Abstract
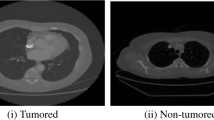
Lung cancer is the leading cause of death from cancer worldwide. Finding pulmonary nodules is a critical step in the diagnosis of early-stage lung cancer. It has the potential to become a tumour. Computed tomography (CT) scans for lung disease analysis offer useful data. Finding malignant pulmonary nodules and determining whether lung cancer is benign or malignant are the main goals. Before further image preparation, image denoising is an important process for removing noise from images (feature extraction, segmentation, surface detection, and so on) maximising the preservation of edges and other intact features. This study employs a novel evolutionary method dubbed the binary grasshopper optimisation algorithm in order to address some of the shortcomings of feature selection and provide an efficient feature selection algorithm, the artificial bee colony (BGOA-ABC) algorithm enhance categorisation. Then, to categorise the chosen features, we employ a hybrid classifier known as a double deep neural network (DDNN) algorithm. A technique used by MATLAB that segments impacted areas utilising improved IPCT (Profuse) aggregation technology employing datasets from the cancer image archive (CIA), the image database resource initiative (IDRI), and the lung imaging database consortium. Various performance metrics are evaluated and associated with different cutting-edge techniques, classifiers that are already in use.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Since lung cancer claims more lives each year than any other kind of cancer, it ranks among the world’s primary causes of rising death rates. Men and women are both affected by this deadly illness. Therefore, in order to save the lives of many lung cancer patients, it is imperative to develop appropriate techniques for the early diagnosis and recognition of this disease. [1]. Many patients’ chances of survival can be increased by early detection and confirmation. When the condition is correctly diagnosed after it has been discovered later, and patient mortality can be decreased. Modern mechanical cleansing techniques can therefore be used in the field of medical image processing to raise the redundancy of use and boost classification accuracy in order to get appropriate and quick results. Thus, early and accurate detection and identification at the earlier stage can undoubtedly increase survival and decrease mortality [2].
The majority of early studies used mammography, computed tomography (CT), and magnetic resonance imaging (MRI). Experts in the field analyse this image using the proper methods to find and classify various lung cancer grades [3]. Chemotherapy, targeted therapy, and radiation therapy are only a few of the clinical and laboratory procedures utilised to eradicate or halt the growth of malignant cells. All of these methods for discovering and diagnosing malignant condition take a lot of time, money, and cause the patient considerable agony. Therefore, to solve these issues, appropriate machine learning techniques are applied to the processing of these CT scan pictures for medical purposes. Because CT scan images have less noise than other medical images like MRI, X-ray, etc., they are better [4].
Lung cancer classification involves applying images to a deep convolutional neural network's input layer, processing them through each of the network's hidden layers, and classifying the images as malignant or non-malignant in the output layer. An input image is fed into the deep learning algorithm DCNN, which then displays the importance of each object in the image [5]. Each object in the image will be further classified from the others once the network has been accurately trained on the broader dataset. Deep learning techniques require the least amount of preprocessing compared to other image processing algorithms. In order to attain the best accuracy, DCNN transforms input images to make them appropriate for processing while minimising allowable image feature loss [6]. Filter size, the number of hidden layers, and the number of extracted feature maps are the design and performance criteria for DCNNs. Higher levels of functional abstraction and detection are made possible by deeper network layers. Due to more convolution operations, computation times increase with network depth. Convolutional filters work best when they are 3 by 3 or 5 by 5. Network performance is reduced by larger convolution kernel sizes [7].
Significant Contribution
-
1.
First, noise and memory constraints from prior research were applied to analyse LIDC-CT DICOM dataset images.
-
2.
The suggested system, which relies on both clinical and image quantitative features for efficient analysis, offers an improved feature representation for computer-aided diagnosis that is more accurate.
-
3.
For effective tumour staging, this technique makes use of a double deep neural network classifier (DDNN).
The essay is organised as follows. Section "Survey" provides a summary of the literature review. Section "Methodology" provides a description of the process. Section "Discussion" contains an outline of the conclusions and future work, while Sect. "Results and Discussion" presents the results and discussion.
Survey
The most popular screening technique for lung cancer early diagnosis is computed tomography (CT). In comparison with plain chest X-rays, a landmark NLST trial from 2011 indicated that CT screening for lung cancer patients was associated with a 20% lower mortality rate. Based on these data, the USA Class B recommendation for annual CT lung cancer screening in adults 55–80 years of age who have smoked for 30 years was released by the Preventive Services Task Force (1-day personal smoking training number) and regardless of whether they were still smoking or had quit within the previous 12 months. The creation and implementation of fresh CT-based lung cancer screening programmes were made easier by this strategy. The process of CT scan identification takes time and might be challenging [8].
