Abstract
The initial strategy to curb the surge of novel coronavirus disease, COVID-19, is prevention and quarantine, which are dependent on early diagnosis. The latest commercial diagnostic methods include AI/ML-based imaging methods and laboratory diagnosis, which differ in their efficiency. The former requires lung imaging and is useful for last stage patients. It was ensured to overcome the limitation of availability of laboratory-based kits, while the latter involves the collection of the suitable sample from an individual (blood sample, nasal or oral swab). Laboratory methods include methods like RT-PCR which is contemporarily contemplated as the benchmark for its quick and efficient SARS-CoV-2 infection detection. Other diagnosis alternatives include Serum Viral Neutralization (SVN) assays involving antigen–antibody reaction with much lower efficiency contrasted to RT-PCR. Apart from these methods, early detection has been key to the treatment of COVID-19, but the lack of sensitive assays to detect low viral titers acts as an impediment. This review presents an overview of detecting COVID-19 with the aid of several diagnostic techniques along with their benefits and limitations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Around the last week of December 2019, a novel coronavirus (nCoV) called SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) appeared in China (Wuhan, to be specific), which initiated the outburst of coronavirus disease 2019 (COVID-19), hence was named Wuhan Coronavirus. A person once infected by the virus can experience seasonal flu-like mild upper respiratory symptoms, to acute progressive respiratory failure, depending on their immunity. Therefore, this illness requires intensive care and an isolated environment (being highly contagious), else it can lead to hospitalization and even death [1,2,3].
Update on the Current Scenario
On 30th January 2020, World Health Organization (WHO) declared disease caused by the “SARS-CoV-2” as “epidemic” considering it public health crisis of international concern, which later on 11th March 2020 was upgraded to a “pandemic”. There have been around 27,406,213 cases reported and 915,920 fatalities as of 29th August 2020, affecting 213 countries and territories worldwide [1,2,3]. Although mankind has confronted bigger pandemics in the past, this pandemic has caused more than a million infections during January-March 2020 alone [4]. This unprecedented severity depends on the transmission rate which has an average reproduction number of ~ 3.28 and death rate [5]. According to WHO, globally, as of 6:35 p.m. CEST, 24th September 2021, there have been 4,724,876 deaths from among 230,418,451 confirmed cases of COVID-19, with about 5,874,934,542 administered vaccine doses by 22nd September 2021. From this, India reported 446,368 deaths from among 33,594,803 confirmed cases of COVID-19 (from 3rd January 2020 to 6:35 p.m. CEST, 24th September 2021) and a total of 818,513,827 administered vaccine doses as of 20th September 2021 [6].
Pathophysiology of SARS-CoV-2?
SARS-CoV-2 constitutes the third coronavirus (CoV) in the current century to become zoonotic, which has crossed the species barrier from animals to human beings, causing a severe respiratory disease after 2003, SARS-CoV and the other in 2012, MERS-CoV (Middle East Respiratory Syndrome Coronavirus). This nCoV is the 7th CoV that had been known to cause trans-infection among human beings.
SARS-CoV-2 is a single-stranded, positive-sense RNA, and belonging to IV group of viruses (Fig. 1). Its viral genome was successfully sequenced from the patient (Genbank: MN908947) on 12th January 2020, and its sequence was brought in public domain via the GISAID (Global Initiative on Sharing All Influenza Data) platform. It is around 30 kilobase pair genome, comprising of fourteen open reading frames (ORFs) that encode structural, replication, and non-structural accessory proteins. The viral genome was found to be analogous to the bat SARS-CoVs and human SARS-CoVs. It was reported through molecular modelling and various analysis that SARS-CoV-2 is bounded by a lipid bilayer membrane (M), which consists of structural membrane and envelope proteins. The viral envelope (E) similar to SARS-CoV gets formed due to the interaction between this membrane and proteins. The characteristic “corona” appearance of this specific viral family is due to its spiky layer (S) on which glycoproteins are present. The virus make inroads in the host cell with the help of spike proteins which bind and adheres to the specific host cell receptors. The nucleocapsid (N) originates by the binding of the RNA genome with the nucleic acid associated protein [2].
Schematic structure of SARS-CoV, MERS-CoV, and SARS-CoV-2 along with its major structural proteins [3]
How Mutation Plays a Vital Role in Diagnostic Selection/Development?
