Abstract
Background
Drug transporters and drug-metabolizing enzymes have been linked to drug-induced hepatotoxicity. Solute carrier organic anion transporter family member 1B1 (SLCO1B1), cytochrome P450 2E1 (CYP2E1), and UDP glucuronosyltransferase 1A1 (UGT1A1) were selected as candidate genes to explore their association with susceptibility to anti-tuberculosis drug-induced hepatotoxicity (ATDH).
Methods
Thirty-four tag single nucleotide polymorphisms (tagSNPs) in SLCO1B1, CYP2E1, and UGT1A1 with 10-kb expansion up- and down-stream were genotyped in 461 patients with ATDH and 466 patients without ATDH in a prospective 1:1 matched case–control study. The frequencies and distributions of genotypes and haplotypes were compared between the groups using three genetic models (dominant, recessive, and additive) to identify associations with susceptibility to ATDH.
Results
Patients with the rs4149034 G/A, rs1564370 G/C, and rs2900478 T/A genotypes of SLCO1B1 had a significantly lower risk of ATDH, while those carrying the rs2417957 T/T and rs4149063 T/T genotypes had an increased risk. The rs4148323 A/A genotype of UGT1A1 was found to significantly reduce the risk of ATDH. Haplotype analysis showed the TGTG, TTTC, and GTTC haplotypes of SLCO1B1 were associated with an increased ATDH risk, whereas the GACC haplotype was related to a reduced risk. The ATG haplotype of UGT1A1 reduced the risk of ATDH. Moreover, treatment outcomes in tuberculosis patients were further affected by genetic variants of SLCO1B1.
Conclusions
Genetic polymorphisms of SLCO1B1 and UGT1A1 were found to be associated with susceptibility to ATDH. Molecular identification of susceptibility genes provides a theoretical foundation for predicting the likelihood of ATDH and predicting treatment outcomes in tuberculosis patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This Chinese population-based prospective case–control study revealed significant associations between genetic polymorphisms of SLCO1B1 and UGT1A1 genes and susceptibility to anti-tuberculosis drug-induced hepatotoxicity. | |
The genetic variants of SLCO1B1 gene have influences on the treatment outcomes in TB patients. |
1 Introduction
China has the third largest burden of tuberculosis (TB) in the world with 78.8–106.0 million newly diagnosed TB patients estimated in 2015, contributing to an incidence of 8.83% worldwide [1]. Chemotherapy of TB is critical, but TB drugs are frequently associated with serious adverse reactions, such as anti-TB drug-induced hepatotoxicity (ATDH). Our preliminary cohort study discovered that the incidence of ATDH in hospitalized patients treated with first-line anti-TB drugs was approximately 12.9%, leading to decreases in the sputum conversion rate, and closure rate of the lung cavities, and then further significantly increasing the rate of treatment failure [2]. Meanwhile, drug-induced liver injury (DILI) if unresolved for over 1 year will transform into chronic DILI, causing a small group of patients to suffer severe adverse consequences such as early-stage liver cirrhosis and acute liver failure and even to require liver transplantation [3, 4].
The underlying causes of ATDH are multifactorial. Our previous study identified several independent risk factors for ATDH including both HBsAg- and HBeAg-positive hepatitis B, systemic lupus erythematosus (SLE), plasma albumin ≤25 g/L, and chronic alcohol abuse. The clinical risk of hepatotoxicity in patients with one or more of these factors was significantly higher after treatment with first-line anti-TB drugs [2], which was consistent with the findings of additional studies [3, 5,6,7,8]. Meanwhile, accumulating evidence shows that differences in the adverse effects of anti-TB drugs among individuals and between different races are mainly determined by genetic factors. It has been shown that genetic variants of drug transporters [9, 10] and drug-metabolizing enzymes [11,12,13,14,15] can cause changes in the functions of related substrates, thus affecting drug absorption, distribution, and metabolism in vivo. However, there has been a lack of consistency in the findings for ATDH-susceptible genes even in the studies comparing patients from the same ethnic group due to the following limitations in study design: (1) the numbers of samples in previous studies, in particular, the number in the case group, have been very small, often only a few dozen. A large sample pool is one of the most critical factors in gene single nucleotide polymorphism (SNP) and phenotypic association studies. Theoretically, the results are more reliable with more case data. However, it is apparently difficult to obtain sufficient sample sizes because the incidence of ATDH in clinical practice is low [16]. (2) Only a few SNP loci were included in previous studies, which often selected repeats of a single or a few SNP loci that could not cover a majority of the target gene.
