Abstract
Aim
Screening amplified genes for targeted therapy with high-throughput technology is very important. The NanoString nCounter system allows multiplexed digital quantification of target molecules through the use of color-coded barcodes with the great advantage that formalin-fixed, paraffin-embedded (FFPE) tissue can be utilized.
Methods
We tested nCounter custom copy number variation (CNV) panels in 220 gastric cancer samples and evaluated the utility of this method as a screening tool for the detection of CNV using HER2. For the validation of results, we compared the nCounter results with immunohistochemistry (IHC), and we further performed in situ hybridization (ISH) in discrepant cases.
Results
The average HER2 gene copy numbers (CNs) by nCounter were 17.25, 2.0 and 2.61 for the HER2 IHC positive (3+), equivocal (2+), and negative cases, respectively. Out of the 16 IHC 3+ cases, 13 (81.3 %) were reported as HER2 CN gain (≥4). Gastric cancers with homogeneous HER2 overexpression or high tumor purity showed HER2 CN ≥10. Among the 192 cases with HER2 IHC negative and without HER2 gene amplification, 29 showed a HER2 CN ≥4 with the nCounter assay. The nCounter assay had a concordance rate of 83.4 % (kappa value, 0.35), a sensitivity of 66.7 %, a specificity of 85.2 %, a negative predictive value of 96 %, and a positive predictive value of 32.6 % compared with HER2 IHC/ISH results. Fresh frozen (FF) samples revealed a higher concordance rate (91.5 %, kappa value, 0.59) than FFPE samples (78.5 %, kappa value 0.27) and showed a high specificity (97.2 %).
Conclusion
The nCounter CNV assay is a reliable and practical method to detect high CN variations. Given the intra-tumoral HER2 heterogeneity and normal cell contamination, additional IHC and/or FISH is necessary and needs caution in interpretation, especially in FFPE tissue samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
With nCounter CNV assay, we analyzed 220 gastric cancer samples to detect HER2 amplifications. |
To evaluate the utility of this method as a screening tool, we validated the results with IHC, FISH and SISH and found a high concordance rate. |
The nCounter assay is able to detect all cancers with high CN gains. Given the intra-tumoral heterogeneity and normal cell contamination, additional IHC is needed to detect all tumors with low HER2 CN gains. |
1 Introduction
Amplification of oncogenes is a major mechanism for gene overexpression and contributes to development and progression of cancer [1–3]. Recent hierarchical stratification of genomic data from a diverse sample of 3000 tumors has subdivided them into two major groups: tumors with somatic mutations, and tumors with copy number (CN) alterations [4]. Beyond the mechanism of tumor development, functional events within a tumor class can suggest a class-specific target therapy [4]. Amplified oncogenes have been given much attention in recent years as they hold much potential for directed therapeutic targets.
Human epidermal growth factor receptor 2 (HER2)-amplified cancer treated with monoclonal antibodies is one of the most successful examples of cancer targeted therapy and has become standard therapy for HER2-positive breast cancer and gastric cancer [5, 6]. Recently, several preclinical data have shown promising therapeutic potential for targeting other amplified oncogenes such as MET, epidermal growth factor receptor (EGFR), and fibroblast growth factor receptors 2 (FGFR2) [7–9], and, in fact, there are current ongoing clinical trials evaluating the efficacy of these drugs across various cancers [10]. Therefore, it is crucial to detect targetable copy number variation (CNV) of oncogenes in order to provide patients with potential treatment options.
To find CNVs in cancers, fluorescent in situ hybridization (FISH) is considered the current standard method used for the detection of gene amplification. However, FISH is expensive and time-consuming as each FISH test can detect the amplification of only one gene [11]. As a screening tool, high-throughput assay seems to be a more practical and reasonable approach to simultaneously detect amplification of target genes. CNVs have been routinely detected by genome-wide comparative hybridization (CGH), array-based CGH, and single nucleotide polymorphism (SNP) arrays [12–14]. Recently, next-generation sequencing (NGS) has made a marked improvement in the detection of CNV [15]. However, accurate CNV calling in FFPE (formalin-fixed, paraffin-embedded) samples by these methods represent many major challenges [15]. One of the high-throughput systems, NanoString nCounter is a DNA-based technology, which allows digital quantification of multiplexed target molecules through the use of color-coded barcodes. It can provide discrete counts of target DNA using low amounts of total DNA without an amplification process, making this method highly suitable for use with FFPE tissue specimens.