An early drift can be deduced from the reports of various lineage strains of SARS-CoV-2, signifying that the virus is mutating. Multiple independent investigations are being carried on the virus which provided the details into the conserved and unique characteristics of SARS-CoV-2’s proteome and genome. This will aid in tracking mutations and further gathering evidence regarding its evolution. It is vital to keep track of these mutations, as these may affect important non-structural and structural components of the virus, influencing the selection of epitopes or contributing to reduced response for the new diagnostic techniques [2], therapeutics and vaccines.
What Makes Diagnostic Techniques Vital?
A plethora of articles and documents have been published about “Coronavirus” research as searched on PubMed (29th Aug 2020) of which most of the results were obtained in the year 2020 [7, 8]. Diagnostic testing is critical as it helps us in contact tracing, hence mediating effective disease management and prevention. Symptomatic infected individuals are the primary source of the spread of the virus. They release millions of infectious viral particles while coughing and sneezing in the form of respiratory droplets in the air. Another major concern is the asymptomatic carriers of the virus as they pose a consequential public health threat by unknowingly spreading the virus to other people [2, 9] and can only be identified through diagnostics. Thus, rapid diagnostic testing for SARS-CoV-2 in these individuals is necessary to take pre-emptive measures, like social distancing and quarantine, which will help alleviate further community flare-up.
Diagnostic Tests
Molecular diagnostics centered on virus sequencing aids in the analytical detection of infected patients. The scientists are contesting to develop and approve diagnostics not only to assess the infection but also to immunize against severe SARS-CoV-2 to expedite a return to normal work routine. These rapid and exact tests that aid us in detecting antibodies after the viral exposure, help us in supervising immunity waning or reoccurrence of the disease. For early detection, imaging diagnosis and laboratory tests play a pivotal role in controlling the current pandemic crisis. To date, all the kits fashioned for COVID-19 detection are either antibody detection kits or RT-PCR (Real-Time, Reverse-Transcriptase Polymerase Chain Reaction) ones [9, 10]. The later kits have high accuracy but need costly equipment, time, and qualified personnel to handle the procedure, while the former does not have these caveats and are comparatively less accurate. Moreover, antibody-based kits detect the infection only after the pathogen has triggered the immune response. Currently, most countries are racing against this invincible enemy in various capacities for a vaccine to restore the balance of lifestyle, economy, and health. Governments across the globe have been dealing with their concerned scenarios faced due to COVID-19 by regulating an enormous amount of research work(s), administrative strategies, and economic graphs. Despite the rapid developments of information and modern advances, precautionary measures, researchers, laboratory trials, governments have been struggling to control this pandemic spread [11].
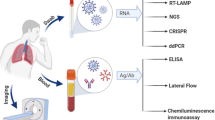
This article provides an overview of the updated information about continuous advancements against the COVID-19 pandemic and highlights the importance of laboratory testing in the prevention and control of the disease. Figure 2 depicts an overall schematic representation of current nCoV diagnostic techniques.
An overview of available nCoV diagnostic techniques and their target molecules. * The last row depicts chest imaging radiograms for CoV detection. Abbreviations—SVN: Serum Virus Neutralization, ELISA: Enzyme-Linked Immuno-Sorbent Assay, CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats, SHERLOCK: Specific High-sensitivity Enzymatic Reporter unLOCKing, RT-LAMP: Reverse Transcriptase Loop-mediated Isothermal Amplification, RPA: Recombinase Polymerase Amplification, RT-PCR: Reverse Transcriptase Real-Time Polymerase Chain Reaction, NGS: Next-Generation Sequencing
Imaging Diagnosis
Imaging diagnosis is a supplementary examination for the identification and routine treatment of CoV diseases. For each alleged infectious patient, a chest radiograph is a requisite. A computed tomography (CT) scan (especially a high-resolution one) can offer doctors supplementary evidence for understanding the condition of the chest in a better way. The chief techniques comprise of Chest radiography and thoracic CT scan. Chest radiography is a technique that focuses on density specificity, which could roughly define lung lesions via transparency in a short period, whereas CT scan possesses spatial specificity that could accurately analyze the transverse section, comprising blood vessels, lesions of lungs, and surrounding tissues. These techniques initially gained importance due to the non-availability of laboratory kits. Although these diagnostics have several advantages like, lower diagnostic time (less than half an hour)) and being inexpensive, but their major disadvantages range from being less accurate than laboratory diagnostics while involving exposure to radiation and at several instances give both false-negative and false-positive results [12].