In the present study, the transport protein solute carrier organic anion transporter family member 1B1 (SLCO1B1), Phase I DME cytochrome P450 2E1 (CYP2E1), and Phase II DME UDP glucuronosyltransferase 1A1 (UGT1A1) were selected as candidate genes. A tagSNP is a representative SNP in a region of the genome with high linkage disequilibrium, which enables the full haplotype information to be sufficiently captured by a small fraction of tagSNPs without genotyping every SNP in a chromosomal region. The tagSNP strategy has been proven to be an effective and valuable tool in the study of genetic polymorphisms, with similar power as haplotype data [17]. In the present study, we genotyped all tagSNPs screened from the target candidate genes using bioinformatic tools to identify associations with susceptibility to ATDH and further to determine the effects of positive SNPs on the prognosis of TB patients in a Chinese Han population.
2 Methods
2.1 Ethical Approval
The present study was approved by the Ethics Committees of both Tongji University School of Medicine and Shanghai Pulmonary Hospital. Written informed consent was received from patients.
2.2 Study Design and Case Population
The TB patients enrolled in this prospective one-to-one matched case–control study were aged from 18 to 65 years, receiving directly observed treatment short-course, and hospitalized at Shanghai Pulmonary Hospital between March 2012 and August 2015. Patients with ATDH after anti-TB therapy served as case group and patients without ATDH served as control group. Patients in the control group were selected and matched with patients in the case group by age (±3 years) and gender. A standard integrated initial therapy with 2–3 months of isoniazid (5 mg/kg), rifampicin (10 mg/kg), ethambutol (15 mg/kg), and pyrazinamide (25 mg/kg) was initiated for all patients, followed by a consolidated treatment with isoniazid, rifampicin, and ethambutol according to the guideline of the Chinese Medical Association. The exclusion criteria were as follows: (1) presence of any basic hepatic diseases such as viral hepatitis, hepatic adipose infiltration, cirrhosis, autoimmune liver disease, and alcoholic liver disease, etc.; (2) had liver dysfunction before anti-TB therapy initiation; (3) confirmed liver injury induced by another drug or unknown cause; (4) having received non-standard treatment regimen initially; (5) drug resistance found in the initial antimicrobial susceptibility test, identification of non-tuberculous mycobacteria, or development of non-liver-related side effects.
During the treatment of TB, patients underwent routine blood examination, and the hepatic enzymes as well as bilirubin levels were evaluated every month. ATDH was diagnosed according to the modified criteria for DILI [18, 19] and the standard diagnostic criteria established by the Chinese Medical Association. The Council for International Organizations of Medical Science (CIOMS)/Roussel Uclaf Causality Assessment Method (RUCAM) scale was used in causality assessment of liver damage [20]. Briefly, ATDH was defined as: (1) increased serumalanine aminotransferase (ALT) that was greater than 5× the upper limit of normal (ULN) or an alkaline phosphatase (ALP) level greater than 2× ULN; or serum ALT level was more than 3× ULN combined with a total bilirubin (TBIL) level greater than 2× ULN; and (2) a total score on the CIOMS/RUCAM scale of more than 8 points [20].
2.3 Bioinformatic Statistics of tagSNPs Screened from SLCO1B1, CYP2E1 and UGT1A1 Genes
The candidate genes SLCO1B1, CYP2E1 and UGT1A1 were loaded into the International HapMap Project SNP database (HapMap Data Rel 28 Phase II + III, August 10, on NIBI B36 assembly, dbSNP b126) for screening of SNPs within expansions 10 kb up- and down-stream of the gene region. All the tagSNPs obtained were then introduced into HaploView software version 4.2 (Broad Institute, Cambridge, MA, USA) with criteria for locus quality control as follows: p value for Hardy–Weinberg equilibrium (HWE-p) >0.001, minor genotype >0.75, minor allele frequency >0.1, and r 2 > 0.8. Finally, a total of 34 tagSNPs were selected, including 21 from SLCO1B1, 9 from CYP2E1, and 4 from UGT1A1 (Table 1).