The earliest indications of lung cancer are lung nodules, which are tiny round or oval opacities on CT scans that have a diameter of less than 30 mm. As the percentage of CT scans examined increases and the resolution rises, reading these massive datasets can cause visual exhaustion and stress, which can lower diagnostic accuracy. Furthermore, because interpretations were so heavily reliant on prior experience, trained radiologists displayed significant diversity in their ability to identify minor lung tumours. When it comes to finding lung nodules, radiologists’ abilities vary greatly. Complex airway and vascular architectures challenge CT scan analysis [9].
Even among seasoned radiologists and doctors, CT screening generates a significant proportion of false positives, which causes grave over diagnosis. The value of CT screening for early lung cancer diagnosis must be minimised because of the significant false-positive rate brought on by benign nodules. Costs and morbidity related to single interventions and interventions can be decreased by minimising false positives and identifying patients who require medicines. The ability to identify benign from malignant nodules becomes increasingly crucial. There are significant challenges in differentiating between benign and malignant nodules as well as indolent and aggressive tumours. In order to assist, computer-aided diagnostic (CAD) systems for physicians with this problem have been extensively studied. [10].
CAD is utilised in all areas of the body, encompassing the kidneys, spine, joints, heart, chest, abdomen, and head, additionally, nuclear medicine, ultrasound, and digital pathology imaging. These CADs often employ pre-existing machine learning techniques, such as region of interest segmentation, feature extraction, and labelling, to get the final conclusion. Traditional machine learning techniques cannot handle natural data in its raw form (raw image pixels), so it is necessary to identify and derive significant value from raw data for learnable descriptions using functional design and technical needs. Contrarily, a class of techniques known as representation learning can automatically determine the optimum representation of unprocessed data and features to support classification, prediction, and detection activities [11].
Lately, deep learning methods have been applied to numerous medical image processing jobs after having great success with object detection operations on picture data. The absence of labelled datasets is the fundamental challenge in the clinical sector, though. The existence of labelled training data, such as millions of labelled images, which enables the use of sophisticated algorithms with thousands of parameters, makes conventional image recognition operations successful in many cases in the Enable training for deep neural networks. These two areas of medicine have significantly less familiarity with brand training. Another issue with clinical imaging methods is the 3D structure of the majority of medical images. Standard computed tomography (CT) or magnetic resonance imaging (MRI) scans have several segments, and the lesion region is quite modest compared to the complete photo stack (and various 3D dimensions) [12].
It is challenging to swiftly gather 3D data from deep neural network designs because the thickness of cross-sectional scan slices is frequently significantly larger than the planar pixel size. It is now more challenging to design deep learning-based systems for 3D medical imaging. Having a greater understanding of how lung cancer develops and progresses, including how long it takes to reach a certain stage, can aid in earlier identification as well as more efficient prevention, treatment, and care. Regular imaging is one of the best approaches to identify early-stage lung cancer. The terminal information gathered as a result of screening can be utilised to create predictions for specific diagnostic and surveillance procedures as well as better approaches to define illness development. More and more people are regarded as having a high risk of developing cancer. However, screenings are now done on a regular basis. However, dangers including radioactive contamination, overtreatment, and the use of preventive medications highlight the need for more precise temporal models to identify cancer risk factors [13].
When creating a screening policy, it is vital to take into account the mean residence time (MST), which is the amount of time that a tumour might be seen on imaging yet not exhibit any clinical signs. It is used to estimate MST are continuous Markov models (CMMs) with MLE (maximum likelihood estimation). Risks including radiation exposure, overdiagnosis, and overtreatment, however, draw attention to the need for more precise temporal models to forecast results. However, typical approaches make the unrealistic assumption that there are no observational mistakes while analysing picture data, which results in inaccurate MST estimations [14]. Additionally, MLE cannot be computed reliably if the data are sparse. Using remedy this weakness, this research suggests a probabilistic modelling method to data from routine cancer screening. In [15, 16], the authors presented various degrees of information and probabilities, and repeated measures of impact were used to estimate the impact of the target class [17]. Assign a positive reduction value and experiment with different patch sizes to increase accuracy overall.
Problem Statement
There are numerous tools, models, and procedures available for classifying lung cancer. The authors of the research [9] employed logistic regression as a machine learning classifier for the categorisation of lung cancer. Data from a total of 80 patients were used. But for their analysis, they only used 50 patient data [18,19,20]. They discovered good accuracy values ranging from 71 to 78%. The following are the primary issues with current research. Due to the limited dataset's tendency to cause overfitting during model implementation, they only employed a very small sample of the 50 patient dataset for their map model. To train the best model for the best accuracy, there is still a difficulty with augmentation and model optimisation. Therefore, the sensitivity and specificity of previous research are not comparable.
To prevent overfitting the algorithm, augmentation and model optimisation are crucial. Small datasets make it more likely for models to identify patterns that do not exist, leading to significant variation and extremely high errors on the test set. This overfitting symptom is typical. Thus, the little dataset and minimal fitting during model training are to blame for the low model accuracy. There are numerous sophisticated categorisation methods available as well. Therefore, in order to deliver improved, high-precision results, we require suitable and cutting-edge algorithms. Additionally, we require efficient tools and languages that require little human effort to operate. The proposed categorisation algorithm is straightforward. It is crucial to execute many algorithms.
