Abstract
The dramatic increase in genetic testing technologies has led to discussion about the roles of genetic counselors and other healthcare providers in variant interpretation. The ability to detect DNA variants greatly surpasses the ability to interpret their clinical impact, limiting the benefit of these technologies. Healthcare providers assisting patients in understanding uncertain genetic test results should be aware of ClinGen and ClinVAR. Challenges include understanding the level of certainty or uncertainty behind the determination of pathogenicity, the variable types of variant assessment performed by clinical laboratories, conveying this to patients, helping patients evaluate potential medical decisions in light of such uncertainty, and of course the vast number of “variants of uncertain clinical significance” which arise from next-generation sequencing tests. This paper discusses the current, and rapidly changing, state of variant interpretation resources and the ways that clinical geneticists, specialist physicians, and genetic counselors can assist in improving variant interpretation resources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the years, clinical genetic testing has evolved from single gene testing, typically via mutation (site)-specific testing or Sanger sequencing, towards array-based and multigene panel testing. However, for these modalities to be highly sensitive and specific, with a limited rate of variants of uncertain clinical significance (VUS), the majority of mutations must be accounted for by a small number of single nucleotide variants and detectable structural rearrangements. Since this is not true for most genes, any clinical provider ordering broad molecular testing must understand the process of variant interpretation, and be aware of available resources. The broadest test to recently enter the clinic is exome genetic testing, which has gained significant popularity [1] since its release in late 2011. Data from multiple clinical and academic laboratories suggest that in a population of patients with diverse clinical manifestations, often including intellectual disability, autism and/or congenital anomalies, there is approximately a 25–30 % diagnostic yield [2–4]. However, both panel and exome testing raise a significant potential issue in that the more genes sequenced, the higher the likelihood of identifying rare or novel variants that are difficult to interpret clinically. A single exome may result in as many as 50 thousand variants [5], and recent data suggest that up to 96 % of identified variants have been reported less than 10 times. In one study, the majority of variants causing severe autosomal recessive disease were uncommon [6]. Personal communication from multiple laboratory directors working on rare diseases reports that greater than 50 % of those variants thought to be pathogenic or likely pathogenic have been seen only once in their laboratory. This supports the need for labs to share data to improve the interpretation of novel and rare variants.
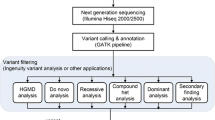
Clinical laboratories have the responsibility to evaluate and prioritize identified variants, and utilize a range of approaches in doing so. ACMG released standards and guidelines in 2008 (updated in 2015) for the interpretation of sequence variants [7••, 8]. Proposals for consensus nomenclature (i.e., “VUS, favor benign”) have also been published [8, 9], but clinical laboratories continue to use different nomenclatures. The original ACMG guidelines [9] are meant to assist clinical laboratories with (A) The validation of next-generation sequencing methods and platforms; (B) The ongoing monitoring of next-generation sequencing testing to ensure quality results; and (C) The interpretation and reporting of variants found using these technologies. The 2015 guidelines [7••] specifically recommend “the use of specific standard terminology: ‘pathogenic,’ ‘likely pathogenic,’ ‘uncertain significance,’ ‘likely benign,’ and ‘benign’ to describe variants identified in Mendelian disorders.” Most importantly, the 2015 guideline outlines the types of variant evidence that should be used (e.g., population data, computational data, functional data, segregation data, etc.) and the process by which variant interpretation should occur [7••].
At the current time, it is not feasible to perform functional testing for all novel or rare variants, and therefore, assessment of variant pathogenicity relies on past reports and in silico predictions based on factors such as allele frequency (which is often based on research data and frequently poorly defined in non-European populations), conservation, protein, and splicing impacts [10–12].). Data suggest these in silico prediction models are imperfect [13, 14], and the lack of a single, centralized, and complete variant interpretation resource that is accurately curated remains one of the greatest barriers to clinical variant interpretation. The difficulty in pairing accurate phenotype data with variants increases the challenge in providing patients with prospective data about the meaning of their genomic variations. Finally, there is no acknowledged standard method for combining disparate data types (e.g., functional assays, population frequency, case–control and family segregation data, histopathology, evolutionary conservation, and in silico prediction algorithms) to determine pathogenicity [15].