2.4 DNA Isolation and Genotyping
A whole blood sample was collected from each patient for genomic DNA extraction. DNA was extracted using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen GmbH, Germany) with the standard protocol given in the manufacturer’s instructions, and genotyped using a custom-by-design 48-Plex SNPscanTM Kit (Cat#: G0104; Genesky Biotechnologies Inc., Shanghai, China). Briefly, a sample of 100–200 ng DNA was first denatured at 98 °C for 5 min in a 10-µL reaction containing 1 × DNA lysis buffer and then mixed well with 10 µL ligation mix composed of 2 µL 10 × ligase buffer, 0.5 µL ligase, 1 µL probe mixture, and 6.5 µL Milli-Q water. The ligation reaction was carried out in an ABI2720 thermal cycler with the following program: 98 °C for 2 min, 5 cycles at 95 °C for 30 sec and 58 °C for 3 h, 94 °C for 2 min, and finishing at 72 °C. Two 48-plex fluorescence polymerase chain reactions (PCRs) were performed for each ligation product. PCRs were prepared in a 20-µL mixture containing 2 µL 10× Takara PCR buffer, 2.4 µL dNTP mix (2.5 mM), 0.8 µL MgCl2 (25 mM), 0.3 µL primer mix set A or set B, 0.8 U Taq polymerase, and 1 µL ligation product. PCR products were separated and detected by capillary electrophoresis in an ABI3730XL sequencer. Raw data were analyzed according to the information obtained for the labeling dye color and fragment size of the allele-specific ligation-PCR product. Genotyping was conducted with blinding of the case or control status. Repeated genotyping was performed with the same assay in a random selection of 4% of samples to ensure the data quality.
2.5 Follow-up of Treatment Outcomes
All patients were followed up for at least 3 months after completion of anti-TB therapy. The effects of different genotypes of susceptible SNPs we discovered on the prognosis of TB patients were analyzed according to the treatment failure rate and sputum conversion rate. An unsuccessful outcome was defined as death or therapeutic failure, including persistent or relapsed positive sputum smear/culture, development of drug resistance, or exacerbation of lesions during the initial therapy.
2.6 Statistical Analysis
The observed and expected gene frequencies as well as the HWE-p values were estimated using GENEPOP. The distributions of each SNP genotype in the case group and control group were analyzed using binary logistic regression, and odds ratios (ORs); 95% confidence intervals (CIs) were estimated with SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The frequencies of alleles and genotypes in the two groups were compared using three different genetic models: dominant, recessive, and additive. The linkage disequilibrium profiles of SNPs were mapped using Haploview software version 4.2 (Broad Institute, Cambridge, MA, USA). The differences in the frequencies of haplotypes between the two groups were analyzed by calculating ORs and 95% CIs. The treatment failure rates of patients with different genotypes of susceptible SNPs were compared by chi-square test (χ 2). The sputum conversion rate of patients over time in the different groups was plotted using the Kaplan–Meier method and compared by the Log-Rank test. A two-tailed p value <0.05 was considered statistically significant.
3 Results
3.1 Patient Characteristics
A total of 927 study patients successfully completed peripheral blood sampling for DNA extraction and genotyping and follow-up. The case group contained 461 patients in total with an average age of 38.2 ± 13.7 years, among whom 283 were male and 178 were female. The control group consisted of 466 patients with an average age of 38.9 ± 13.5 years, among whom 287 were male and 179 were female.