Here we tested nCounter custom CNV panels in 220 gastric cancer samples and evaluated the efficacy as a screening tool to detect CNV. We chose to use HER2 for validation since the correlation between HER2 immunohistochemistry (IHC) and in situ hybridization (ISH) is well established [16]. We correlated HER2 results in nCounter CNV analysis with HER2 IHC. For discordant cases, HER2 ISH was performed for confirmation of HER2 amplification status.
2 Materials and Methods
2.1 Patient Selection
Between November 2013 and July 2015, 550 patients were enrolled in the NEXT-1 trial (NCT#02141152) and VIKTORY trial (NCT#02299648), which were based on metastatic or advanced solid cancer patients. We performed NGS (Ion Torrent or Illumina HiSeq) and nCounter CNV analysis to detect molecular alterations for trial enrollment. From them, we selected gastric cancer patients with available NanoString nCounter CNV results (n = 220). Eighty-five samples (38.6 %) were fresh frozen (FF) tissue and 135 samples (61.4 %) were submitted as FFPE blocks. HER2 IHC results were available for all 220 patients. The study was approved by the institutional review board (IRB) at Samsung Medical Center (2014-1-136).
2.2 Genomic DNA Isolation
A pathologist (M.H) evaluated hematoxylin and eosin stained slides of samples to determine the tumor cell population. Manual microdissection was performed if tumor cell percentages were less than 70 % in surgical samples. For biopsy samples, manual microdissection was not available due to small tissue size. Genomic DNA was extracted using Qiagen DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) or QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions as previously described [3]. RNase A (Qiagen #19101) was used for all samples. Before fragmentation step, we measured concentration as well as 260/280 and 260/230 nm ratios (ND1000, Nanodrop Technologies, Thermo-Fisher Scientific, MA, USA).
2.3 NanoString nCounter Assay
For detection of CNVs, a panel of 20 gene probes including AURKA, CCND1, CCNE1, CDK4, CDK6, CDKN1A, CDKN2A, EGFR, ERBB2, ERBB3, FGFR1, FGFR2, IGF1R, KLF5, KRAS, MDM2, MITF, MYC, PIK3CA, and TNIK were designed using NanoString nCounter technology and subsequently analyzed on the NanoString nCounter platform [17]. For custom CodeSets, nCounter Standard chemistry in which the biotin label and molecular barcode are covalently attached to the target specific probe was used. Three probes were designed for each gene. Each assay contained six positive dsDNA control probes, 8 negative control probes, and 10 invariant control probes (INVs) designed for autosomal genomic regions predicted not to contain common CNVs.
The NanoString nCounter assay was perfor med according to NanoString’s standard protocol. Briefly, after quantification, 100–200 ng of gDNAs were fragmented with 1 μL of 5 U/μL AluI Fragmentation Enzyme. Afterwards, fragmented DNAs were denatured to produce single strands. The custom CNV CodeSet was then hybridized with the capture and reporter probes in 30 μL total volume and incubated overnight at 65 °C for at least 16 h (average 17 h). Hybridized DNA-CodeSet complexes were purified using the fully automated nCounter Prep station, and reporters were counted using the nCounter Digital Analyzer. For normalization of results obtained from FF tissue sample, we used results obtained from human gDNA (Promega #G3041, Madison, WI, USA). For normalization of DNAs from FFPE tissue block, combinations of DNAs extracted from 20 randomly selected normal gastric mucosae from FFPE gastrectomy specimens were used. Final data analysis was performed using nSolver software version 2.5.
CN was determined by averaging over three probes per region according to the manufacturer’s protocol. If the average CN was below 1.4, the gene was considered to have one copy; if it was between 1.5 and 2.4, it was considered to have two copies; and if it was between 2.5 and 3.4, it was considered to have three copies.
2.4 IHC for HER2
We performed IHC for HER2 in all cases with FFPE tissue blocks. After deparaffinization and rehydration, 4 µm unstained slides were stained for HER2. IHC for HER2 (PATHWAY HER-2/neu (4B5) rabbit monoclonal antibody, Ventana Medical Systems, Inc., Tucson, AZ) were performed in all cases with a BenchMarkXT automated stainer (Ventana Medical Systems, Inc., Tucson, AZ, USA) as previously described [18]. For FFPE samples, we tried to use the same blocks for both nCounter assay and HER2 IHC/ISH. However, different tumor blocks were used in 8 out of 135 FFPE samples because of inappropriate amounts of remaining tissue. Two pathologists (KMK, MEH) evaluated the results.