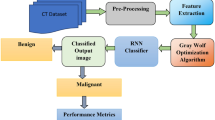
Methodology
Adaptive Nonlocal Means Filter
The fundamental purpose of image preprocessing is to adjust the pixel values, contrast, and greyscale to improve the image quality. The adaptive non-local averaging filter is more popular than the normal non-local averaging filter because it performs better when noise levels are higher than 0.2 dB. The impulse noise of the image is determined by this filter using spatial processing. By comparing surrounding pixel values with a threshold, it may identify noisy pixels [21]. Noise is defined as pixels that are higher than the threshold. The average of the noise-classified surrounding pixels takes the place of this noisy pixel as shown in Fig. 1.
A weighted average of all the pixels across the filtered value for pixel I in a is calculated using the entire image typical non-local mean (NLM) filter. The NLM can be expressed as follows given a 2D noisy image with the format x = x(i)|iIpos, where Ipos are all pixel coordinates.
where I is the search window, which is often a square neighbourhood with a specified size that is centred on pixel i. The weights are made in the following way:
where Ni is a fixed-size, pixel-centred square neighbourhood. Gaussian weights are used to determine how similar two pixels, I and j, are search field in Ωi: the normalising factor to determine j(i,j) = 1 is the Euclidean distance of pixel grey values x(Ni) and x(Nj) between two neighbours Ni and Nj in Z i. By modifying the weighted decay of the exponential function, the parameter h regulates the level of filtering. In general, h is non-local average denoising, which directly impacts the quality of the images. H is determined for the proposed ANLM filter as
where w is the Merror weight, Merror is the decomposed error distribution map, which depicts the error area with enhanced filtering strength, and hwhole is the smoothing parameter of the entire image to preserve as much detailed underlying data features as feasible [22]. A wise choice of w can strike a balance between enhancing the base material and smoothing out the entire image. The breakdown error distribution map Merror construction process will be covered individually. The ANLM filter can be reduced to an existing NLM filter when w is 0. The decomposition error distribution map is used in the new ANLM filter developed in this paper. There are two elements to the smoothing parameter h. The weight of the decomposed error distribution map is adjusted using the second part, while the first part serves as a constant for the entire image [23]. Therefore, when used to denoise images of a particular material, the proposed ANLM filter can maintain properties of related materials while removing aspects like artefacts. The simulation results of preprocessing are shown in Fig. 2.

:
Improved Profuse Clustering Technique-Based Segmentation
An improved profuse clustering algorithm and deep learning neural networks to predict cancer. The next critical step is to use extended preservation clustering techniques to isolate cancer-infected areas in enhanced lung CT (IPCT) images. To identify problem areas, a new segmentation technology analyses lung computed tomography (CT) images pixel by pixel and divides the image into smaller images [24]. The new Profuse technology functions in two stages: first, it analyses the picture pixels that are there, and then, it groups related super pixels into one group to find unexpected image pixels. Using spectral or pixel eigenvalues, the image segmentation method continually evaluates each pixel or piece of data in order to forecast data similarity. Pixel similarity values include quantitative evaluations of the images that are utilised to effectively create clusters. Look at each pixel in a quality-enhanced lung CT image as a point in feature space. To divide the impacted area, create an undirected graph. V is shown as a G(V, E) vector or data point in a closed picture association graph, and E is shown as an edge connecting two data points [25]. Wij(i), E represents the precise weight values for connections between edges. The similarity between the points of data E calculates I and j. By dividing vertices into equal high and low values, the graph partitioning problem is solved after initialising a closed video-associative undirected graph. The pertinent normalised cut of the image is thus, using A and B as two subset vertices,
; the following steps should be taken to complete the n-cut method after defining the vertex weights.
Compare the similarity between data points or pixels in different areas of the image using the ncut process. Then, using equation, the overall similarity between pixels is determined (6).
By estimating the lowest cut value, which is determined as, the NP-hard problem of similarity calculation is solved.
It is possible to think of D as a n*n d-diagonal matrix. The dimension of a symmetric matrix is n*n, and it is denoted as W. Using this illustration, the minimum cut is determined as follows:
In Eq. (8), yi1, -b, where -b is a constant and yTD1 is equal to 0. As previously stated, we determine the similarity between data points or pixels by minimising the NP-hard problem and ncut. Run the batch process in the number of batches specified. During clustering, the k-means problem is solved by using the k-means kernel algorithm, which operates on multiple data points in the scene and accurately captures nonlinear pixels in scene space. k-means kernel function definition
In Eq. (9), the kernel function for a specific pixel cluster of the image is (xi). The kernel problem is expanded by computing the inverse of the cluster members. After resolving the issue, you can place comparable pixels in one cluster and dissimilar pixels in another to help forecast the affected areas clustering problem. The IPTC algorithm, among other things, operates in accordance with an undirected graph connected to each image pixel. In lung pictures, this efficiently predicts edges. Simulation result of proposed improved profuse clustering technique based segmentation as shown in Fig. 3.

