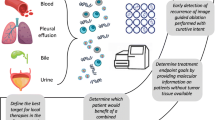
Abstract
Circulating DNA in the bloodstream has been studied since the 1940s, leading to its identification as a possible early marker for the presence of a primary tumor. Recently, it has been more successfully employed in liquid biopsies to determine the early presence of a metastatic tumor arising after chemotherapy, radiotherapy, and surgery. The appearance of such circulating tumor DNA permits the identification of the metastatic tumor before it is detected by either palpation or radiological analysis. Nevertheless, the liquid biopsy may possibly be affected by the removal of circulating tumor DNA via the kidneys and spleen as it is released. Furthermore, the liver removal of cell-free DNA has not yet been considered to be involved in this process. Here, we review the literature on the removal of free single- and double-stranded DNA and nucleosomal, vesicular, and exosomal DNA via the liver and examine its possible impact on circulating DNA levels. The removal of all forms of DNA by the liver, together with that removed by the kidneys and spleen, may delay the timing of positive results from liquid biopsies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The liver is capable of removing free single- and double-stranded DNA and nucleosomal, vesicular, and exosomal DNA. |
The removal of all such forms of DNA by the liver, together with that removed by the kidneys and spleen, may delay the timing of positive results from liquid biopsies. |
1 Introduction
DNA fragments circulating in the blood stream have been studied for over 40 years as possible early diagnostic markers for the presence of a primary tumor, but with little success [1]. However, circulating DNA (cirDNA) has been more successfully employed in noninvasive liquid biopsies, blood samples of 2–5 ml taken from patients. These are currently used to determine the presence of genetic markers for fetal chromosome disorders, e.g., Down syndrome in the first trimester of pregnancy, replacing invasive amniocentesis [2]. Importantly, Thierry et al. [3] presented data showing increased circulating tumor DNA (ctDNA) with increasing tumor burden in a SW620 xenografted mouse model. This led to the early detection of the presence of a tumor DNA released by both primary tumors [4, 5] and secondary metastatic tumors after chemotherapy, radiotherapy, and surgical removal of the primary tumor [6, 7].
Furthermore, the appearance of ctDNA permits the identification of the metastatic tumor before its detection by either palpation or radiological analysis, leading to early treatment [3, 8,9,10,11]. In addition, an early indication of a tumor presence can lead to an early treatment decision [12,13,14,15,16].
To date, the US FDA has approved the liquid biopsy next-generation sequencing-based FoundationOne Liquid CDx test involved with the identification of BRCA1 and BRCA2 mutations in patients with ovarian cancer eligible for rucaparib treatment. In addition, the FDA also approved ALK rearrangements in patients with non-small-cell lung cancer offered alectinib treatment and mutations in the PIK3CA gene in patients with breast cancer treated with alpelisib. Furthermore, the Guardant360 CDx test for EGFR exon 19 deletions, mutant L858R, and mutant T790M were approved as markers for patients with non-small-cell lung cancer treated with osimertinib.
However, it appears that the liver [17,18,19,20,21,22,23,24,25,26,27], kidneys [28, 29], and spleen [30, 31] can remove part of the cirDNA containing possible tumor markers, possibly further affecting the timing of the appearance of detectable ctDNA in liquid biopsy.
Here, we consider the indications that the liver may remove a sufficiently important fraction of cirDNA, including ctDNA, so as to have an impact upon the timing of the identification of the presence of secondary metastatic tumors and hence, a delay in new treatment.
2 Released DNA—Free DNA
The cirDNA can be derived through various cellular and tissue processes, including apoptosis, necrosis, pyroptosis, ferroptosis, NETosis, sepsis, mitochondrial DNA release, hemopoietic cell release, vesicles, e.g. exosomes and virtosomes, the release of transposons and retrotransposons, and the presence of bacterial and viral DNA in healthy individuals [1, 32, 33]. Of this variety of cfDNA sources, apoptosis is considered to be the main source [34].