3.2 Associations of SLCO1B1, CYP2E1, and UGT1A1 Polymorphisms with Susceptibility to ATDH
The allele frequency at each locus was in HWE (Table 1). Five tagSNPs from SLCO1B1 were detected to be closely related to ATDH susceptibility. Patients with the rs4149034 G/A (OR 0.642, p = 0.003), rs1564370 G/C (OR 0.736, p = 0.029), and rs2900478 T/A (OR 0.652, p = 0.012) genotypes had a significantly lower risk of ATDH, whereas patients carrying the rs2417957 T/T (OR 2.197, p = 0.002) and rs4149063 T/T (OR 1.704, p = 0.032) genotypes had an increased risk of developing ATDH. In the Dom model, both rs4149034 G/A + G/G (OR 0.728, p = 0.023) and rs2900478 T/T + A/A (OR 0.667, p = 0.014) genotypes were associated with a decreased risk of ATDH. In the Rec model, the rs2417957 T/T genotype (OR 2.067, p = 0.003) was found to increase the risk of ATDH. In the Add model, alleles of rs2417957 minor T (OR 1.328, p = 0.006) and rs4149063 minor T (OR 1.256, p = 0.023) were associated with an increased risk of ATDH, whereas the rs2900478 minor A allele (OR 0.712, p = 0.025) was related to a reduced risk of ATDH.
The UGT1A1 gene rs4148323 A/A genotype was found to significantly reduce the risk of ATDH (OR 0.371, p = 0.020). The rs4148323 A/A genotype (OR 0.394, p = 0.023) in the Rec model and the minor A allele (OR 0.791, p = 0.040) in the Add model were significantly associated with a reduced risk of ATDH. No association was observed between any tagSNP of CYP2E1 and susceptibility of ATDH (Tables 2, 3).
3.3 Associations of Haplotypes of SLCO1B1, CYP2E1, and UGT1A1 Genes with Susceptibility to ATDH
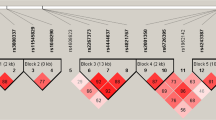
The linkage disequilibrium distributions of 21 loci of SLCO1B1, 10 loci of CYP2E1, and 4 loci of UGT1A1 are shown in Fig. 1. Twenty-one loci of SLCO1B1 were divided into three linkage disequilibrium blocks, with eight SNPs (rs327543, rs852549, rs4149022, rs12580258, rs4149023, rs16923519, rs11045802, and rs7138177) grouped in Block 1, 4 SNPs (rs2417957, rs4149045, rs4149050, and rs1564370) grouped in Block 2, and 4 SNPs (rs4149063, rs2900478, rs11513225, and rs12578392) grouped in Block 3. Ten loci of CYP2E1 were divided into two linkage disequilibrium blocks, 2 SNPs (rs10857736 and rs3813867) in Block 1 and 6 SNPs (rs915908, rs4646976, rs743535, rs2249695, rs1952467, and rs4240522) in Block 2. Three loci of UGT1A1, rs4148323, rs4148326, and rs12479045, were centered in one block.
Haplotypes were assigned according to linkage disequilibrium distribution, and relative distribution frequencies of haplotypes in the case and control groups were analyzed. Four haplotypes of SLCO1B1 were associated with ATDH susceptibility, and among them TGTG (OR 1.286, p = 0.015), TTTC (OR 1.260, p = 0.022), and GTTC (OR 2.249, p = 0.03) were discovered to increase the risk of ATDH, while GACC (OR 0.708, p = 0.023) reduced the risk. Haplotype ATG (OR 0.793, p = 0.045) of UGT1A1 was found to potentially decrease the risk of ATDH. None of the haplotypes of CYP2E1 was found to be associated with ATDH (Table 4).
3.4 Impact of SLCO1B1, CYP2E1, and UGT1A1 Variants on Treatment Outcomes in TB Patients
In the present study, 91 of 927 cases (9.8%) were labeled as unsuccessful treatment. The treatment failure rate of patients carrying the SLCO1B1 rs4149034 G/A genotype (7.8%; 36/462) was significantly lower (OR 0.630, p = 0.039) than that of patients carrying the A/A + G/G genotypes (11.8%; 55/465). No additional SNP genotype was found to be significantly associated with the treatment failure rate.