:
Classification
Binary Grasshopper Optimisation Algorithm
We use the proposed hyperparameter optimisation approach to tune the hyperparameters in accordance with the cancer classification after denoising the LIDC-IDRI dataset. The pixel values of each convolutional layer are modified during training. Model parameters and hyperparameters are the weights that were produced as a result, and they have an impact on how the model behaves during training.
The basic objective of binary grasshopper optimisation techniques is to solve optimisation issues posed by the distinctive motion, navigation and motion usage of grasshoppers in the search space. Equation (10) is the mathematical representation of GOA.
In this equation, Xi is referred to as the ith locust position, Si as social interaction, Gi as ith locust gravity, and Ai as wind advection. called random numbers in the [0, 1] range are r1, r2. These assign arbitrary weights to the distinctive components of (10). The acquired social interaction is defined in Eq. 2 in terms of the social power denoted by s(r)(r). Equation 2 is used to express this. The distance between the ith and jth locusts is referred to as dij.
where l is referred to as the tensile length scale and (3) is referred to as the tensile strength. In order to solve optimisation issues, Eq. 12 through 10 are employed to solve various elements of locust motion. Equations are then used to express the enlarged mathematical equations. (13):
where the d dimension’s upper and lower bounds are referred to as ubd and lbd, respectively. The personnel area’s comfort, repellency, and reduction factors are represented by the letters c. GOA chooses 1 as a factor that is crucial to establish the ideal balance between navigation agent localisation and random work since it has a strong inclination to advance swiftly towards the current optimum. We can employ GOA optimisation in conjunction with GAN methods using the determined optimal parameter values to prevent the overfitting issue within the performance range.
The ABC Algorithm for Artificial Bee Colonies
The ABC algorithm, or artificial bee colony, is a relatively new technique. ABC is based on the foraging behaviours of bee colonies. In the ABC algorithm, a colony of bees consists of three distinct species: scout bees, hired bees, and observer bees. There are twice as many colonists as there is food available. The number of food sources indicates the locations where the optimisation problem might have a solution, and the amount of honey in the food source indicates the quality (fitness) of the related solution. The task assigned to the hired bees is to produce the food supply and notify the viewers of the juice quality of the food source they are creating. The number of onlookers matches the number of bees working. Based on information on the bees used, viewers choose which food source to use. When their food supply is exhausted, employers start scouting the area in search of fresh food supplies. The likelihood that the spectator bees would select a certain source increased with the size of the juice. The ABC algorithm works as follows:
Each recruited bee was given a string of binary bits that represented feature selection and had a length equal to the number of features in the dataset. The connected feature is selected if the nth bit of the binary string is set to “1”, as opposed to “0”, which denotes that it is not. The number of hired and observed bees is equal to the number of features (m) in the dataset because features are regarded as food sources.
Each recruited bee was given a string of binary bits that represented feature selection and had a length equal to the number of features in the dataset. The connected feature is selected if the nth bit of the binary string is set to “1”, as opposed to “0”, which denotes that it is not. The number of hired and observed bees is equal to the number of features (m) in the dataset because features are regarded as food sources.
Observer bees gather data from hired bees and compute the likelihood of choosing features using Eq. 15.
Using the projected accuracy of the feature pointed by the hired bee and the feature chosen by the observer bee, the observer then calculates a new solution, vi. The used bee points to a feature subset made up of the previously pointed features and the newly chosen features if the new solution vi is greater than xi. The previously utilised bee features are kept, and the newly chosen features are ignored if vi is not greater than xi. Use equation to calculate the new solution vi (16).
where xi represents the feature’s predictive accuracy that was given to the hired bees, and xj represents the feature’s predictive accuracy that was chosen by the audience. I is a real random number with uniform distribution that falls between [0, 1]. The viewer tries to use and generate a new feature subset configuration each time a bee employed in this manner is given a new feature subset. The juice content was accumulated to better construct functional subgroups after employing all available features to form them. If the hired bee doesn’t advance, he or she turns into a scout. According to equation, scouts are given a new set of binary functions (17).
where x maxj and x minj stand for the population dimension’s bottom and upper limits, respectively. The bees carried out the same action repeatedly until the ideal functional subset was created.