This paper will discuss the current, and rapidly changing, state of variant interpretation resources, discuss ways that clinical geneticists, specialist physicians, and genetic counselors can assist in improving variant interpretation resources, and discuss roles for the genetic counselor in the variant interpretation process.
History of Variant Interpretation Resources
In recognition that microarray testing was determining an increasing number of copy number variations (CNVs) that were clinically difficult to interpret, the International Standards for Cytogenomic Arrays (ISCA) Consortium was launched in 2007 for CNVs [16•, 17•], followed by the NIH GO Grant in 2009 which supported the expansion of mechanisms for clinical cytogenetic laboratories to contribute test data to public databases and to develop models for expert data curation. Because technological advances were quickly allowing genome-wide analysis to become commonplace in the care of patients, in 2012, the International Collaboration for Clinical Genomics (ICCG) was launched to include both structural and sequence variants. However, the ability to detect DNA variants continues to greatly surpass the ability to interpret their clinical impact, limiting the clinical benefit of these technologies. Improving genomic interpretation requires a coordinated effort from both the clinical and research communities.
ICCG, an organization of laboratories, clinicians, and researchers dedicated to improving the quality of genomic testing through data sharing and collaboration, continued its work by becoming a founding member of the Clinical Genome Resource (ClinGen, which will be described below) and collecting genotype and phenotype data from clinical laboratories. ClinGen worked closely with a team of bioinformatics experts at the National Center for Biotechnology Information (NCBI), part of the National Library of Medicine, a division of the NIH, to develop the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) to house the data.
In 2013, the National Human Genome Research Institute (NHGRI) funded a large consortium project, “ClinGen,” which is a collaboration between the NHGRI U41 (Geisinger, UCSF, Partners/Harvard), and an NHGRI U01 (Stanford, Baylor, UNC, Geisinger, ACMG, and NCBI). The overarching goal of the consortium is to develop a centralized database for the curation and utilization of consensus-based information on molecular genetic structural and sequence variants that is traceable, searchable, and designed to assist in providing efficient and effective clinical care. ClinGen supports widespread sharing of anonymized genotypic and phenotypic data from laboratories and patients by providing curated data to ClinVar. A key aspect of all of the grants that are part of ClinGen is the concept that data sharing of variant interpretation allows the awareness, and potentially resolution, of conflicting variant interpretations, which will ultimately improve medical care to patients with genomic variation.
To share data effectively, the ClinGen resource defines standard approaches to the interpretation of human genomic data through collaborative activities with regulatory and professional organizations. Specifically, ClinGen draws representation from professional organizations including the American College of Medical Genetics and Genomics (ACMG) patient advocacy organizations such as UNIQUE and the Genetic Alliance, and experts from disease-specific research laboratories and specialty clinics. The concept of genomic data sharing is supported by the American Medical Association (AMA) [18] and the National Institutes of Health (NIH) [19]. Currently, there is no requirement that clinical laboratories submit data, but the Association of Molecular Pathology (AMP) and CAP have been discussing a pilot project as part of the quality control process. On the research front, NIH and NHGRI have had a policy on genomic data sharing for several years, which has required the deposit of de-identified genomic research results into dbGAP [19]. However, dbGAP data include variable phenotypic data (which limits interpretation), require IRB and institutional approval for use, and as a result have historically had limited use in the clinical laboratory. The NIH recently strengthened the data sharing policy and requirements for grant proposals submitted to NIH on or after January 25, 2015, and for intramural projects generating genomic data on or after January 25, 2015 [20]. NIH is also developing recommendations about informed consent for broad data sharing for research and clinical testing as part of the ClinGen project.