Of the 927 cases, 447 patients had positive sputum smear/culture results, and among them 27 (6.04%) remained positive through the end of post-therapy follow-up. The sputum conversion rate and median time in carriers of the SLCO1B1 rs4149034 G/A genotype were 95.3% (203/213) and 1.5 months (95% CI 1.336–1.664 months), and these values were significantly higher (Log rank p = 0.033) than the 92.7% (217/234) and 2.0 months (95% CI 1.799–2.201 months) for those carrying A/A + G/G genotypes (Fig. 2). Moreover, a sputum conversion rate of 90.7% (39/43) among patients carrying the SLCO1B1 rs2147957 T/T genotype with a median time of 2.5 months (95% CI 1.795–3.205 months) was significantly lower (Log rank p = 0.026) than the sputum convesion rate of 94.3% (381/404) among those carrying C/C + T/C genotypes, for whom the median time was 1.5 months (95% CI 1.350–1.650 months; Fig. 3). No other individual SNPs were found to be associated with the sputum conversion rate in TB patients.
Comparisons of sputum smear and culture conversion rates in patients carrying different genotypes of the rs4149034 SNP of SLCO1B1. SNP single nucleotide polymorphism, SLCO1B1 solute carrier organic anion transporter family member 1B1, CYP2E1 cytochrome P450 2E1, UGT1A1 UDP glucuronosyltransferase 1A1
Comparisons of sputum smear and culture conversion rates in patients carrying different genotypes of the rs2147957 SNP of SLCO1B1. SNP single nucleotide polymorphism, SLCO1B1 solute carrier organic anion transporter family member 1B1, CYP2E1 cytochrome P450 2E1, UGT1A1 UDP glucuronosyltransferase 1A1
4 Discussion
In this present study, we screened and selected all tagSNPs spanning SLCO1B1, CYP2E1, and UGT1A1 with 10-kb expansion up- and down-stream by bioinformatic statistical methods and genotyped 927 samples from a Chinese Han population in a prospective 1:1 matched case–control study. We further analyzed the impact of different genotypes of susceptible SNPs on treatment outcomes in TB patients and demonstrated remarkably significant associations of gene polymorphisms of SLCO1B1 and UGT1A1 with ATDH susceptibility and the further effects of variants of SLCO1B1 on treatment outcomes in Chinese TB patients.
Organic anion-transporting polypeptides (OATPs) belong to a large group of uptake membrane transporters from the solute carrier family (SLC). In addition to transport of bile acid and other endogenous substances, OATPs are also involved in transporting of a variety of exogenous substances, particularly drugs, playing a key role in drug absorption, distribution, and elimination [9, 10, 21]. OATP1B1 is an important member of the OATP family, specifically present in the basolateral membrane of hepatocytes. It is currently known that a variety of drugs, including methotrexate, sorafenib, statins, rifampicin and rifabutin among the first-line anti-TB drugs, can be absorbed and transported from the hepatic portal system into the liver cells as a substrate for OATP1B1 to be metabolized and eliminated [21,22,23]. Therefore, the expression and transport function of OATP1B1 in the liver tissue potentially influence the drug concentration in blood, bioavailability, and side effects. OATP1B1 is encoded by SLCO1B1 on chromosome 12p. Our study revealed that the risk of ATDH was reduced by genotypes of SLCO1B1 including rs4149034 G/A, rs1564370 G/C, and rs2900478 T/A, but increased significantly by the rs2147957 T/T and rs4149063 T/T genotypes by up to 2.197- and 1.704-fold, respectively. Although the mechanisms by which different SNP genotypes of SLCO1B1 alter the risk of ATDH remain unknown, it has already been made clear that rifampicin is a substrate for OATP1B1 that is transported into liver cells for metabolism. As an important anti-TB drug with known hepatotoxicity, rifampicin is also a powerful catalyst that induces a variety of metabolizing enzymes such as cytochrome P450 in the liver. When co-administered with drugs including isoniazid and pyrazinamide, rifampicin will react with intermediate metabolites from the drugs in synergistic action, leading to increased toxicity and damage of liver cells [24]. Weiner et al. [22] found that the SLCO1B1 c.463 C>A gene polymorphism resulted in a lower rifampicin concentration in blood. Thus, we conclude that different genotypes may result in upregulation or downregulation of SLCO1B1, leading to functional changes in OATP1B1 binding and transporting of drugs and therefore altering the blood concentrations and metabolite levels of anti-TB drugs. Decreased expression of SLCO1B1 may result in the reduced transport capacity of OATP1B1, leading to weakened clearance of rifampicin and its toxic metabolites and thereby causing hepatic injury. Further studies should be conducted to investigate how the variants of the loci affect the transport function of OATP1B1 and the corresponding changes in the concentrations of anti-TB drugs and metabolites that lead to hepatic injury.