One apoptotic cell can shed millions of nuclear DNA fragments into the extracellular environment, ~36 × 106 fragments with a mean of 167 base pairs [35], of which two fragments can be mutant [36]. Hence, for example, if 108 cells are present in 1 g of tumor, the bulk of the cirDNA will be of non-tumor origin, leading to the conclusion that there are only a very small number of mutant fragments present in the cirDNA [37]. This would represent the lower limit of being able to identify ctDNA released from an as-yet non-palpable and non-radiologically identified tumor of < 1.0 g. Moreover, asymptomatic or very small tumors will release very low amounts of cell-free DNA (cfDNA), considered insufficient for an accurate DNA analysis [37].
The ctDNA so released can be present in a number of forms, namely, pieces of either single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA) or nucleosomes.
3 Transported DNA—Extracellular Vesicles and Exosomes
It is now clear that the cfDNA circulates in blood in both extracellular vacuoles (EVs) and a non-vacuolar form as well as nucleosomes. For the vacuolar cfDNA, it has been estimated that, in healthy individuals, as much as 90% of cfDNA can be present in EVs, leaving some 10% in a non-vacuolar form [38]. The cfDNA found with the exosomal fraction may also be attached primarily to the external surface of exosomes (ExV). However, this calculation omits the nucleosomal DNA since nucleosomes would have been excluded from the assessment during the isolation of the Exs. Nevertheless, Exs would appear to contain a substantial proportion of the cfDNA including ctDNA.
A parallel analysis does not appear to have been performed on ctDNA from patients with cancer, though it is possible to suggest that the result will be similar since much of the cfDNA is considered to be released from both healthy and cancer cells in the form of vesicles [39]. Jeppesen et al. [40] questioned the cfDNA content of ExVs, arguing that highly purified ExVs do not contain cfDNA but that it is preferentially released in an alternative group of larger microvesicles. Hence, the cfDNA, in all its formats, found in either serum or plasma, will be present as either circulating tumor cells or vacuolar cfDNA or non-vacuolar cfDNA or large oncosomes or apoptotic vesicular cfDNA [38, 41]. It is now questionable that ExVs contain cfDNA as opposed to the DNA attaching to the outer exosomal surface as discussed [40, 42]. Nevertheless, ExVs have been shown to contain a variety of molecules, including genomic DNA (100 base pairs; 17 kilobase pairs).
ExV DNA will be important in the identification of a specific cancer and the timing of the appearance of metastases via the liquid biopsy procedure [43,44,45,46]. Thus, given that nucleosomes are not present in ExVs, the extraction of cfDNA from either plasma or serum would be preferential and can be done via a total DNA extraction procedure from either the whole serum or plasma rather than just the isolated ExVs/EVs. Specific DNA extraction kits are commercially available for this purpose. Basing liquid biopsy solely on ExVs may be unwise because of the omission of nucleosomal-derived cf/ctDNA.
4 Hepatic Clearance
4.1 The Liver and Blood Flow
Approximately 1.5 L of blood flows through the liver every minute. Since the average human blood volume is ~ 5 L, the total volume of blood passes through the liver in 3–4 min [47] and therefore will flow through the liver some 350 times daily. This means that the total ctDNA will be continuously exposed to the liver, presenting it with a regular opportunity to remove ctDNA and other cirDNA that has not already been removed by the spleen [31], the gastrointestinal tract, placenta [30], or the kidneys [28, 29]. The major liver cell types responsible for the removal of cirDNA, including ctDNA, are Kupffer (K) cells and sinusoidal endothelial (E) cells, and—to a lesser extent—the parenchymal hepatocyte cells that make up the largest liver cell population.