Current Variant Interpretation Resources
Several databases, curated with different levels of quality, exist for variant interpretation. Some of these databases include population frequencies (see Table 1) and can be useful in ascertaining if a variant is rare or novel in specific populations. Typically, variants at greater than 1 % allele frequency are considered less likely to be pathogenic (and may be filtered out of variant interpretation pathways before any manual review occurs), although certain notable exceptions (e.g., Factor V Leiden, CF) would be missed if this cutoff were used exclusively in determining pathogenicity. A significant downside to the currently available data is that it is primarily focused on European Caucasians, and allele frequencies for other populations may not be available. When using these databases, one should be aware of the demographics of the included subjects, including age and health status when known. The presence of an allele in a population does not rule out pathogenicity, since limited clinical phenotype data were collected on most participants, and the health status of participants is often limited to a single disease that was the focus of the original research.
Other databases are locus- or disease-specific databases, primarily containing variants noted in patients with disease, which are assumed to be pathogenic. Examples of broad databases are listed in Table 2, and include, but are not limited to OMIM [21], HGMD [22], LOVD [23], and ClinVar [24]. Other locus-specific databases exist for specific diseases, genes, or phenotypes (e.g., CFTR2 [25], BIC [26], InSIGHT [27]), and are listed in Table 3. It is worth noting that the curation processes vary significantly from database to database [28–31], and some of these databases contain inaccurate variants that are pulled from the published literature without a primary review to determine how patients were ascertained or defined, and how the pathogenicity was validated [6, 28]. The ACMG guidelines [7••] state that “when using databases, clinical laboratories should: (1) determine how frequently the database is updated, whether data curation is supported and what methods were used for curation; (2) confirm the use of HGVS nomenclature and determine the genome build and transcript references used for naming variants; (3) determine the degree to which data is validated for analytical accuracy (e.g., low pass next-generation sequencing—NGS versus Sanger-validated variants) and evaluate any quality metrics that are provided to assess data accuracy, which may require reading associated publications; and (4) determine the source and independence of the observations listed.”
Finally, given the current lack of comprehensive and curated databases, and most importantly, the high frequency of novel variants [1], there is a significant need for better automated prediction models for variant pathogenicity. One final area of focus for the ClinGen project is to develop machine-learning algorithms to improve variant interpretation, and to develop approaches to experimentally validate predicted functional effects of novel variants. Machines can be ‘trained’ on a gene-specific level with known pathogenic variants, good phenotypic data, and protein modeling in the hopes that novel missense variants can be more accurately interpreted.
Genetic Counseling Clinical Practice Implications
Genetic counselors as well as other healthcare providers providing genomic results are faced with a number of challenges, including understanding the level of certainty or uncertainty behind pathogenicity calls, the variable types of variant research performed by clinical laboratories, conveying this uncertainty and variability to patients, helping patients evaluate potential medical decisions in light of such uncertainty, and of course the vast number of ‘VUS’ which arise from multigene NGS tests.
In the pre-testing context (both research and clinical settings), the responsibility of ordering clinicians is to convey to patients the potential range of results that may result from genomic testing, including the potential for VUS results, and to select the most appropriate testing option for the circumstance. For many patients, there may be an inherent tendency to consider any “change” in their DNA as a possible cause of disease. It is important to discuss that VUS are common findings and that sharing their data will help resolve some of this uncertainty. In some cases, this entails offering patients the choice between single gene, panel tests, and exomes based on their expectation for anxiety if multiple VUS results are discovered, and discussing the follow-up process if one is identified. Clinicians should also evaluate panel tests versus exome testing for coverage of the specific genes of interest, since exome sequencing interrogates many more genes than panels, but lower coverage in exome testing in relevant genes may mean missing some relevant variants [32].
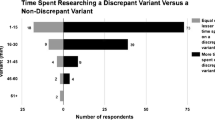
At the time of results disclosure, ordering clinicians have various roles. Some genetic counselors and clinicians feel they have a responsibility to review laboratory results and do an independent assessment of evidence for the pathogenicity of reported variants. Others may feel this responsibility lies exclusively with the laboratory. In the short term when there exists more variability in pathogenicity interpretation, it may be prudent for clinicians to spend some time reviewing reports and considering the interpretation of variants, particularly those with uncertain interpretation, more carefully. There should be open communication between the clinical team and the laboratory, and the clinical laboratory should be provided with all of the phenotypic data available. The first step to re-evaluating a variant is a discussion with the clinical laboratory.