Chest Radiograph
Chest radiographs are commonly referred as chest X-ray, is a projection radiography technique that utilizes ionizing radiation in the form of X-rays to yield chest images. It is generally a secondary line of diagnosis recommended, as it is not sensitive enough to detect COVID-19 during earlier stages. It is used as a screening method by medicos who are dealing with shortages or when an individual’s physical situation does not permit for conveyance to a CT scanner. In the later stages of the disease, a chest radiograph can help to identify numerous mottled opacities all over the pulmonary area. Eventually, these opacities merge and in serious cases, it may appear as a “whited out pulmonary region” on the film. In several fatal cases, pleural fluid has been reported on a chest radiograph along with the presence of ground-glass opacification (GGO) and consolidation [5, 13].
Computed Tomography
Computed tomography (CT) is the highly recommended imaging technique, that diagnose COVID-19 in its initial stage. It assesses the nature and extension of lesions with precision, which can be readily observed on a chest radiograph. The imaging characterizes lesions based on a variety of factors including dispersal, bulk, pattern, shape, extent, and associated signs. CT scan of a patient suffering from nCoV include several two-sided, marginal, sub- segmental, patchy, or segmental GGO and areas that have merged and are mostly spread along the bronchovascular region and sub-pleural space. At times, there is the presence of accompanying interlobular septal stiffening in the areas of GGO, delivering a crazy-paving view. Air bronchograms with bronchial wall thickening and the areas of consolidation are often extant with rare pleural effusion cases. There is no swelling observed in lymph nodes that may indicate cancer. According to “Expert Recommendations from the Chinese Medical Association Radiology Branch,” based on the degree of lesion, observed on chest CT,COVID-19 patients, can be categorized into three stages: Early, Advanced, and Severe [13].
Early-stage: Single or Multiple patches (scattered or groped) of GGO can be observed on Chest CT, chiefly in the central and lower lungs, along with the bronchovascular bundles. Sometimes, a crazy-paving style can be observed in the zones of GGO due to the presence of intra- and inter-lobular septal thickening. The compulsive progression during the preliminary stage is widening and blocking of the alveolar septal capillary, an emanation of fluid in the alveolar cavity, and it can also reach inter-lobular interstitial oedema.
Advanced stage: During this stage, more lesions that are analogous to prior lesions can be observed on chest CT. The lesions increment both in number and magnitude, marking new disease zones. Air bronchograms can be found in the areas of consolidation. The pathological progression during the advanced stage is the accrual of a cell-rich exudate in the pulmonary cavity, vascular expansion and emanation of fluid in the interstitium. A fusion state gets formed as the fibrous emanation links each alveolus via the inter-alveolar space.
Severe stage: During the later stages of the disease, further consolidation of COVID-19 results in, non-consolidated areas of the lung look as patchy GGO on chest CT. The lungs are observed as a “whited out lung” when most of the lungs are involved, and the thickening of pleura can be observed along with a minor quantity of pleural effusion [14].
Laboratory Diagnosis
Real-Time Reverse Transcriptase Polymerase Chain Reaction
Detection of presence of the viral genome as well as its quantification can be obtained by the single technique RT-PCR. The resemblance between single-stranded COVID-19 viral RNA genomic sequences with SARS-CoV and numerous other bat coronaviruses have aided us in the swift progression of RT-PCR assays for nCoV utilizing the above-mentioned viruses as references. Initial upper, as well as lower respiratory tract specimens, are collected for recognition of the Human Corona Virus. High priority samples for SARS-CoV-2 include nasopharyngeal swabs, whereas oral and pharyngeal swabs, pulmonary aspirates, and sputum are considered low priority specimens. Approved viral isolation kits are used to extract RNA from clinical specimens [2].
The isolated RNA is reverse transcribed to complementary DNA (cDNA), who’s copy number is increased with the help of a real-time quantitative PCR [9]. WHO declared numerous SARS-CoV-2 probe sets and primers hitherto constructed in some developed nations [1]. Primers that target various other segments of the viral genome including the RNA-dependent RNA polymerase (RdRp) gene, the envelope (E) gene, and multiple other genes are mentioned in Table 1. The highest sensitivity was reported by targeting the E gene, tailed by the RdRp gene for approval. Several laboratories have a different approach as they have combined PCR tests entailing multiple primers and probe sets that can be found at specific or various regions in the SARS-CoV-2 genome. Multiple genes are targeted concurrently via primer sets or different regions can be detected in a particular target such as the E gene through proper channeling of these assays [2, 3, 5].