K cells are resident macrophages localized within the lumen of the liver sinusoids and adhering to the E cells lining the blood vessel walls. Kinoshita et al. [48] grouped K cells into four populations based upon surface markers. Those bearing either F4/80+ CD68+ or F4/80+ CD11b+ showed higher phagocytic activity, whereas those bearing either F4/80+ CD11b+ or F4/80+ CD68- surface markers, on lipopolysaccharide stimulation, yielded a higher intensity of tumor necrosis factor and interleukin-12. Sinusoidal E cells constitute a permeable interface between the blood cells and the hepatocytes.
4.2 DNA Removal from Blood by the Liver
As further discussed in the following sections, the liver has been demonstrated to actively remove circulating cfDNA, i.e., single-stranded free DNA (ssfDNA) and double-stranded free DNA (dsfDNA), from the blood stream of experimental mammals.
In addition, nucleosomes may also be removed. If this cfDNA removal is extrapolated to humans, there should be a direct effect on the serum/plasma DNA levels circulating in both healthy and unwell individuals, including those with cancer, given the speed of the circulation of the total blood volume through the liver [47].
The cfDNA fragments form a range of sizes, with the bulk being in the range of 60–400 bp in the case of dsfDNA derived from patients with colorectal cancer, with a good proportion being in the range of 60–150 bp [49].
Thus, although current methods permit the identification of even single DNA fragments, and that, initially, the level of relevant DNA is small, any removal from the blood stream by the liver of such tumor-related cfDNA may result in a delay to the first indication of a metastasis.
In considering the role of the liver in the removal of serum/plasma cfDNA forms, the importance of K cells and E cells in this process are discussed.
4.2.1 Nucleosome Removal
Gauthier et al. [18] studied the fate of isolated mononucleosomes in normal C57BI/CJ mice [18]. The amounts injected were given as nucleosomal DNA levels. Thus, post-injection, nucleosomes were rapidly removed from the circulation when doses of < 11 µg DNA were employed. However, increasing the number of nucleosomes injected (11–85 µg DNA) led to a reduction in the rate of removal because of a saturation of K cells. Indeed, the shapes of the curves for amounts > 11 µg are typical of a nonlinear kinetic mechanism for saturation, indicating that K cells alone are not enough to eliminate all DNA, resulting in an increase in the elimination half-life.
When working with the lower level of injected nucleosomes, the liver accounted for the removal of 71.0–84.7% from the circulation within 10 min, with only a small percentage (0.52 ± 0.15%) of the nucleosomes localized in the kidneys. Interestingly, concurrent injection of dsfDNA and nucleosomes resulted in a sixfold reduction in nucleosomal clearance, implying that the dsfDNA was preferentially removed with respect to the nucleosomes.
The hepatic removal of the nucleosomes was unlikely due to the non-parenchymal cell population, K cell phagocytosis being cited as a likely mechanism [17]. The role of the K cells in nucleosome clearance was confirmed by Du Clos et al. [19] through the presence of blood nucleosomal material after a reduction of K cells and splenic macrophages through treatment with dichloromethylene bisphosphonate.
An explanation for the results obtained by Gauthier et al. [18] could be that the K cells completely phagocytosed the nucleosomes at the lower levels employed but that, above 11 µg DNA, the higher levels of nucleosomes caused a saturation of the phagocytic activity of the K cells.
4.2.2 Removal of Single-Stranded Free and Double-Stranded Free DNA
DNA emanating from mitochondria and bacteria can be considered as dsfDNA. However, there will be a limited involvement of ssfDNA in either plasma or serum since it is likely to be linked to either lipoprotein or protein structures in one form or another. A number of the early studies on the uptake of DNA were devoted to identifying the fate of both dsfDNA and ssfDNA to determine the mechanisms to be used in introducing genetic material into cells in vivo.