:
Double Deep Neural Networks Classification
Using training a neural network to categorise photographs into a specified list of files or types, image classification is the foundation for image recognition by double deep neural networks. It is used to ascertain whether something is acknowledged in its most basic form. In our situation, we may categorise medical photos to identify whether cancer is present and limit the identification to a simple YES/NO (YES cancer, NO). Before being used to categorise images, neural networks need to be trained. This method compares the actual output to the expected outcome after feeding a list of input data to the network. The input layer in our example can have as many input nodes as the array size because the input data are a CT image dummy that the network is shown in Fig. 4. This may result in a crowded input layer, sluggish training, and an overloaded network. An array or a single node 0/1 is the network’s output. In our scenario, the export layer generates a decimal value of 0.0 (non-cancer) or 1.0 (cancer) between 0.0 and 1.0.
An example of a neural network type used for image classification is a double deep neural network (DDNN). All dataset picture sets are loaded into the DDNN, and before each layer passes the images to the next, it reads the features of the images [26]. A manually created feature extractor is used by the general DDNN model for image identification to collect data and information from input photos while eliminating unimportant variables. The classifier that trains feature vectors into classes comes after this extractor.
It has been suggested to use a dual deep neural network to identify and categorise lung nodules as benign or cancerous [27]. The suggested DDNN model has a number of combinatorial layers where the picture attributes from one layer are passed on to the subsequent layer. More features are extracted by the following layer and are sent there for immediate delivery. Filtered by removing the undesired features, the features gathered from the softmax layer (used to compare the features of the previous layer with the input picture to eliminate unwanted features from the last layer of the classifier) are created a trained model [28].
The feature extractor rules are implemented in the first layer’s convolutional layer, and as a pooling layer, the second layer eliminates some of the undesirable features from the first layer as shown in Fig. 5. A completely linked layer below integrates all of the previous layers’ features to change the model. In test images, softmax compares and eliminates features of unimportant factors.
Results and Discussion
Dataset Description
Data from LIDC-IDRI were gathered from TCIA, the Cancer Imaging Archives. With pixel sizes ranging from 0.48 to 0.72 mm and slice thicknesses between 1.25 and 2.5 mm, the data comprised 1,018 CT images from 1,010 patients. 30% of department budget goes towards testing, and 70% goes towards training. Here is a complete explanation of LIDC-IDRI [18].
Measures of Performance
In simulation, we use MATLAB 2021(a) tools to carry out the preprocessing, segmentation, feature extraction, and classification operations using the algorithms mentioned above after segmenting the images. Based on the explanation above, we evaluated the accuracy of various existing classification techniques and the classification of lung pictures [29]. The image was enhanced with a wide variety of noise densities that ranged from 0.1 to 0.9 in 0.1 increments. Traditional to evaluate denoising performance objectively, three metrics were used: mean square error (MSE), peak signal-to-noise ratio (PSNR), and structural similarity index (SSIM). MSE should be low and PSNR high for optimal image quality comparison. The similarity between two photographs X and Y is measured using SSIM, which is calculated as Eq. (18),
where \({\mu }_{x}\) the average of x is, \({\mu }_{y}\) is the average of y, \({\sigma }_{x}^{2},{\sigma }_{y}^{2} is \, the \, variance \, of \, x, \, y\) and \({\sigma }_{xy} \, is\) covariance of x and y. \(c1 = {\left(k1L\right)}^{2}, c2 = {\left(k2L\right)}^{2}\) division of two variables with a weak numerator that is stable. L is pixel value with dynamic range. In this experiment, k1 and k2 are both equal to 0.01. The effectiveness of the dual deep neural network technique is examined using the following measures to assess its superiority: accuracy, sensitivity, specificity, and AUC curves.
The learning parameters for each architecture are displayed in Table 1. For comparison, all architectures share certain parameters.
Peak Signal-to-Noise Ratio (PSNR)
Table 1 displays the suggested PSNR approach for Gaussian noise as well as the fundamental PSNR values. According to Table 5, which compares the PSNR measurements of several denoising methods, the ANLM method outperforms the others for noise levels ranging from 5 to 90% [30]. It can be seen that even in situations where there is a lot of noise, the proposed denoising technique still results in high-quality images. The suggested adaptive nonlocal means filter algorithm’s average PSNR is 39.74, compared to 37.17 for weighted median filter, 31.06 for switching mean median filter, and 33.42 for bilateral filter. The mean PSNR of the suggested method is higher than that of competing algorithms, demonstrating conclusively that there is very little difference between the processed and original images.
Structural Similarity Index Measure (SSIM)
The average SSIM for the proposed adaptive nonlocal means filter method in this study is 0.9204, whereas the average SSIM for bilateral filter, switching mean median filter, and weighted median filter is 0.9013, 0.8917, and 0.8625, respectively. The proposed algorithm typical SSIM is very near 1. The proposed algorithm processed structure is more similar to the original structure than those of other algorithms. To put it another way, the final graphics are superior. The suggested method performs significantly better than traditional phantom image processing techniques as the noise level rises, and the quality measure matrix increases along with it.