The sensitivity can be improved by preventing the loss of viral RNA during sampling and nucleic acid extraction via these combined assays. Mutations in the viral genome can also be responsible for hindering the sensitivity. Positive controls in these assays can be obtained from laboratory manufactured RNA stemmed from transcripts, and these RNAs can also be useful in generating standard curves. The occurrence and standard of nucleic acid in samples can be substantiated using RNA polymerase as an internal control and molecular grade nuclease-free water as a negative amplification control. If a patient is tested negative, the sample from the infected individual aids as a negative extraction control to observe cross-contamination athwart samples. It also helps in the ratification of test reagents [2, 3].
Advantages of RT-PCR
RT-PCR is projected as the vanguard diagnostic technique for COVID-19. It can be used to analyze numerous specimens in a very short period and can analyze hundreds and thousands of samples within 24 h with a high testing sensitivity (~ 95%). It helps in the timely detection of low viral loads as the estimated limit of recognition of the RT-PCR coronavirus test is < 10 copies/reaction. An individual can be diagnosed as positive for CoV as the presence of viral RNA is indicated by gene amplification. The patient after observation of gene amplification should correlate with clinical inspections, history of the patient, and epidemiological data [2].
Disadvantages of RT-PCR
Apart from expensive instrumentation, it has the possibility to give rise to cross-reactivity (co-infection due to other viruses or bacteria) of primers with nucleic acids and could produce false-positive results. The infective agent detected cannot be considered as the definite cause of the disease in such cases. Rapid viral mutations might appear in the probe-target regions and primers, potentially giving rise to false-negative results. One should not be complacent, even after getting negative results for SARS-CoV-2 infection, and for exclusive confirmation, the results should be confirmed multiple times with changed primer sets being targeted against the same gene and corresponded with medical data to accurately administer infection status in the individual [2, 3, 9].
Serological Assays
Enzyme-Linked Immuno-Sorbent Assay (ELISA)
ELISA is frequently being employed as a helping hand in diagnostics for therapeutic purposes, for instance with convalescent plasma therapy, it is used to evaluate the neutralizing activity of the spike protein’s receptor-binding domain of nCoV, against specific IgM and IgG immunoglobulins present in the donor’s plasma. After the completion of the transfusion, ELISA was further used to identify IgG, IgM, and counteracting antibody titers in the sera of infected individuals for assessing the reaction to the treatment [9]. Since orthodox ELISA is a time-consuming and troublesome method, many diagnostic firms utilize pre-coated assays to detect human CoV within the specimen being tested. The sensitivity and specificity of ELISA kits depend specifically upon the onset of symptoms and the type of viral protein targeted. Multiple immunoassays and molecular tests were promptly developed, albeit several are yet to achieve formal approval and clinical validation [5].
Multiple investigations, so far have utilized ELISA to gain knowledge of the temporal expression of antibody in SARS-CoV-2 infected individuals and have not much concentrated on immunoassays targeting full spike protein antigen [15]. Amanat et al. [16] found that spike protein antigen reactivity demonstrated excellent results, with IgG3, IgA, and IgM being the dominant isotypes in the patient’s specimen. Similarly, Okba et al. [17] obtained 116 banked infected specimens of the patients, their investigation proved, cross-reactivity in few samples from other human CoV, non-human CoV respiratory viruses, non-respiratory viruses, and nuclear antigens. They further examined commercial ELISA using spike protein subunit-1 antigen for identifying IgG and IgA antibodies, in which the former depicted higher specificity while the latter depicted higher sensitivity.
Zhao et al. [18] also developed a similar receptor binding domain sandwich ELISA but targeted it to only estimate IgM reactivity and a nucleocapsid ELISA directed to estimate IgG reactivity. The total antibody assay had excellent performance with 93% sensitivity and 99% specificity, whereas the IgM and IgG ELISA displayed 99% specificity: and 83% and 65% sensitivity, respectively. A likewise study by Adams et al. [4] reported similar results with IgG having high sensitivity from 10 days after the initial symptoms occur. Similarly, MacMullan et al. [19] analyzed saliva specimens, with a high degree of sensitivity (84.2%) and cent per cent preciseness in a set of 149 clinical samples, due to uncertainty about which immunoglobulins are most persistent over time, they targeted IgG, IgA, and IgM. Few more studies on the efficacy of ELISA for COVID-19 diagnosis in various nations can be seen in Table 2.