Uptake of DNA via the lysosomal system was identified early by Wattiaux et al. [21], who injected 100 ng 35S-dsfDNA intravenously into Wistar rats (300–350 g) that were subsequently killed at intervals. Analysis of isolated livers indicated that ~ 60% of the 35S-dsfDNA was present after 5 min, with a reduction to ~ 45% at 30 min and to ~ 20% at 2 h. After homogenization and differential centrifugation followed by iso-picnic centrifugation, radioactivity was found to be associated with the sedimentable fractions. Since radioactivity was associated with the hydrolytic activity of caspase C, it implied the presence of 35S-dsfDNA in the lysosomal and endosomal fractions. Treatment with Triton WR1339 permitted the distinction between the two fractions, indicating one to be the lysosomal fraction and the other the precursor endosomal fraction. These authors did not identify the cell types involved, but since there was also labeling of the nuclear fraction, it was considered likely that the hepatocytes were also involved. A delayed entry, but not a blockage, of the 35S-dsfDNA into the lysosomes through a parallel treatment with an artificial cationic lipid, N-(1-(2,3-dioleoyloxy)propyl)-N,N,N,-trimethylammonium-sulphate (DOTAP), resulted in a greater percentage of the 35S-DNA entering the nuclei. Thus, a proportion of the DNA entering hepatocytes can be destroyed by the lysosomal system. Using both transmission electron microscopy autoradiography (TEM-ARG) and biochemical analyses, Emlen and Mannik [22] showed that the uptake of ssfDNA in liver perfusion studies resulted in the attachment of the ssfDNA to specific DNA-binding sites on the surfaces of both K cells and parenchymal E cells [22]. No radioactivity was associated with the hepatocytes. The TEM-ARG of the K cell presented showed a number of small vesicles along the periphery of the cell with silver grains that could be associated with them rather than just the cell membrane surface. This would imply that the ssfDNA had entered the lysosomal pathway for destruction. However, on flushing the livers with an anti-DNA enzyme, deoxyribonuclease (DNAse), the radioactivity associated with the liver was rapidly eliminated, suggesting a proportion of the DNA was cell surface associated. This could involve DNA binding to surface receptors for DNA on both E and K cells. Since blood contains DNAase [23], the surface-bound DNA could be readily broken down. A comparative study of the uptake and removal of ssfDNA and dsfDNA by the liver showed that the ssfDNA was more readily taken up and its breakdown products excreted at a faster rate than that of dsfDNA [22, 24,25,26], all different fragment sizes being removed at similar rates [22].
Importantly, Chia et al. [24] also determined whether or not the catabolic pathways for DNA metabolism could become saturated following daily intravenous injections of mice with 100 µg of either ssfDNA or dsfDNA for 5 days. This was followed by injection of radio-labeled homologous DNA followed by the determination of blood and organ DNA levels. No differences were observed between the treated and untreated control animals, i.e., the liver did not become saturated with respect to the breakdown of DNA. In contrast, Emlen and Mannik [22] showed that the ssfDNA uptake by the liver was saturable in parallel with the clearance of the blood. They argued that a sudden release of a large amount of DNA into the blood could result in the saturation of the liver system and lead to the presence of high DNA amounts in the blood. However, this does not appear to be the case for cfDNA.
Using plasmid DNA (pDNA), Kobayashi et al. [27] also found that the liver system could be saturated. In addition, they showed that pDNA was taken up essentially by the E cells, with such uptake not affected by K-cell blockade with gadolinium chloride.
4.2.3 Extracellular Vesicle Removal
Of interest is the possible role that K cells might play in the removal of at least some of the EVs and hence the DNA and the mitochondria either contained therein or, in the case of the ctDNA, possibly surface attached.
Imai et al. [20], who used B16BL6 Exs labeled with PKH26, a lipophilic fluorescent dye, showed that they were taken up by macrophages in the liver and spleen but not in the lungs, where they were taken up by E cells. Interestingly, if the liver K cells were depleted by an injection of clodronate-containing liposomes followed by an injection of B16BL6 Exs labeled with PKH26, clearance of the EVs from the blood was much slower than in untreated mice, i.e., in the absence of most of the K cells. Injections of 1.25, 2.5, and 5 µg of Exs in untreated mice resulted in their total clearance. The serum concentration of Exs in macrophage-depleted mice was 285 times that of the untreated mice.