Signal-to-Noise Ratio (SNR)
The mean ratio of signal to noise (SNR) of the algorithm in this work is 0.6454. This demonstrates that for all input SNR stages, the proposed approach outperforms previous algorithms in terms of SNR. The suggested method performs significantly better than traditional phantom image processing techniques as the noise level rises, and the quality measure matrix increases along with it.
The adaptive nonlocal means filter and the bilateral filter both outperform the median, weighted median, and switching mean median filters for low noise densities and acquired lung cancer images with a noise density of 0.9 to show the suggested filter’s effectiveness. The selective adaptive non-local mean filter performs better than other filters with low SNR value, high PSNR, and high SSIM, as shown in Table 1.
Here, the suggested tumour predictor as well as the results of DDNN simulation parameters tabulated as Table 2. For the initial iteration, the validation test data are divided into four distinct folds of (a) the confusion matrix and (b) the ROC. Next, we compare our suggested design to other well-known transfer learning models including GoogleNet, SqueezeNet, ShuffleNet, DenseNet, and MobileNetV2 by computing various performance metrics mentioned in Table 3. The effectiveness of the proposed framework in comparison with other popular algorithms is highlighted in Table 3.
Table 3 suggests algorithm for detecting normal, benign, and malignant CT images shows 98.95% accuracy, 98.15% sensitivity, 98.76% accuracy, and 98.8% high F1-score. Table 4 shows comparable results for detecting cancerous images. It demonstrates that the presented network exhibits higher detection rates than conventional techniques when capsules and saliency are combined with optimised transition learning. Based on the architecture's ability to place restrictions on computational bottlenecks, GoogleNet can operate at its best performance. Consequently, maintaining a stable design is simple. SqueezeNet, with 94.13% accuracy, 99.06% specificity, and 62.69% sensitivity, is the second-best architecture. New modules are used in both architectures to implement 1 × 1 filters. The performance of the architecture can be enhanced by incorporating a 1 × 1 filter, which can ease the strain of computational bottlenecks.
DenseNet performed as the next classifier, achieving 93.52% accuracy, 89.83% specificity, and 59.7% sensitivity. We discover that propagating the input layer to succeeding layers is a useful property since it enables the earlier levels to pick up more details and increase precision. However, because it may cause slower execution times, it might not be appropriate for usage on a single GPU machine set up with 121 layers. ShuffleNet’s performance is below average when compared to others because it only achieves 92.91% accuracy, 98.83% specificity, and 55.22% sensitivity. With 92.91% accuracy, 97.65% specificity, and 62.64% sensitivity, MobileNetV2 is the final architecture. It can still attain accuracy of more than 90%, though. This demonstrates that all architectures make use of new modules created for maximum accuracy while maintaining several crucial parameters. With an AUC of 86.84%, GoogleNet has the highest area under the ROC curve. According to the ROC curve, DenseNet’s AUC is 86.12%, SqueezeNet’s is 85.04%, ShuffleNet’s is 82.34%, and MobileNetV2’s is 82.11%.
From Table 4, compared to other models, the trained model is fairly compact (8.33 MB in size). This demonstrates that using the Proposed BGOA-ABC-DDNN has a considerably lower computational cost than using other models. The DenseNet model has a size of 42.5 MB, which is appropriate for heavyweight models and is 15.9 MB larger than the MobileNetV2 model. For cloud-based discovery processes, the ShuffleNet model has a size of 282.8 MB and GoogleNet has a size of up to 635.2 MB.
The time complexity of the proposed model is tabulated in Table 5. The proposed time complexity model is O(N2), whereas the conventional DenseNet time complexity is O(N/2).
Analysis of Computation Time
Computation time: This is the amount of time the system needs to spend in order to produce the desired outcome. This work uses the MATLAB 2018a environment on a computer with 4 GB RAM that is based on an Intel Core i3 II generation processor. The proposed task's execution time for each process is displayed in Table 6. The BGOA-ABC-DDNN classifier required a total of 44.64 s to simulate. 35.67 s is estimated to be the BGOA-ABC-DDNN classification run time.
Table 7 shows the comparative accuracy suggested and conventional techniques. When compared to the existing method, the approach suggested in this table produced the highest accuracy for LIDC-IDRI data. These findings demonstrate that BGOA-ABC-DDNN, the suggested method, is the most effective option for diagnosing cancer.
Discussion
The proposed adaptive NLM method outperforms the others for noise levels ranging from 5 to 90% [30]. It can be seen that even in situations where there is a lot of noise, the proposed denoising technique still results in high-quality images. The suggested adaptive nonlocal means filter algorithm’s average PSNR is 39.74, compared to 37.17 for weighted median filter, 31.06 for switching mean median filter, and 33.42 for bilateral filter. The proposed BGOA-ABC-DDNN algorithm for detecting normal, benign, and malignant CT images shows 98.95% accuracy, 98.15% sensitivity, 98.76% accuracy, and 98.8% high F1-score. It demonstrates that the presented network exhibits higher detection rates than conventional techniques when capsules and saliency are combined with optimised transition learning. Based on the architecture's ability to place restrictions on computational bottlenecks, GoogleNet can operate at its best performance. Consequently, maintaining a stable design is simple. SqueezeNet, with 94.13% accuracy, 99.06% specificity, and 62.69% sensitivity, is the second-best architecture. GoogleNet has the highest AUC, or area under the ROC curve, being 86.84%; according to the ROC curve, DenseNet’s AUC is 86.12%, SqueezeNet’s is 85.04%, ShuffleNet’s is 82.34%, and MobileNetV2’s is 82.11%.