Although the sensitivity of all these assays varies depending on the time of specimen acquired, the overall sensitivity remains consistent. This method has number of disadvantages like being labor-intensive, less accurate than RT-PCR, and is not useful for point-of-care testing, however, it can determine antibody titers and is helpful in selective isotype detection. The utilization of these alternatives as clinical approaches is yet to be confirmed. Determination of final role of these technologies in SARS-CoV-2 diagnosis is challenging at current times, as it is restricted by resource limitations, preventing investigation of drawbacks in experimental methods. Thoroughly, investigated assays and tested across variety of studies will provide compulsive evidence about utility of these technologies [5, 15].
Serum Virus Neutralization (SVN) assay
This assay is a serum-based assay that assesses the ability of antibodies of a patient to neutralize the infectivity of human coronavirus. It aids in measuring their ability to attenuate infection. SVN is one of the best tools for evaluating defensive antibodies and is contemplated as one of the most dependable tools, as it can update us about the recovering plasma. That can further be used as a passive antibody treatment for infection from CoV, especially in fatal cases of individuals. It has not been much in practice, therefore, we are still not having an abundance of data, but current observations portray that transfusion of recovering plasma in the infected patients can curb the replication of CoV and protect people from further CoV infection. Though serological assays are neither used frequently nor for repetitive diagnosis yet are considered as frontiers when it comes to special symptoms like these [20, 21].
The blood specimens can be garnered from patients recovering from CoV and utilized to arrange plasma. A consent form informing in written is required from both the recovering patient who will donate and the recipient. Various cell lines can be implemented for the transduction of CoV. Cell lines of kidneys from humans as well as monkeys etc. can be used. Serial dilutions of serum collected from recovering patients are added to identify strains of the virus. The combination of serum and viral strain is inoculated into a liable cell monolayer. It is later gestated for adsorption of SARS-COV-2. A microscopic examination helps in measuring the cytopathic effect. It is carried out after a 120-h incubation period, or colonial growth, after incubating it for a day. A decrease in the activity of coronavirus can be observed in the defusing antibody load which is the highest dilution of serum [22].
Advantages of SVN
One of the major aspects of serological neutralization tests is that they are highly vigorous and can be smoothly replicated again. The other salient feature is that they might help in recognizing CoV neutralizing antibodies in recovering plasma models, which help us determine the best aspirants for therapy. The neutralizing action of the assay when combined with antibody and viral load, can be concurrently supervised. They can be monitored via balancing plasma models in patients getting recovery plasma. It also helps to predict the clinical efficiency for regulating patient and donor aspects and institute suitable techniques and protocols [2].
Disadvantages of SVN
The laboratory analysis by SVN has not yet been able to meet up its true potential because of improper ease of access as the live CoV strain is controlled and supervised strictly. Although it is a low-cost assay, it is labour-intensive and necessitates vigilant in-house calibration, along with quality regulation [2].
Some Other Promising Techniques for SARS-COV-2 Detection
Scientists are focusing on developing approaches for quick nucleic acid detection. After the detection of specific nucleic acids, they can be used for unfolding new approaches for COVID-19 identification.
Isothermal Nucleic Acid Amplification (INAA)
This technique compared to PCR don’t involve thermal cycling. It intensifies nucleic acid target sequence in an efficient and exponential manner for identification. Numerous approaches have been emerged for detecting nucleic acids to date. A few commercially viable and critically acknowledged techniques based on INAA, include Loop-mediated Isothermal Amplification (LAMP), which consorts RT and LAMP to identify RNA, recombinase polymerase amplification (RPA), helicase-dependent amplification (HDA), and nucleic acid sequence-based amplification (NASBA). These assays facilitate primer binding through the integration of isothermal means and later aids in amplification. A polymerase that facilitates strand displacement is utilized for parting the annealed strand to the target sequence for recognition. Photometry helps to detect the multiplied gene products. Various commercial molecular diagnostic platforms and big Pharma labs employ INAA, it is deemed to be the quickest accessible molecular laboratory point-of-care (real-time) testing method for the recognition of nCoV [10, 23].