The CYP enzyme system is one of the most important Phase I-metabolizing enzymes, metabolizing about 60–70% of clinical drugs. CYP2E1 coded by gene CYP2E1 on chromosome 10q, is a member of the CYP450 family mainly distributed in the liver. Whether genetic polymorphisms of CYP2E1 are related to ATDH susceptibility is still controversial. Singla et al. [15] reported that the heterozygous genotype of c1/c2 of CYP2E1*5B was associated with ATDH susceptibility (OR 7.47, 95% CI 1.263–34.32, p = 0.04, n = 17), and Bose [25] found that the DraI C/D genotype of CYP2E1 was associated with the occurrence of ATDH (OR 3.22, 95% CI 1.28–8.08, p = 0.00157, n = 41). Sheng et al. [26] demonstrated an association of the c1/c1 genotype of gene CYP2E1 with ATDH via meta-analysis. However, several other studies showed no evidence of a relationship between CYP2E1 and ATDH [12, 13]. In the present study, the frequencies and distributions of certain genotypes and haplotypes of 10 tagSNPs from CYP2E1 within 10-kb expansion up- and down-stream were verified in a larger sample pool; however, none of the SNPs was found to be associated with ATDH susceptibility.
Uridine 5′-diphospho-glucuronosyl transferase (UGT) is one of the most important Phase II metabolizing enzymes, mainly catalyzing endogenous (bilirubin, steroids, etc.) or exogenous compounds such as drugs and phenols that combine with cofactor uridine diphosphate glucuronic acid and increase the heteropolarity of lipophilic substrates, promoting excretion of the compound in urine or bile. Thus, the glucuronidation reaction can reduce the activity or toxicity of the potential compounds while accelerating the elimination of the compounds, playing an important role in metabolic detoxification [23, 27, 28]. As a member of the UGT family, UGT1A1 is an enzyme that is encoded by the UGT1A1 gene on chromosome 2q and mainly distributed in the liver. A few studies have demonstrated a correlation between UGT1A1 polymorphisms and ATDH susceptibility. One study in only 17 patients with ATDH showed that UGT1A1*27 and UGT1A1*28 were associated with an increased risk of ATDH (OR 13.859; 95% CI 1.085–177.056) [29]. However, Chen [30] found no association between UGT1A1 and ATDH in a study with only 87 cases. Our study revealed that the UGT1A1 rs4148323 A/A genotype significantly reduced the risk of ATDH, and its minor A allele significantly decreased the risk of ATDH in the Add model. The ATG haplotype of UGT1A1 was potentially associated with a decreased risk of ATDH. Therefore, we propose that the presence of mutant A or haplotype ATG might increase the expression of UGT1A1 to enhance the activity of the UGT1A1 enzyme, resulting in accelerated elimination of toxic metabolites from anti-TB drugs through glucuronidation reaction to protect the liver.
The SLCO1B1 and UGT1A1 genes were confirmed to be associated with susceptibility to ATDH in the present study. Our previous cohort study demonstrated the negative impact of ATDH on the treatment failure rate, sputum conversion rate, and closure rate of the lung cavities in TB patients [2]. Therefore, we further investigated the effects of different genotypes of SLCO1B1 and UGT1A1 on treatment outcomes in TB patients. The results showed possible impacts of genetic variants of SLCO1B1 on the prognosis of TB patients. Patients carrying the SLCO1B1 rs4149034 G/A genotype experienced a significantly lower treatment failure rate, shorter median time to sputum negative conversion, and an increased sputum conversion rate over time. On the other hand, patients carrying the SLCO1B1 rs2147957 T/T genotype had a longer median time to sputum negative conversion and a significantly decreased sputum conversion rate over time. We can infer that the two polymorphic loci rs4149034 G/A and rs2147957 T/T may have caused a decrease and increase, respectively, in the incidence of ATDILI in TB patients, leading to an impact on the treatment outcomes. These results may be practically beneficial and useful in predicting outcomes of TB treatment after chemotherapy.