5 Conclusions
Methods are available that permit the detection of single mutant ctDNA (mut-ctDNA) fragments via liquid biopsy [50, 51]. There may be low levels of such ctDNA in a liquid biopsy, and there can be a long period between removal of the primary tumor and the first appearance of metastases-generated ctDNA fragments [1]. Part of this delay can be due to the newly initiated metastasis releasing ctDNA that includes some mut-ctDNA fragments and that will be available for removal by the liver. Therefore, as more ctDNA is released, either as ctDNA or as circulating exosomal DNA, a proportion will be continuously removed by the liver, including the mutated fragments, so delaying the appearance of enough mut-ctDNA to permit the identification of a tumor presence and hence to commence treatment. The degree to which this is the case could depend upon the growth rate of the metastatic tumor.
References
Gahan PB. A Brief history and the present and future status of CNAPS. In Circulating Nucleic acids in early diagnosis, prognosis and treatment monitoring: an introduction. Ed. Gahan PB. 2015. Springer Science + Business Media
Hui L. Non-invasive prenatal testing for fetal aneuploidy: charting the course from clinical validity to clinical utility. Ultrasound Obstet Gynecol. 2013;41:2–6.
Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–76.
Mouliere F, Messaoudi SE, Gongora C, Lamy PJ, del Rio M, Lopez-Crapez E, et al. Personalized medicine by analyzing circulating DNA: application to the management care of colorectal cancer patients. Ann Oncol. 2013;24:i7.
Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11:3475.
Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92.
Coombes R. Cancer drug resistance needs urgent attention, says research chief. BMJ. 2019;365:l1934.
Garcia-Murillas I, Chopra N, Comino-Méndez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019;5:1473–8.
Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–81.
Nabet BY, Esfahani MS, Moding EJ, Hamilton EG, Chabon JJ, Rizvi H, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell. 2020;183:363-376.e13.
Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10:1842–53.
Thomsen MBH, Nordentoft I, Lamy P, Vang S, Reinert L, Mapendano CK, et al. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Sci Rep. 2017;7:11702.
Snyder A, Morrissey MP, Hellmann MD. Use of circulating tumor DNA for cancer immunotherapy. Clin Cancer Res. 2019;25:6909–15.
Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–65.
Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Alaiwi SA, et al. Author Correction: Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat Med. 2020;26:1663.
Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–7.
Gauthier VJ, Mannik M, Striker GE. Effect of cationized antibodies in performed immune complexes on deposition and persistence in renal glomeruli. J Exp Med. 1982;156:766–77.
Gauthier VJ, Tyler LN, Mannik M. Blood clearance kinetics and liver uptake of mononucleosomes in mice. J Immunol. 1996;156:1151–6.
Du Clos TW, Volzer MA, Hahn FF, Xiao R, Mold C, Searles RP. Chromatin clearance in C57Bl/10 mice: interaction with heparan sulphate proteoglycans and receptors on Kupffer cells. Clin Exp Immunol. 1999;117:403–11.
Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, et al. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles. 2015;4:26238.
Wattiaux R, Jadot M, Dubois F, Misquith S, Wattiaux-De CS. Uptake of exogenous DNA by rat liver: effect of cationic lipids. Biochem Biophys Res Commun. 1995;213:81–7.
Emlen W, Mannik M. Kinetics and mechanisms for removal of circulating single-stranded DNA in mice. J Exp Med. 1978;147:684–99.
Tamkovich SN, Cherepanova AV, Kolesnikova EV, Rykova EY, Pyshnyi DV, Vlassov VV, et al. Circulating DNA and DNase activity in human blood. Ann N Y Acad Sci. 2006;1075:191–6.
Chia D, Dorsch CA, Levy L, Barnett EV. The metabolism of nucleic acids in mice. Immunology. 1979;36:323–9.