2.5 FISH and SISH for HER2
FISH or silver in situ hybridization (SISH) was performed in all available HER2 2+ cases on IHC with FFPE tissue blocks. In HER2 IHC negative group, ISH was performed in 11 discordant cases. In HER2 IHC positive group, ISH was performed in 2 discordant and 7 concordant cases. FISH was performed using dual-color DNA-specific probes from PathVision™ (Abbott/Vysis: LSI HER2 SpectrumOrange™ and CEP 17 SpectrumGreen™) as previously described [19]. We counted the hybridization signals in 20 nuclei per sample under a fluorescent microscope (Zeiss Axioskop) using filter sets recommended by Vysis (DAPI/Spectrum Orange dual bandpass, DAPI/Spectrum Green dual bandpass). Automated SISH was performed on the Ventana Benchmark XT (Ventana Medical Systems, Inc., Tucson, AZ) according to the manufacturer’s protocols for the INFORM HER2 DNA and Chromosome 17 probes as previously described [18].
All overlapping nuclei were excluded, and only nuclei with a distinct nuclear border were evaluated. HER2 gene was considered amplified when the ISH signal ratio of HER2/CEP17 was greater than or equal to 2.0 [20]. Polysomy of chromosome 17 was defined as more than 3 CEP17 signals on average.
2.6 Statistical Analysis
Statistical analysis was performed using SAS version 9.4 (SAS Institute, Cary, NC). Utilizing IHC as the reference standard, sensitivity was defined as the ratio of HER2 IHC positive cases among HER2 amplified cases by nCounter assay. The specificity was defined as the ratio of HER2 IHC negative cases among HER2 non-amplified cases. The positive predictive value (PPV) was defined as the ratio of HER2 amplified cases among HER2 IHC positive patients. The negative predictive value (NPV) was defined as the ratio of HER2 non-amplified cases among HER2 IHC negative patients. The cut-off of amplification by nCounter CNV assay was defined as 4 and the ratio of HER2 amplification by FISH is >2.0. For IHC equivocal (2+) cases, ISH results were used for the final results. Cohen’s kappa test was also performed. Association between the results of IHC and nCounter assay was determined by Fisher’s exact test.
The distribution of HER2 CN by nCounter assay according to IHC results was evaluated by ANOVA test. For analysis of discordant cases, the association between IHC heterogeneity and the nCounter assay results was evaluated by the two-sided Fisher exact test. The association between tumor population and nCounter results was evaluated by the two-sided Wilcoxon rank sum test. A p value <0.05 was considered significant.
3 Results
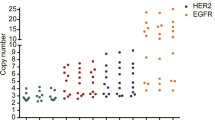
A total of 220 gastric cancer samples were analyzed. In the nCounter CNV assay, the estimated HER2 CN ranged from 1 to 68. When we defined amplification as CN ≥4 by nCounter, 43 (20 %) cancers were amplified out of 220 total cases. The results of HER2 IHC were 3+ in 16 samples (7.3 %), 2+ in 12 samples (5.5 %), 1+ in 47 samples (21.4 %), and 0 in 145 samples (65.9 %). Overall, the results of HER2 CN by nCounter assay compared to IHC/ISH results are summarized in Table 1. The distribution of HER2 CN by using nCounter assay was significantly different (p < 0.0001) according to IHC scores (Fig. 1). For FFPE samples (n = 135), the average HER2 CN seen in IHC positive, equivocal, and negative cases was 18.5 (range 3–68), 3.0 (range 2–5), and 3.0 (range 2–7), respectively. For FF samples (n = 85), the average HER2 CN seen in IHC positive, equivocal, and negative cases was 16.0 (range 2–52), 1.9 (range 1–2), and 1.9 (range 1–6), respectively.