5 Conclusion
In conclusion, both genotype and haplotype analysis in the present study revealed that SLCO1B1 and UGT1A1 gene polymorphisms were closely related to ATDH susceptibility in a Chinese Han population. Moreover, genetic variants of SLCO1B1 were associated with the treatment outcomes in TB patients. Molecular identification of susceptibility genes provides a theoretical foundation for planning individualized treatment schemes and better predicting the likelihood of adverse effects in patients. Further studies are needed to better understand the molecular mechanisms by which susceptibility to ATDH is regulated by the expression of ATDH-susceptibility genes and proteins.
References
World Health Organization. Global tuberculosis report 2016. (WHO/HTM/TB/2016.13).
Sun Q, Zhang Q, Gu J, et al. Prevalence, risk factors, management, and treatment outcomes of first-line antituberculous drug-induced liver injury: a prospective cohort study. Pharmacoepidemiol Drug Saf. 2016;25:908–17.
Medina-Caliz I, Robles-Diaz M, Garcia-Muñoz B, Spanish DILI registry, et al. Definition and risk factors for chronicity following acute idiosyncratic drug-induced liver injury. J Hepatol. 2016;65(3):532–42.
Chen M, Suzuki A, Borlak J, Andrade RJ, Lucena MI. Drug-induced liver injury: interactions between drug properties and host factors. J Hepatol. 2015;63(2):503–14.
Sharma SK, Balamurugan A, Saha PK, Pandey RM, Mehra NK. Evaluation of clinical and immunogenetic risk factors for the development of hepatotoxicity during antituberculosis treatment. Am J Respir Crit Care Med. 2002;166:916–9.
Shakya R, Rao BS, Shrestha B. Incidence of hepatotoxicity due to antitubercular medicines and assessment of risk factors. Ann Pharmacother. 2004;38:1074–9.
Breen RA, Miller RF, Gorsuch T, et al. Adverse events and treatment interruption in tuberculosis patients with and without HIV co-infection. Thorax. 2006;61:791–4.
Yee D, Valiquette C, Pelletier M, Parisien I, Rocher I, Menzies D. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am J Respir Crit Care Med. 2003;167:1472–7.
Kim SH, Kim SH, Lee JH, et al. Polymorphisms in drug transporter genes (ABCB1, SLCO1B1 and ABCC2) and hepatitis induced by antituberculosis drugs. Tuberculosis (Edinb). 2012;92(1):100–4.
Chen R, Wang J, Tang S, et al. Association of polymorphisms in drug transporter genes (SLCO1B1 and SLC10A1) and anti-tuberculosis drug-induced hepatotoxicity in a Chinese cohort. Tuberculosis (Edinb). 2015;95(1):68–74.
Guaoua S, Ratbi I, Laarabi FZ, et al. Distribution of allelic and genotypic frequencies of NAT2 and CYP2E1 variants in Moroccan population. BMC Genet. 2014;15:156.
Sharma SK, Jha BK, Sharma A, et al. Genetic polymorphisms of CYP2E1 and GSTM1 loci and susceptibility to anti-tuberculosis drug-induced hepatotoxicity. Int J Tuberc Lung Dis. 2014;18(5):588–93.
Tang SW, Lv XZ, Zhang Y, et al. Cytochrome P450 2E1 gene polymorphisms/haplotypes and anti-tuberculosis drug-induced hepatitis in a Chinese cohort. PLoS One. 2013;8(2):e57526.
Ng CS, Hasnat A, Al Maruf A, et al. N-Acetyltransferase 2 (NAT2) genotype as a risk factor for development of drug-induced liver injury relating to antituberculosis drug treatment in a mixed-ethnicity patient group. Eur J Clin Pharmacol. 2014;70(9):1079–86.