RT-LAMP
This technique has demonstrated successful detection of CoV in medical samples from infected patients by targeting multiple genes to improve copies/reaction number (c/r), refer Fig. 3. DNA strand displacement and intensification of the target can be achieved through multiple loop primers. A milestone of detection of 200 c/r and 20 c/r was achieved by targeting the S gene and the ORF1ab gene, respectively, displaying analogous results to RT-PCR amplification. It can be considered as a decisive testing method as it demonstrates 100% sensitivity as well as 100% specificity. The average time it consumed for detection was < 30 min, making it a promising approach in COVID-19 diagnosis [9, 23].
LAMP schematic performance is depicted [9]. [A] The beginning of LAMP when the forward inner primer binds to the A2 (C) region while the forward primer (A1) binds to A1 (C), displacing the complementary strand. [B] depicts the backward inner primer binding to B2 (C) while the backward primer (B3) binds B3 (C), displacing the complementary strand. [C] shows that a complementary sequence initiates loop formation and lastly, [D] is where the loop structures are formed that permit the use of multiple loop primers
Recombinase Polymerase Amplification (RPA)
It is a single tube, INAA technique that detects nucleic acid without the need to generate cDNA by adding a reverse transcriptase enzyme to a recombinase polymerase amplification reaction. It incorporates isothermal approaches for intensification and channels reverse transcription ensued by recombinase activity that arbitrates primer (N gene is targeted) binding to a similar sequence in double-stranded DNA. It is considered a decisive approach as it delivers 100% analytical sensitivity and specificity through successive intensification by polymerase arbitrated primer extension. RPA can be considered as significantly advantageous over RT-PCR as it is quicker and portable, and further has much more scale than the latter [2].
Comparative Advantages and Disadvantages of Mentioned INAA Techniques
Among the mentioned isothermal nucleic acid amplification techniques, LAMP shows several advantages like being cost-effective, tolerant to biological substances, can be easily detected even by an inexpensive turbidity-meter and lastly, they utilize 4 to 6 primers spanning 6 to 8 distinct sequences depicting high specificity. Their only limitation is that they cannot perform multiplex amplification and their primer designing is complex too. Whereas RPA is extremely quick (~ 20 min), saves power when operated at room temperature (37 °C) as no initial heating step is required, have a simple primer design, and are robust to biological substances. HDA shares similar advantages with RPA, with its only drawback being expensive enzymes. NASBA is specifically designed to detect RNA and conserves power at 41 °C, but is less efficient in amplifying RNA targets out of the range of 120 to 250 base pairs [24].
Next-Generation Sequencing (NGS)
SARS-CoV-2 genome consists of ~ 30,000 nucleotides. NGS empowers us to completely sequence these nucleotides of the genome of the nCoV. We can successfully detect CoV through NGS as it offers us the identification of the virus and its scrutinization. We can also observe the evolutionary pattern of the virus and its source through NGS. All these sequences are submitted and stored into the database exclusive to CoV (GISAID EpiCoVTM Database). More than 17,000 coronavirus sequences have been identified and reserved because of the NGS efforts around the world to date.
Using medical samples, RNA is obtained and purified to eradicate human cytoplasmic and ribosomal rRNA. After the cDNA synthesis, the RNA is fragmented, amplified, and sequenced to produce CoV typing with very high precision in less than a day. A set of highly precise, universal CoV primers is used for achieving this grade of sensitivity. Viral load, the rate of depletion of human rRNA, and read count per sample influence the quantity of virus-specific reads acquired and aids in proper scrutiny of the SARS-CoV-2 genome.
According to the global NGS data collected, it depicts SARS-CoV-2 genome is comparatively stable, numerous mutations have been observed in people with noticeable indications that were absent in the strain that originated from Wuhan, China. An 81 bp deletion in the viral genome was reported in a patient from the United States of America. These mutations can insinuate a more or less virulent strain of the virus. Although NGS is one of the most thorough, sensible methods for recognizing nCoV, it is quite expensive, and therefore cannot be used for large-scale testing as it requires multiple sample preparation steps making it labour-intensive [25].