Once the clinicians determine which results will be returned to patients, they must determine the manner in which these results will be conveyed. Several studies in different settings suggest that patients and research subjects are interested in receiving as much information as possible from whole exome sequencing (WES) [33]. Indeed, with regulations for release of healthcare results [34], most institutions have developed policies that patients are entitled to release of their laboratory reports through health portals within a set period of time. Therefore, clinicians need to be cognizant that patients may review full reports and they should have the knowledge and referral resources to help patients to understand any material that may be present in these reports. Clinicians should be comfortable discussing the specific classification of any noted variants, and the clinical implications and/or level of certainty surrounding such recommendations. If follow-up testing or interpretation is to be performed, clinicians should describe what it will entail and the potential results. The clinician and lab must determine how they may operationalize any potential duty to re-evaluate and re-contact for VUS. Some laboratories may do this on a regular basis (potentially for an additional fee), while others may not. Clinicians and researchers may not have the resources to re-contact patients manually, and may suggest that patients remain in contact on a regular basis as a more practical approach to re-evaluating variants. Other centers are investigating the use of electronic healthcare portals and laboratory software to automate and update variant interpretations [35–37], and this is likely to become more typical within the next decade.
The Virtuous Cycle: What Can Healthcare Providers Do to Impact Data Quality
In order to improve the ability to prospectively provide high-quality variant interpretation to patients, healthcare providers of all types who offer genomic testing have the responsibility to contribute to the phenotyping data and to facilitate data sharing and conflict resolution. Without this participation, clinical genomics will suffer from “the free rider problem,” leading to a completely preventable hindrance on the quality of genomic data and, therefore, it’s clinical utility. Several approaches can be considered. First, collect and provide strong phenotype data on symptomatic patients, using a consistent ontology [17•, 38] and provide such data to clinical laboratories. Second, encourage patients themselves to engage in this process, which is feasible through genomeconnect.org (a secure patient portal connected to the ClinGen portal). Third, individual providers can offer their participation in variant curation teams within their areas of clinical expertise, strive to select laboratory partners who provide variant data to open databases, and find ways to encourage for-profit laboratories to balance proprietary data and business practices against the significant population value that accrues through data sharing. Finally, on a more global basis, the entire genomics profession needs to identify and work to minimize genomic health disparities by improving allele frequency data in underrepresented populations so that people of all backgrounds can utilize prospective genomic data in an equitable manner [39].
Conclusion
With the increased use of multigene panels, exome sequencing, and genome sequencing, it is impossible for a single laboratory to become an expert on all variants they may encounter during testing. To solve this issue, clinical laboratories, genetic counselors, physicians, and other healthcare providers must be encouraged to share genomic data and phenotypic data. This should be discussed with patients, and their informed consent should be collected when possible. Clinicians must work closely with clinical laboratories to improve our interpretation and understanding of genetic variants.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Amendola LM, Dorschner MO, Robertson PD, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25(3):305–15.
Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–11.
Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18):1880–7.
Farwell KD, Shahmirzadi L, El-Khechen D, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2014;. doi:10.1038/gim.2014.154.
Facio FM, Sapp JC, Linn A, et al. Approaches to informed consent for hypothesis-testing and hypothesis-generating clinical genomics research. BMC Med Genomics. 2012;5:45.
Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3(65):65ra4.
•• Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. Recommendations from a working group of experts who met in 2013 from the American College of Medical Genetics/Genomics (ACMG), the Association of Molecular Pathology (AMP), and the College of American Pathologists (CAP), for the interpretation of sequence variants from genotyping, single genes, panels, exomes, and genomes.
Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–91.
Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10(4):294–300.
PolyPhen-2; http://genetics.bwh.harvard.edu/pph2/. Accessed 19 Mar 2015.
SIFT; http://sift.jcvi.org/. Accessed 19 Mar 2015.
