell lung cancer resistant to EGFR tyrosine kinase inhibitors: focus on novel therapies. Lung Cancer. 2022;170:41–51.
Champagnac A, Bringuier P-P, Barritault M, Isaac S, Watkin E, Forest F, et al. Frequency of MET exon 14 skipping mutations in non-small cell lung cancer according to technical approach in routine diagnosis: results from a real-life cohort of 2,369 patients. J Thorac Dis. 2020;12(5):2172–8.
Du X, Shao Y, Qin H-F, Tai Y-H, Gao H-J. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac Cancer. 2018;9:423–30.
Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865–75.
Kashima J, Kitadai R, Okuma Y. Molecular and morphological profiling of lung cancer: a foundation for “next-generation” pathologists and oncologists. Cancers (Basel). 2019;11:599.
Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res. 2019;8(3):286–301.
Zonta E, Garlan F, Pécuchet N, Perez-Toralla K, Caen O, Milbury C, et al. Multiplex detection of rare mutations by picoliter droplet based digital PCR: sensitivity and specificity considerations. PLoS ONE. 2016;11: e0159094.
Mao X, Liu C, Tong H, Chen Y, Liu K. Principles of digital PCR and its applications in current obstetrical and gynecological diseases. Am J Transl Res. 2019;11:7209–22.
Wang Q, Yang X, He Y, Ma Q, Lin L, Fu P, et al. Droplet digital PCR for absolute quantification of EML4-ALK gene rearrangement in lung adenocarcinoma. J Mol Diagn. 2015;17:515–20.
Lambros L, Caumont C, Guibourg B, Barel F, Quintin-Roué I, Marcorelles P, et al. Evaluation of a fast and fully automated platform to diagnose EGFR and KRAS mutations in formalin-fixed and paraffin-embedded non-small cell lung cancer samples in less than one day. J Clin Pathol. 2017;70:544.
Madic J, Jovelet C, Dehri I, Mallory AC. 6-Color crystal digital PCRTM for the high-plex detection of EGFR mutations in non-small cell lung cancer. In: Santiago-Cardona PG, editor. Lung cancer: methods and protocols. New York: Springer, US; 2021. p. 127–44. https://doi.org/10.1007/978-1-0716-1278-1_10.
Williamson DFK, Marris SRN, Rojas-Rudilla V, Bruce JL, Paweletz CP, Oxnard GR, et al. Detection of EGFR mutations in non-small cell lung cancer by droplet digital PCR. PLoS ONE. 2022;17: e0264201.
Xu Q, Zhu Y, Bai Y, Wei X, Zheng X, Mao M, Zheng G. Detection of epidermal growth factor receptor mutation in lung cancer by droplet digital polymerase chain reaction. Onco Targets Ther. 2015;8:1533–41. https://doi.org/10.2147/OTT.S84938.
Decraene C, Silveira AB, Bidard F-C, Vallée A, Michel M, Melaabi S, et al. Multiple hotspot mutations scanning by single droplet digital PCR. Clin Chem. 2018;64:317–28.
Watanabe M, Kawaguchi T, Isa S, Ando M, Tamiya A, Kubo A, et al. Multiplex ultrasensitive genotyping of patients with non-small cell lung cancer for epidermal growth factor receptor (EGFR) mutations by means of picodroplet digital PCR. EBioMedicine. 2017;21:86–93.
Olmedillas-López S, Olivera-Salazar R, García-Arranz M, García-Olmo D. Current and emerging applications of droplet digital PCR in oncology: an updated review. Mol Diagn Ther. 2022;26:61–87.
Yung TKF, Chan KCA, Mok TSK, Tong J, To K-F, Lo YMD. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–84.
Visser E, de Kock R, Genet S, van den Borne B, Soud MY-E, Belderbos H, et al. Up-front mutation detection in circulating tumor DNA by droplet digital PCR has added diagnostic value in lung cancer. Transl Oncol. 2023;27:101589.
Rajagopal A, Yurk D, Shin C, Menge K, Jacky L, Fraser S, et al. Significant expansion of real-time PCR multiplexing with traditional chemistries using amplitude modulation. Sci Rep. 2019;9:1053.
NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for non-small cell lung cancer V.4. 2023© National Comprehensive Cancer Network, Inc. 2023. Available from: https://NCCN.org. Accessed 18 Oct 2023.
Cabrera K, Gole J, Leatham B, Springer MJ, Smith M, Herdt L, et al. Analytical performance and concordance with next-generation sequencing of a rapid, multiplexed dPCR panel for the detection of DNA and RNA biomarkers in non-small-cell lung cancer. Diagnostics (Basel). 2023;13(21):3299.
Henzler C, Schomaker M, Yang R, Lambert AP, LaRue R, Kincaid R, et al. Optimization of a microfluidics-based next generation sequencing assay for clinical oncology diagnostics. Ann Transl Med. 2018;6(9):162.
Herdt LR, Berroteran P, Rajagopalan M, Brown BA, Schwartz JJ. NSCLC digital PCR panel returns low-input sample results where sequencing fails. Diagnostics (Basel). 2024;14(3):243.
Shen C-I, Chiang C-L, Shiao T-H, Luo Y-H, Chao H-S, Huang H-C, et al. Real-world evidence of the intrinsic limitations of PCR-based EGFR mutation assay in non-small cell lung cancer. Sci Rep. 2022;12:13566.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7.
Acknowledgments
This work would not have been possible without the collective efforts and collaboration of the Molecular Diagnostics Laboratory and the Advanced Research and Diagnostics Laboratory at the University of Minnesota.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This research was funded by ChromaCode Inc.
Ethics Approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the University of Minnesota (protocol code STUDY00017413 on 31 October, 2022).
Conflicts of Interest/Competing Interests
Abdulaziz F. Al Mana, Kristen Culp, Abby Keeler, Orlando Perrera, Malini Rajagopalan, Jacky Lucien, Brad Brown, and Bharat Thyagarajan have no competing interests or conflicts of interest that are directly relevant to the content of this study. Although ChromaCode Inc. employees participated in the study’s design and execution, the authors at the University of Minnesota had the final say in deciding the study’s design and collecting, analyzing, and interpreting of the data, writing the manuscript, and decision to publish the results.
Consent to Participate
Patient consent was waived as the University of Minnesota Institutional Review Board determined that this study was human research, not engaged.
Consent for Publication
Not applicable.
Availability of Data and Material
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Code Availability
Not applicable.
Authors’ Contributions
Conceptualization: KC, BT; methodology: AK, KC, MR, AAM, BB, OP; data curation: AK, KC, AAM; formal analysis and investigation: AAM, KC, BB, MR, BT; writing - original draft preparation: AAM, KC; writing - review and editing: BT, MR, BB, OP, LJ; software: LJ, BB; funding acquisition: BT; resources: MR, BT; supervision: BT. All authors have read and agreed to the published version of the manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Al Mana, A.F., Culp, K., Keeler, A. et al. Performance of a Rapid Digital PCR Test for the Detection of Non-Small Cell Lung Cancer (NSCLC) Variants. Mol Diagn Ther (2024). https://doi.org/10.1007/s40291-024-00732-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s40291-024-00732-y