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Assay
DNA or RNA can be sensed using the CRISPR assay through nucleic acid pre-amplification. This assay is pooled with CRISPR-Cas enzymology for better sensitivity and specificity of the sequences. It is a lately found recognition technique that is being used for the detection of nCoV. It works on a system that is specific to RNA detection as it works on a CRISPR RNA (crRNA) directed recognition technique. The Cas13a enzyme is a crucial feature of this method as it binds to specific RNAs sequence while unbound RNA is removed by non-specific trans-endonuclease. It is crucial for signal intensification and nucleic acid recognition. We can achieve more sensitive results by pairing the Cas13a assay with RPA. SHERLOCK- which is abbreviated for Specific High-Sensitivity Enzymatic Reporter unLOCKing is an approach that constitutes pairing of the enzyme with any isothermal exponential amplification method. When these two techniques are combined, it permits colorimetric, cross flow, and additional monitored approaches to facilitate the quick identification of SARS-CoV-2 and a lot of other targets as well [26].
Cas13a Assay
The Cas13a protein must be recombinantly expressed, it is a must that we purify it after the expression for optimal results. The purified Cas13a possesses an endonuclease activity and is necessary for crRNA targeting sequences in the ORF1ab and S gene of the nCoV. Trans-cleavage of reporter probes occurs due to target site recognition, which increases fluorescence output signals, validating the existence of SARS-CoV-2 RNA. CRISPR has been modified successfully for lateral-flow assays and promises extensive functions as a nCoV indicator in the clinical as well as R&D sectors. We can develop a paper dipstick test through crRNA/Cas13a that produces signals in about ½ an hour to 1 h [26]. Cas13a is a potential tool, and these constructive and sequential developments in life sciences tender huge hopes for proper disease regulation in the future.
Table 3 below summarizes the vital aspects of chief laboratory assays discussed in this review for recognizing nCoV elements.
Conclusion
This pandemic has proven the worth of various rapid diagnostics. More extensive testing is required to inspect the prevalence of the disease in the population and its virulence and fatality in different age groups since the current public health emergency presents us with expanding outbreak. It is the need of the hour to accurately recognize and distinguish between symptomatic and asymptomatic people. Thorough epidemiological statistics will better determine the critical infection and mortality rates among diseased populations.
As seen in Table 3, each diagnostic method has its own importance, hence we must support constant research, enhancing prevailing antibody tests to resolve whether immunity is being sustained and/or averts repeated infection, and explore the efficiency of inert antibody treatments for SARS-CoV-2 infection. To predict the seriousness and advancement of the disease, scientists could request permissions to study the deposited samples that might be a biological bulk reservoir of the virus, to identify related biomarkers. The novel disease biomarkers may provide insights about the infection mechanism and the reason why some individuals are susceptible to CoV infection. Blood and plasma collection centers should be run under precise supervision and should allow access to infected patient’s samples for testing efficacy of novel diagnostics. The hunt for improved diagnostics in terms of cost, efficiency and mass testing for COVID-19 still continues. In this context, we would like to mention that Akermi et al. have proposed an electrolysis-based DNA chip-based method [26].
The CoV pandemic demonstrates how rapidly data needs to be distributed, how social contacts and good communication are vital in today’s world. Instituting contact athwart laboratories globally aids to develop master protocols. We can corroborate orientation panels that can be accessed by other researchers with ease, helping us synchronize the information. It aids us with assemblage and use of findings and supervisory organization. Having options for diagnosis also lessens the pressure on any particular manufacturer or pharma corporation, as different manufacturers may use distinct materials, which helps us relieve tricky choices. We can also limit testing to the most liable patients and fatal conditions.
Today, we are betting on long-lasting immunity against SARS-CoV-2 after an infection or via vaccination. It is vital to note that by now more than half a dozen vaccine candidates are being used for mass immunization. While serologic testing seems promising, there are still data gaps that must be elucidated to give significant endorsements for its utilization in diverse clinical circumstances.