Conclusions
In this paper, we present a novel approach to disease detection, diagnosis, and prediction in order to improve disease management and preventive measures. In this paper, we propose a hybrid deep neural technique with an adaptive control process for cancer detection. The primary goal of this paper is to classify malignant lung tumours and determine whether lung cancer is malignant or benign. Preprocessing, segmentation, feature extraction, feature selection, and classification are all used to detect cancer. A preprocessing step is first performed using ANLM filters. The IPCT segmentation method is then used to isolate and segment cancer nodules from lung images. The proposed BGOA-ABC-DDNN algorithm for identifying normal, benign, and malignant LIDC-IDRI dataset CT images achieves 98.95% accuracy, 98.15% sensitivity, 98.76% accuracy, and 98.8% F1-score. In this case, we use DDNN for softmax regression and the DDNN output class estimation procedure for tumour classification. Simulation results show that the BGOA-ABC-DDNN method outperforms all other classifications and new methods in all performance measures. In the future, performance can be improved by incorporating different types of medical images, and other metaheuristic techniques can be added to boost system performance.
Availability of Data and Materials
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Abbreviations
- CT:
-
Computed tomography
- BGOA-ABC:
-
Binary grasshopper optimisation algorithm artificial bee colony
- CIA:
-
Cancer image archive
- LIDC-IDRI:
-
Lung imaging database consortium, and the image database resource initiative
- MRI:
-
Magnetic resonance imaging
- M-CNN:
-
Multilayer convolutional neural network
- CAD:
-
Computer-aided diagnostic
- MRT:
-
Mean residence time
- CMM:
-
Continuous Markov models
- MLE:
-
Maximum likelihood estimation
- RGB:
-
Red–green–blue
- NLM:
-
Non-local mean
- ANLM:
-
Adaptive nonlocal means
- DDNN:
-
Double deep neural network
- MSE:
-
Mean square error
- PSNR:
-
Peak signal-to-noise ratio
References
M. Palaniappan, M. Annamalai. Advances in signal and image processing in biomedical applications. (2019). https://doi.org/10.5772/intechopen.88759
S. Kolli, V. Praveen, J. Ashok, A. Manikandan. Internet of things for pervasive and personalized healthcare: architecture, technologies, components, applications, and prototype development. (2023). https://doi.org/10.4018/978-1-6684-8913-0.ch008
T. Guo, J. Dong, H. Li, Simple convolutional neural network on image classification. In Proceedings of the 2nd International Conference ICBDA, Beijing, China, 10–12 March 2017.
A. Ivanov, A. Zhilenkov, The prospects of use of deep learning neural networks in problems of dynamicimages recognition. In Proceedings of the EIConRus, Moscow, Russia, 29 January–1 February 2018.
H.P. Chan, L.M. Hadjiiski, R.K. Samala, Computer-aided diagnosis in the era of deep learning. Med. Phys. 47(5), e218–e227 (2020)
A.R. Venmathi, S. David, E. Govinda, K. Ganapriya, R. Dhanapal and A. Manikandan, An automatic brain tumors detection and classification using deep convolutional neural network with VGG-19. in 2nd International Conference on Advancements in Electrical, Electronics, Communication, Computing and Automation (ICAECA), Coimbatore, India, 2023, pp 1–5 (2023). https://doi.org/10.1109/ICAECA56562.2023.10200949
L. Ficsor, V.S. Varga, A. Tagscherer, Z. Tulassay, B. Molnar, Automated classification of inflammation in colon histological sections based on digital microscopy and advanced image analysis. Cytometry A 73A(3), 230–237 (2008)
Y. LeCun, Y. Bengio, G. Hinton, Deep learning. Nature 521(7553), 436–444 (2015)
M. Anthimopoulos, S. Christodoulidis, L. Ebner, A. Christe, S. Mougiakakou, Lung pattern classification for interstitiallung diseases using a deep convolutional neuralnetwork. IEEE Trans. Med. Imaging 35(5), 1207–1216 (2016)
S. Kolli, M. Ranjani, P. Kavitha, D.A.P. Daniel and A. Chandramauli, Prediction of water quality parameters by IoT and machine learning. in 2023 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 2023, pp. 1–5, https://doi.org/10.1109/ICCCI56745.2023.10128475
H. Yu, Z. Zhou, Q. Wang, Deep learning assisted predict of lung cancer on computed tomography images using the adaptive hierarchical heuristic mathematical model (2020) [online]. Available: https://doi.org/10.1109/ACCESS.2020.2992645