The average HER2 gene copy numbers by the nCounter CNV assay in IHC negative (score 0 and 1+), equivocal (2+), and positive (3+) cases were 2.61, 2.0 and 17.25, respectively. The distribution of HER2 copy number by using nCounter assay was significantly different (p < 0.0001) according to IHC scores
Overall, the nCounter assay had a concordance rate of 83.4 % (kappa value 0.35) a sensitivity of 66.7 %, a specificity of 85.2 %, a NPV of 96 %, and a PPV of 32.6 % compared with HER2 IHC/ISH results (Table 2). There was a statistically significant association between the results of IHC and nCounter assay (p < 0.0001). Next, the concordance rate was also calculated according to the sample storage type (FFPE vs. FF) and is summarized in Table 1. FF samples revealed a higher concordance rate (91.5 %, kappa value 0.59) than FFPE samples (78.5 %, kappa value 0.27). While the sensitivity was lower in FF samples (54.5 %) than in FFPE samples (80 %), the specificity was higher in FF samples (97.2 %) than FFPE samples (78.4 %).
3.1 Analysis of the 16 HER2 IHC 3+ Cases
We found 16 HER2 IHC 3+ cases in which CN values detected by the nCounter CNV analyses varied from 2 to 68 (CN2, n = 1; CN3, n = 2, CN4, n = 4; CN5, n = 2; CN ≥10, n = 7) and 13 of them (81.3 %) were reported as HER2 CN gains (≥4). Three cases harboring a CN less than 4 (low) by nCounter were interpreted as discordant, and they are highlighted in gray in Table 3. Two of the three discordant cases were from FF biopsy samples. The remaining one was from a FFPE tumor in which HER2 IHC was performed in different tumor block other than used for nCounter analyses. For gastric cancer cases with CN equal to 4 or 5 by nCounter (highlighted by green in Table 2; Fig. 2), HER2 IHC showed heterogeneous overexpression, and the tumor purity was relatively low. In cancers with CN >10 by nCounter, all cases showed HER2 overexpression mostly with high percentage of positivity on IHC. However, there were no statistically significant associations between tumor percentage or IHC heterogeneity and CN detection by nCounter (p > 0.13) due to the small number of discordant cases.
The immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH) images of two HER2 IHC 3+ cases. a Regional heterogeneity was not identified on IHC, and the tumor cell percentage was 75 %. In this case, estimated number of HER2 copies was 68 in nCounter assay, b and the HER2/CEP17 ratio was 35 by FISH. c Regional heterogeneity was observed on IHC with 60 % of the tumor being 3+ and 40 % being 1+. Tumor cell percentage was 60 % of the whole specimen. Estimated number of HER2 copies was 5 in nCounter assay, d and the HER2/CEP17 ratio was 2.7 by FISH
3.2 Analysis of the 12 HER2 IHC 2+ Equivocal Cases
We found 12 HER2 IHC 2+ equivocal cases, in which 9 cases were available for FISH or SISH. Five cases (55.6 %) showed HER2 amplification while 4 cases (44.4 %) showed no amplification. Interestingly, we found four 2+ cases with chromosome 17 polysomy without (n = 2) or with (n = 2) HER2 amplification (Fig. 3). The precise results of all nCounter CNV assay, IHC, and FISH results in HER2 IHC 2+ cases are summarized in Table 4.
In the nCounter CNV assay, one case with ≥4 HER2 CN was confirmed to have HER2 amplification by in ISH (HER2/CEP17 ratio 3.0). Three cases with CN 2 by nCounter assay showed low-grade gene amplification (HER2/CEP17 ratio 2.4, 2.17 and 2.25) by ISH. All but one case showed heterogeneous expression of HER2 on IHC.
3.3 Analysis of the 192 HER2 IHC Negative Cases
Among the 192 IHC negative (0 or 1+) gastric cancer cases, 29 showed a HER2 CN ≥4 with the nCounter assay. For 29 cases with ≥4 HER2 CN, 27 (93.1 %) were from FFPE tumor blocks and only two cases (6.9 %) were from FF sample. By types of acquisition, 19 were from operation specimens and 10 were from biopsy samples. ISH was performed in all but two available cases and all of them showed no amplification of HER2 gene (Table 5).
4 Discussion
The purpose of this study was to test whether the nCounter CNV analysis could be used as a screening tool for CNV detection in a clinical setting. Previously, we tested the nCounter CNV analysis in 96 FFPE gastric cancer samples and validated HER2, MET, and KRAS results using IHC, FISH and real time PCR [3]. In the present study, we chose HER2 because IHC results represent gene amplification [16, 18] and further extended to a larger scaled cohort with diverse samples in terms of storage status and sizes of the tissue specimen. In 220 advanced gastric cancer patients, we compared the nCounter CNV analysis and IHC results and explored discordant cases thoroughly. Overall, we found a fair level of concordance (concordance rate 83.4 %, kappa value 0.35), with a sensitivity of 66.7 %, a specificity of 85.2 %, a NPV of 96 %, and a PPV of 32.6 %.