Chused TM, Steinberg AD, Talal N. The clearance and localization of nucleic acids by New Zealand and normal mice. Clin Exp Immunol. 1972;12:465–76.
Dorsch CA, Chia D, Levy L, Barnett EV. Persistence of DNA in the circulation of immunized rabbits. J Rheumatol. 1975;2:161–6.
Kobayashi N, Kuramoto T, Yamaoka K, Hashida M, Takakura Y. Hepatic uptake and gene expression mechanisms following intravenous administration of plasmid DNA by conventional and hydrodynamics-based procedures. J Pharmacol Exp Ther. 2001;297:853–60.
Coritsidis GN, Beers PC, Rumore PM. Glomerular uptake of nucleosomes: evidence for receptor-mediated mesangial cell binding. Kidney Int. 1995;47:1258–65.
Buzder T, Yin X, Wang X, Banfalvi G, Basnakian AG. Uptake of foreign nucleic acids in kidney tubular epithelial cells deficient in proapoptotic endonucleases. DNA Cell Biol. 2009;28:435–42.
Doerfler W, Schubbert R. Uptake of foreign DNA from the environment: the gastrointestinal tract and the placenta as portals of entry. Wien Klin Wochenschr. 1998;110:40–4.
Olsen I, Harris G. Uptake and release of DNA by lymphoid tissue and cells. Immunology. 1974;27:973–87.
Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc. 2018;93:1649–83.
Hu Z, Chen H, Long Y, Li P, Gu Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit Rev Oncol Hematol. 2021;157:103166.
Heitzer E, Auinger L, Speicher MR. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med. 2020;26:519–28.
Sanchez C, Roch B, Mazard T, Blache P, Al Amir Dache Z, Pastor B, et al. Circulating nuclear DNA structural features, origins, and complete size profile revealed by fragmentomics. JCI Insight. 2021;6(7).
Schwarzenbach H, Gahan PB. Exosomes in immune regulation. Non Coding RNA. 2021;7:4.
Fiala C, Diamandis EP. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018;16:166.
Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE. 2017;12:e0183915.
Jurj A, Zanoaga O, Braicu C, Lazar V, Tomuleasa C, Irimie A, et al. A comprehensive picture of extracellular vesicles and their contents. Molecular transfer to cancer cells. Cancers (Basel). 2020;12:298.
Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019;177:428-445.e18.
Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7:1505403.
Pluchino S, Smith JA. Explicating exosomes: reclassifying the rising stars of intercellular communication. Cell. 2019;177:225–7.
Wu M, Wang G, Hu W, Yao Y, Yu X-F. Emerging roles and therapeutic value of exosomes in cancer metastasis. Mol Cancer. 2019;18:53.
Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. PNAS Natl Acad Sci. 2017;114:E9066–75.
Kalluri R, LeBleu VS. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb Symp Quant Biol. 2016;81:275–80.
Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–9.
Rocha FG. Liver blood flow: physiology, measurement, and clinical relevance. In: Jarnagin WR, Blumgart LH, editors. Chapter 4 in Blumgart's surgery of the liver, pancreas and biliary tract. 5th ed. W.B. Saunders. 2012.
Kinoshita M, Uchida T, Sato A, Nakashima M, Nakashima H, Shono S, et al. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J Hepatol. 2010;53:903–10.
Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE. 2011;6:e23418.
Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–8.
Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124:345–58.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No sources of funding were used to conduct this study or prepare this manuscript.
Conflict of interest
Sonia Khier and Peter B. Gahan have no conflicts of interest that are directly relevant to the content of this article.
Availability of data and material
Not applicable.
Ethics approval
Not applicable.
Consent
Not applicable.
Author contributions
Both authors contributed equally to the work.
Rights and permissions
About this article
Cite this article
Khier, S., Gahan, P.B. Hepatic Clearance of Cell-Free DNA: Possible Impact on Early Metastasis Diagnosis. Mol Diagn Ther 25, 677–682 (2021). https://doi.org/10.1007/s40291-021-00554-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-021-00554-2