References
World Health Organization (2020) Coronavirus disease 2019 (COVID-19): Situation report, 57. https://apps.who.int/iris/handle/10665/331481. Accessed 25 Sept 2021
D’Cruz RJ, Currier AW, Sampson VB (2020) Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Front Cell Dev Biol 8:468. https://doi.org/10.3389/fcell.2020.00468
Yüce M, Filiztekin E, Özkaya KG (2021) COVID-19 diagnosis—a review of current methods. Biosens Bioelectron 172:112752. https://doi.org/10.1016/j.bios.2020.112752
Adams E, Ainsworth M, Anand R et al (2020) Antibody testing for COVID-19: a report from the National COVID Scientific Advisory Panel. Wellcome Open Res 5:139. https://doi.org/10.12688/wellcomeopenres.15927.1
Ejazi SA, Ghosh S, Ali N (2021) Antibody detection assays for COVID-19 diagnosis: an early overview. Immunol Cell Biol 99:21–33. https://doi.org/10.1111/imcb.12397
WHO (2021) WHO Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/?gclid=CjwKCAiAyc2BBhAaEiwA44-wW-1MZaSsA6ro9Kvl0lZEaKY0EvZXPwSN6MqMy92rf27DlqQl_UeSUxoCV4QQAvD_BwE. Accessed 22 Feb 2021
Rajkumar RP (2020) COVID-19 and mental health: a review of the existing literature. Asian J Psychiatr 52:102066. https://doi.org/10.1016/j.ajp.2020.102066
Lazarus JV, Palayew A, Rasmussen LN et al (2020) Searching PubMed to retrieve publications on the COVID-19 pandemic: comparative analysis of search strings. J Med Internet Res. https://doi.org/10.2196/23449
Sen TA, Nerurkar SN, Tan WCC et al (2020) The virological, immunological, and imaging approaches for COVID-19 diagnosis and research. SLAS Technol 25:522–544. https://doi.org/10.1177/2472630320950248
Akermi S, Sinha S, Johari S et al (2021) Computational Intelligence Methods for the Diagnosis of COVID-19. In: Studies in computational intelligence. Springer, pp 207–223. https://pesquisa.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/resource/pt/covidwho-891249?lang=en. Accessed 25 Sept 2021
Brodeur A, Gray D, Suraiya AI, Bhuiyan J (2020) A literature review of the economics of COVID-19. https://doi.org/10.1111/joes.12423
Koo HJ, Lim S, Choe J et al (2018) Radiographic and CT features of viral pneumonia. Radiographics 38:719–739. https://doi.org/10.1148/rg.2018170048
Zalzala HH (2020) Diagnosis of COVID-19: facts and challenges. New Microbes New Infect. 38:100761. https://doi.org/10.1016/j.nmni.2020.100761
Jin YH, Cai L, Cheng ZS et al (2020) A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 7:4. https://doi.org/10.1186/s40779-020-0233-6
Espejo AP, Akgun Y, Al Mana AF et al (2020) Review of current advances in serologic testing for COVID-19. Am J Clin Pathol 154:293–304. https://doi.org/10.1093/ajcp/aqaa112
Amanat F, Stadlbauer D, Strohmeier S et al (2020) A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. https://doi.org/10.1038/s41591-020-0913-5
Okba NMA, Müller MA, Li W et al (2020) Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. https://doi.org/10.3201/eid2607.200841
Zhao J, Yuan Q, Wang H et al (2020) Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034. https://doi.org/10.1093/cid/ciaa344
MacMullan MA, Ibrayeva A, Trettner K et al (2020) ELISA detection of SARS-CoV-2 antibodies in saliva. Sci Rep 10:1–8. https://doi.org/10.1038/s41598-020-77555-4
Guo L, Ren L, Yang S et al (2020) Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71:778–785. https://doi.org/10.1093/cid/ciaa310
Shen C, Wang Z, Zhao F et al (2020) Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA 323:1582–1589. https://doi.org/10.1001/jama.2020.4783
Nie J, Li Q, Wu J et al (2020) Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9:680–686. https://doi.org/10.1080/22221751.2020.1743767
Yan C, Cui J, Huang L et al (2020) Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 26:773–779. https://doi.org/10.1016/j.cmi.2020.04.001
Zaghloul H, El-shahat M (2014) Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. World J Hepatol 6:916–922. https://doi.org/10.4254/WJH.V6.I12.916
Korber B, Fischer WM, Gnanakaran S et al (2020) Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812-827.e19. https://doi.org/10.1016/j.cell.2020.06.043
Kellner MJ, Koob JG, Gootenberg JS et al (2019) SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 14:2986–3012. https://doi.org/10.1038/s41596-019-0210-2
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Quraishi, M., Upadhyay, S.K. & Nigam, A. COVID-19 Diagnostics: A Panoramic View on Its Present Scenario, Challenges and Solutions. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 92, 709–721 (2022). https://doi.org/10.1007/s40011-022-01375-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-022-01375-x