K. Bommaraju, A. Manikandan, S. Ramalingam. Aided system for visually impaired people in bus transport using intel galileo Gen-2: Technical note. International Journal of Vehicle Structures and Systems, 9(2), 110–112 (2017). https://doi.org/10.4273/ijvss.9.2.09
S. Kolli, M. Hasan, B. Hazela, A. A. J. Pazhani, Secure the smart grid by machine learning. in International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 2023, pp. 1–4, https://doi.org/10.1109/ICCCI56745.2023.10128269
K. Srinivas, J. Aswini, P. Patro, D. Kumar, Functional overview of integration of AIML with 5G and beyond the network, in International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 2023, pp. 1–5, https://doi.org/10.1109/ICCCI56745.2023.10128466
M. Sandler, A. Howard, M. Zhu, A. Zhmoginov, L.C. Chen, Mobilenetv2: inverted residuals and linear bottlenecks. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 4510–4520
S.G. Armato III., G. McLennan, L. Bidaut, M.F. McNitt-Gray, C.R. Meyer, A.P. Reeves et al., The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38(2), 915–931 (2011)
A. Asuntha, A. Srinivasan. Deep learning for lung Cancer detection and classification. Multimedia Tools and Applications 1–32 (2020)
X.Z. Zhao, L.Y. Liu, S.L. Qi, Y.Y. Teng, J.H. Li, W. Qian. Agile convolutional neural network for pulmonary nodule classification using CT images. International Journal of Computer Assisted Radiology and Surgery 13, 585–595 (2018)
X.L. Liu, F. Hou, A. Hao. Multi-view multi-scale CNNs for lung nodule type classification from CT images. Pattern Recognition 77, 262–275 (2018)
D. Kumar, A. Wong, D.A. Clausi. Lung nodule classification using deep features in CT images. in Proceedings of the 2015 12th Conference on Computer and Robot Vision (CRV), Halifax, NS, Canada, 3–5 June 2015, pp. 133–138
W. Shen, M. Zhou, F. Yang, C.Y. Yang, J. Tian. Multi-scale convolutional neural networks for lung nodule classification. in information processing in medical imaging (Springer, Cham, Switzerland, 2015) pp 588–599
Y. Xie, J. Zhang, Y. Xia, Semi-supervised adversarial model for benign–malignant lung nodule classification on chest CT. Medical Image Analysis 57, 237–248 (2019)
S.R. Sannasi Chakravarthy, H. Rajaguru. Lung cancer detection using probabilistic neural network with modified crow-search algorithm. Asian Pacific Journal of Cancer Prevention 20(7), 2159 (2019)
S. Kolli, A.P. Krishna, M. Sreedevi. A meta heuristic multi-view data analysis over unconditional labeled material: An intelligence OCMHAMCV: Multi-view data analysis. Scalable Computing: Practice and Experience 23(4), 275–289 (2022)
S. Kolli, M. Sreedevi. Adaptive clustering approach to handle multi similarity index for uncertain categorical data streams. Journal of Advanced Research in Dynamical and Control Systems 10(04) (2018)
R. Ali, A. Manikandan, J. Xu. A novel framework of adaptive fuzzy-GLCM segmentation and fuzzy with capsules network (F-CapsNet) classification. Neural Computing and Applications (2023). https://doi.org/10.1007/s00521-023-08666-y
M. Annamalai, P. Muthiah. An early prediction of tumor in heart by cardiac masses classification in echocardiogram images using robust back propagation neural network classifier. Brazilian Archives of Biology and Technology 65. (2022). https://doi.org/10.1590/1678-4324-2022210316
A. Manikandan, M. Ponni Bala, Intracardiac mass detection and classification using double convolutional neural network classifier. Journal of Engineering Research 11(2A), 272–280. (2023). https://doi.org/10.36909/jer.12237
D. Balamurugan, S.S. Aravinth, P. Reddy, A. Rupani, A. Manikandan. Multiview objects recognition using deep learning-based wrap-CNN with voting scheme. Neural Processing Letters 54, 1–27 (2022). https://doi.org/10.1007/s11063-021-10679-4
A. Manikandan. A survey on classification of medical images using deep learning. Journal of Image Processing and Intelligent Remote Sensing (JIPIRS) 1(01), 5–14. (2021). https://doi.org/10.55529/jipirs.11.5.14
Acknowledgements
Not applicable.
Funding
No funding was received by any government or private concern. “No funding was obtained for this study”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kolli, S., Parvathala, B.R. A Novel Assessment of Lung Cancer Classification System Using Binary Grasshopper with Artificial Bee Optimisation Algorithm with Double Deep Neural Network Classifier. J. Inst. Eng. India Ser. B (2024). https://doi.org/10.1007/s40031-024-01027-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40031-024-01027-w