Singla N, Gupta D, Birbian N, Singh J. Association of NAT2, GST and CYP2E1 polymorphisms and anti-tuberculosis drug-induced hepatotoxicity. Tuberculosis (Edinb). 2014;94(3):293–8.
Daly AK. Drug-induced liver injury: past, present and future. Pharmacogenomics. 2010;11(5):607–11.
Zhang K, Qin ZS, Liu JS, Chen T, Waterman MS, Sun F. Haplotype block partitioning and tag SNP selection using genotype data and their applications to association studies. Genome Res. 2004;14:908e16.
Abboud G, Kaplowitz N. Drug-induced liver injury. Drug Saf. 2007;30:277–94.
Aithal GP, Watkins PB, Andrade RJ, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther. 2011;89:806–15.
Danan G, Benichou C. Causality assessment of adverse reactions to drugs. a novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol. 1993;46:1323–30.
Hennig S, Naiker S, Reddy T, et al. Effect of SLCO1B1 polymorphisms on rifabutin phamacokinetics in African HIV-infected patients with tuberculosis. Antimicrob Agents Chemothor. 2015;60(1):617–20.
Weiner M, Peloquin C, Burman W, et al. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother. 2010;54(10):4192–200.
Bins S, Lenting A, El Bouazzaoui S, et al. Polymorphisms in SLCO1B1 and UGT1A1 are associated with sorafenib-induced toxicity. Pharmacogenomics. 2016;17(14):1483–90.
Tostmann A, Boeree MJ, Aarnoutse RE, de Lange WC, van der Ven AJ, Dekhuijzen R. Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. Gastroenterol Hepatol. 2008;232:192–202.
Bose PD, Sarma MP, Medhi S, Das BC, Husain SA, Kar P. Role of polymorphic N-acetyl transferase2 and cytochrome P4502E1 gene in antituberculosis treatment-induced hepatitis. J Gastroenterol Hepatol. 2011;26:312–8.
Sheng YJ, Wu G, He HY, Chen W, Zou YS, Li Q, Zhong L, Huang YM, Deng CL. The association between CYP2E1 polymorphisms and hepatotoxicity due to anti-tuberculosis drugs: a meta-analysis. Infect Genet Evolut. 2014;24:34–40.
Kim JY, Cheong HS, Park BL, Kim LH, Namgoong S, Kim JO, Kim HD, Kim YH, Chung MW, Han SY, Shin HD. Comprehensive variant screening of the UGT gene family. Yonsei Med J. 2014;55(1):232–9.
Gagné JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol. 2002;62(3):608–17.
Chang JC, Liu EH, Lee CN, et al. UGT1A1 polymorphisms associated with risk of induced liver disorders by anti-tuberculosis medication. Int J Tuberc Lung Dis. 2012;16(3):376–8.
Chen R, Wang J, Tang SW, et al. CYP7A1, BAAT and UGT1A1 polymorphisms and susceptibility to anti-tuberculosis drug-induced hepatotoxicity. Int J Tuberc Lung Dis. 2016;20(6):812–8.
Acknowledgements
We specially acknowledge the technical assistance from Genesky Biotechnologies Inc. Shanghai, China. We also thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by the National Nature Science Foundation of China (no. 81400006), the Shanghai Pujiang Program (no. 16PJD041), and the National Science and Technology Major Program (no. 2014ZX09507008).
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
The present study was approved by the Ethics Committees of both Tongji University School of Medicine and Shanghai Pulmonary Hospital.
Informed consent
Written informed consent was received from patients.
Rights and permissions
About this article
Cite this article
Sun, Q., Liu, Hp., Zheng, Rj. et al. Genetic Polymorphisms of SLCO1B1, CYP2E1 and UGT1A1 and Susceptibility to Anti-Tuberculosis Drug-Induced Hepatotoxicity: A Chinese Population-Based Prospective Case–Control Study. Clin Drug Investig 37, 1125–1136 (2017). https://doi.org/10.1007/s40261-017-0572-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40261-017-0572-6