Based on sample acquisition types, FFPE samples showed a concordance rate of 78.5 % with a sensitivity of 80 % and a specificity of 78.4 %. FF samples showed a higher concordance rate (91.5 %; kappa value 0.59) with a lower sensitivity (54.5 %) and a higher specificity (97.2 %) compared to FFPE samples (78.5 %; kappa value 0.27). Interestingly, although we found high specificity in FF samples, the sensitivity was relatively low (54.5 %). This would be caused by different tissue samples used for IHC and nCounter CNV assay and the underlying intra-tumoral heterogeneity present in those discrepant cases. In this study, the cut-off of nCounter CNV assay was set as 4 as the ratio of HER2 amplification by FISH was >2.0. HER2 CN by nCounter CNV assay in most false-positive cases was 4 or 5 especially in FFPE samples. Given the strict cut-off, the concordance rate was relatively low in the present study. However, all 7 samples with high HER2 CN (≥10) showed a 100 % correlation with IHC results. Based on these observations, we concluded that nCounter CNV assay is a reliable method to detect high levels of amplification. However, caution is needed in cases with low CN alteration, especially in FFPE tissue samples.
The discordant cases were analyzed in detail. For the 16 HER2 IHC positive (3+) cases, we found 3 false-negative cases by nCounter CNV assay, of which two were FF biopsy samples and one was a FFPE tissue. For all the cases, different tumor blocks or tissue fragments were used for IHC and are likely caused by intra-tumoral heterogeneity. We also speculate that false-negative cases may be caused by a small population of HER2 amplified cells within tumors and normal cell contamination. Review of IHC slides of the false-negative cases by the nCounter CNV assay showed intra-tumoral heterogeneity in most cases.
Heterogeneous HER2 staining is a common biologic feature in gastric cancer and is estimated to be present in up to 30 % of HER2-positive gastric cancer cases [21]. Given the intra-tumoral HER2 heterogeneity and normal cell contamination, in cases that show low CN alterations by the nCounter assay, an additional IHC and/or ISH is necessary for confirmation.
For IHC equivocal (2+) cases, FISH or SISH confirmation is required according to the current treatment guideline [20]. Five out of nine available cases showed low-grade HER2 gene amplification. In addition, four cases revealed chromosome 17 polysomy consistent with the previous finding that polysomy of chromosome 17 is an important underlying cause of weak HER2 overexpression (2+) in non-amplified cases [18].
Finally, the nCounter assay was positive in 29 IHC-negative cases when defining amplification as ≥4 HER2 CN. In particular, FFPE samples often showed a higher CN compared to FF samples. The PPV in FFPE samples was 22.9 %. Not only does formaldehyde lead to cross linkages between DNA or RNA and proteins, but it also results in fragmentation of DNA [22]. The background signal on the FFPE samples was higher compared to the FF samples. Fragmentation of DNA is influenced by the fixation procedure and AluI can be negatively affected by remaining contaminations of the extraction method or formalin. This is a plausible explanation for the increased false-positive results in this FFPE cohort. The 260/280 ratio and 260/230 ratio of discordant cases were acceptable in most of the tested samples. We did not measure the fragment size of samples because the measurement of fragment size was not mandatory in the manual. However, measurement of fragment size in future study can elucidate the causes of discordance in FFPE samples.
Somatic CN alteration is the hallmark of many cancers. An efficient screening test to help illuminate the CN alterations seen within a particular cancer has great potential clinical utility, particularly in this era of targeted therapy. Although FISH has become the gold standard for detecting many of these CN alterations, it is time consuming and inefficient, especially when looking at multiple different genes. On the other hand, IHC is available in most pathology laboratories and offers more advantages in terms of cost effectiveness. Furthermore, IHC and ISH are able to detect tumor heterogeneity and amplified tumor cells with low percentage, which allows for accurate results. However, there are not many available IHC antibodies with proven quality and high-throughput technologies that make it possible to detect a large number of DNA alterations simultaneously, hence facilitating genome-wide screening. So, there have been several new attempts to detect CNV including NGS and droplet digital PCR [15, 23]. Compared with these methods, the nCounter CNV assay offers many advantages, including a low required volume, the utilization of FFPE tissue blocks, and no amplification step is needed.