Mutation Taster; http://www.mutationtaster.org/. Accessed 19 Mar 2015.
Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2014;14(4):533–7.
Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;15:2125–37.
Tavtigian SV, Greenblatt MS, Goldgar DE, et al. Assessing pathogenicity: overview of results from the IARC Unclassified Genetic Variants Working Group. Hum Mutat. 2008;29(11):1261–4.
• Riggs ER, Church DM, Hanson K, et al. Towards an evidence-based process for the clinical interpretation of copy number variation. Clin Genet. 2012;81(5):403–12. Overviews an evidence based review process for determining the clinical significance of structural variations throughout the genome. Discusses the process of choosing and weighing different types of evidence.
• Riggs ER, Jackson L, Miller DT, et al. Phenotypic information in genomic variant databases enhances clinical care and research: the International Standards for Cytogenomic Arrays Consortium experience. Hum Mutat. 2012;33(5):787–96. Summarizes the experience of the International Standards for Cytogenomic Arrays (ISCA) in using phenotype and genotype data and its application to research that impacts clinical care.
http://www.ama-assn.org/ama/pub/news/news/2013/2013-06-18-new-ama-policies-annual-meeting.page. Accessed 19 Mar 2015.
http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html. Accessed 19 Mar 2015.
http://gds.nih.gov/03policy2.html. Accessed 19 Mar 2015.
http://omim.org/. Accessed 19 Mar 2015.
http://www.hgmd.org/. Accessed 19 Mar 2015.
http://www.lovd.nl/3.0/home. Accessed 19 Mar 2015.
https://www.ncbi.nlm.nih.gov/clinvar/. Accessed 19 Mar 2015.
http://www.cftr2.org/. Accessed 19 Mar 2015.
http://research.nhgri.nih.gov/bic/. Accessed 19 Mar 2015.
http://insight-group.org/. Accessed 19 Mar 2015.
Cotton RG, Auerbach AD, Beckmann JS, et al. Recommendations for locus-specific databases and their curation. Hum Mutat. 2008;29(1):2–5.
Gout AM, Ravine D, Harris PC, et al. Analysis of published PKD1 gene sequence variants. Nat Genet. 2007;39(4):427–8.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3898141/. Accessed 19 Mar 2015.
Vail PJ, Morris B, van Kan A, et al. Comparison of locus-specific databases for BRCA1 and BRCA2 variants reveals disparity in variant classification within and among databases. J Community Genet. 2015. doi:10.1007/s12687-015-0220-x.
Consugar MB, Navarro-Gomez D, Place EM, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2015;17:253–61.
Hitch K, Joseph G, Guiltinan J, et al. Lynch syndrome patients' views of and preferences for return of results following whole exome sequencing. J Genet Couns. 2014;23(4):539–51.
https://www.federalregister.gov/articles/2014/02/06/2014-02280/clia-program-and-hipaa-privacy-rule-patients-access-to-test-reports. Accessed 19 Mar 2015.
Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32(5):532–6.
Aronson SJ, Clark EH, Varugheese M, et al. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012. doi:10.1038/gim.2012.19.
Verma SS, de Andrade M, Tromp G, et al. Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet. 2014;5:370.
Kohler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(Database issue):D966–9674.
Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1000 participants’ exomes. Am J Hum Genet. 2013;93(4):631–40.
Acknowledgments
This manuscript is original work inspired by the author’s participation in the ClinGen Consortium, funded by the NHGRI, in conjunction with additional funding from the NICHD and NCI, through the following grants and contracts: 1U41HG006834-01A1 (Rehm), 1U01HG007437-01 (Berg), 1U01HG007436-01 (Bustamante), HHSN261200800001E.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical collection on Genetic Counseling and Clinical Testing.
Rights and permissions
About this article
Cite this article
Ormond, K.E., Rashkin, M. & Faucett, W.A. Standardizing Variant Interpretation in Genomic Sequencing: Implications for Genetic Counseling Practice. Curr Genet Med Rep 3, 137–142 (2015). https://doi.org/10.1007/s40142-015-0073-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-015-0073-y