In conclusion, we observed a fair level of concordance between the custom nCounter CNV assay and IHC for the detection of HER2 amplification. For CNV screening, nCounter CNV assay seems a reliable method; however, caution should be taken in the interpretation of results.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi:10.1016/j.cell.2011.02.013.
Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12(7):791–801.
Kim S, Lee J, Hong ME, Do IG, Kang SY, Ha SY, et al. High-throughput sequencing and copy number variation detection using formalin fixed embedded tissue in metastatic gastric cancer. PLoS One. 2014;9(11):e111693. doi:10.1371/journal.pone.0111693.
Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–33. doi:10.1038/ng.2762.
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–97. doi:10.1016/S0140-6736(10)61121-X.
Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–26.
Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19(9):2572–83. doi:10.1158/1078-0432.CCR-12-3898.
Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29(36):4803–10. doi:10.1200/JCO.2011.35.4928.
Zhang L, Yang J, Cai J, Song X, Deng J, Huang X, et al. A subset of gastric cancers with EGFR amplification and overexpression respond to cetuximab therapy. Sci Rep. 2013;3:2992. doi:10.1038/srep02992.
Lee J, Kim KM, Kang WK, Ou SH. Innovative personalized medicine in gastric cancer: time to move forward. Clin Genet. 2014;86(1):37–43. doi:10.1111/cge.12408.
Selinger CI, Rogers TM, Russell PA, O’Toole S, Yip P, Wright GM, et al. Testing for ALK rearrangement in lung adenocarcinoma: a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol. 2013;26(12):1545–53. doi:10.1038/modpathol.2013.87.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–21.
Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29(3):263–4. doi:10.1038/ng754.
Ylstra B, van den Ijssel P, Carvalho B, Brakenhoff RH, Meijer GA. BAC to the future! Or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH). Nucleic Acids Res. 2006;34(2):445–50. doi:10.1093/nar/gkj456.
Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24(12):2022–32. doi:10.1101/gr.175141.114.
Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5(1):63–9.
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25. doi:10.1038/nbt1385.
Cho J, Jeong J, Sung J, Sung CO, Kim KM, Park CK, et al. A large cohort of consecutive patients confirmed frequent HER2 positivity in gastric carcinomas with advanced stages. Ann Surg Oncol. 2013;20(Suppl 3):S477–84. doi:10.1245/s10434-012-2818-0.
Cho EY, Srivastava A, Park K, Kim J, Lee MH, Do I, et al. Comparison of four immunohistochemical tests and FISH for measuring HER2 expression in gastric carcinomas. Pathology. 2012;44(3):216–20. doi:10.1097/PAT.0b013e3283513e8b.
Ruschoff J, Hanna W, Bilous M, Hofmann M, Osamura RY, Penault-Llorca F, et al. HER2 testing in gastric cancer: a practical approach. Mod Pathol. 2012;25(5):637–50. doi:10.1038/modpathol.2011.198.
Kim KM, Bilous M, Chu KM, Kim BS, Kim WH, Park YS, et al. Human epidermal growth factor receptor 2 testing in gastric cancer: recommendations of an Asia-Pacific task force. Asia Pac J Clin Oncol. 2014;10(4):297–307. doi:10.1111/ajco.12263.
Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2(6):e537. doi:10.1371/journal.pone.0000537.
Belgrader P, Tanner SC, Regan JF, Koehler R, Hindson BJ, Brown AS. Droplet digital PCR measurement of HER2 copy number alteration in formalin-fixed paraffin-embedded breast carcinoma tissue. Clin Chem. 2013;59(6):991–4. doi:10.1373/clinchem.2012.197855.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
SA, MH, MVV, YP, SK, SP, WK, SJ, MW, JL, and KM have declared no conflicts of interest.
Funding
This research was supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDHI), funded by the Ministry of Health and Welfare, Republic of Korea (HI14C2750 to YSP, HI14C3418 to JL) and National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A3015504 to KMK) and 20 by 20 project of Samsung Medical Center (GF01140111).
Ethical approval and informed consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
S. Ahn and M. Hong contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ahn, S., Hong, M., Van Vrancken, M. et al. A nCounter CNV Assay to Detect HER2 Amplification: A Correlation Study with Immunohistochemistry and In Situ Hybridization in Advanced Gastric Cancer. Mol Diagn Ther 20, 375–383 (2016). https://doi.org/10.1007/s40291-016-0205-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-016-0205-4