Abstract
In the present study, ten genotypes of Lablab purpureus L. Sweet bean were screened for genetic divergence by using 5 primers, i.e., OPA 01, OPA 04, OPA 13, OPB 09, and OPB 10. Analysis of diversity such as PIC, MI, MRP, Dendrogram, and RP was done. Out of the 5 primers used, the average number of Polymorphic bands detected was 7.6 and the average level of polymorphism was 74.23%. OPA 01 showed the highest level of polymorphism (92.85%) while OPB-09 showed the least level of polymorphism, i.e., 55.55%. The polymorphic information content ranged from 0.24 (OPA-04) to 0.318 (OPA-01) at an average PIC value of 0.275. The mean of resolving power per RAPD primer is 3.65. The MI value ranged from 1.32 to 2.79 with a mean of 1.79/primer. The mean of MRP per primer was 0.47. The clustering of genotype was performed following the UPGMA method. The optimal tree with the sum of length branch is 13.30325.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dolichos lablab or Lablab purpureus is an inhabitant of Southeast Asia or India [1]. It is likely to be originated from Asia and has been used for planting for a long time. A wild variety of Lablab is grown in India. D. lablab was taken to the African tropical part and from there spread across Indonesia, Sudan, Caribbean, Malaysia, Papua New Guinea, Mainland China, Philippines, West Africa, Egypt, and South America.
Flowers grow in clusters, on an inflorescence that is unbranched at an angle between the main stem and leaf. Based on the variety, the color of the flower may be white, blue, or purple. Seed pods are 5 cm [2] to 10 cm in length [3], flat, smooth, pointed, and contain 2 to 4 seeds. Seeds may be cream, white, pale brown, red, dark brown, black, or mottled depending on the variety.
The stems are twining and Cylindrical, hairy or glabrous, often 2–3 m, but usually to 10 m long. Others are dwarf and bushy. Leaves are alternate and trifoliate, with leaflets ovate, measuring 5–14 cm × 4–14 cm. On the other hand, very broad leaflets, lateral ones are lopsided, 7.5–15 cm long and nearly as broad, rather abruptly acuminate.
Lablab purpureus has originated in India but very less work has been reported for the genetic divergence for improving yield. During the last 12 years, different molecular techniques have been screened in knowing the profiles of genotypes of various plant species or individuals, and various genotypes have been discovered for creating genotypes banks. However, DNA markers, such as RFLP, AFLP, and RAPD, have major advantages in comparing with isozyme and morphological markers due to their availability in large numbers provide higher discrimination of cultivars and are unaffected by environment factors [4]. In the present study, RAPD [5, 6] has been studied not only for knowing the variability within groups of plants but also for obtaining fingerprint [5, 7, 8]
Comparing with other studies, RAPD markers are a well-known method for detecting genetic diversity. Despite lesser reproducibility, this method is generally more suitable due to its cost-effectiveness by using a nanogram amount of sample. This technology was used to evaluate the genetic divergence between and within populations.
Various examples of the application of RAPD molecular markers are there in the study of the genetic divergence of Lablab purpureus [9,10,11,12,13,14,15,16]. The work cited by the above workers has analyzed the genetic variability among various genotypes that are different from the present genotypes taken by the authors. Moreover, the present work establishes the efficiency of different DNA markers for analysis of genotypes through RAPD markers which have been ignored by previous workers. A similarity matrix among the targeted genotypes along with phylogenetic analysis is considered a unique feature in the present work. Therefore, present investigation was carried out to analyze genetic diversity and genetic relatedness of unexplored Lablab purpureus bean genotypes at the molecular level, to construct a dendrogram, and a similarity band matrix and generate efficiency and polymorphism data of RAPD marker in genetic divergence analysis among Lablab purpureus.
Material and Methods
Genotypes Used For Study
In the present Investigation, 10 genotypes (VERSEM- 847, 739, 799, 757, 776, 733, 730, 746, 843, 702) were used for studying the genetic diversity of Lablab purpureus.
Extraction of DNA
Mature leaves were used for DNA extraction by the CTAB method with little improvement in the procedure [17]. By using mortar and pestle, 150 mg of leaf was crushed with extraction buffer (1 ml). The extraction buffer was then added in vials having crushed leaves. The tubes were incubated for 45 min at 65 °C with usual mixing. An equal volume of chloroform: isoamyl alcohol (24:1) was added and shaken for 5 min following centrifugation at 14,000 rpm for 15 min. To the supernatant, a 2/3 volume of isopropanol was added and centrifuged at 14,000 rpm for 10 min. Then, the supernatant was removed and the pellet was washed with ethanol (70%) which contained DNA, suspended in 100 μl of sterile D.W. with 10–15 μl of RNAse and incubated for 30 min at 65 °C. A qualitative assessment of the DNA was done by running a sample on 0.8% agarose gel. The agarose gel (0.8%, 30 ml) was casted in 1X TBE (Tris Borate EDTA) buffer containing 0.5 µg/ml of ethidium bromide. The sample (5 µl of DNA) was loaded in every well. Electrophoresis was done at 100 V for 1 h. The gel was visualized under the Gel documentation unit (Syngene). The presence of single compact band indicates the intact and high molecular weight of isolated DNA and quantification was made by recording its absorbance at 260 nm and 280 nm using a UV–VIS spectrophotometer.
RAPD Analysis
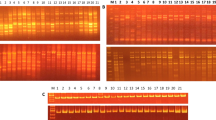
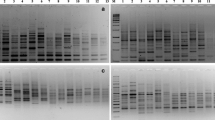
RAPD was done by using 5 primers. The PCR reaction mixture had a volume of 25 μl of 50 ng of DNA, 1 × Taqbuffer, 1.5 mM MgCl2, 0.2 mMdNTP, 20 pmol of each primer and 1.25 U TaqDNA polymerase (‘Fermentas’). The annealing temperature of the cycling constraint was readjusted for every microsatellite primers depending on Tm calculated in the composition of sequence: Tm = 4·(G + C) + 2·(A + T) − 3·C. The PCR was performed in ‘Bio-Rad Thermocycler.’ After amplification, the PCR samples were casted on 1.2% Agarose gel (Hi-media, molecular grade), which was prepared in 0.5 × TBE buffer having 0.5 µg/ml of the EtBr. The amplified samples were electrophoresed for 2–3 h at 100 V with cooling. After analysis, the gel was visualized under the Gel documentation unit (Fig. 1).
Data Analysis and Polymorphic Information Content
The image of the gel was taken by a digital camera, in the Gel documentation unit. Bands of RAPD were separated based on their molecular weight (length of amplified polynucleotide). 1 kb DNA Ladder was loaded with each primer sample to determine the molecular size. The distance travelled by fragments which are amplified from the well was translated to molecular sizes in relation to molecular weight marker. Band presence was scored as ‘1’ and band absence as ‘0’. Dimly seen bands were not scored but a major band equivalent to a dimly band was measured for scoring. To verify the presence of bands and to determine the reproducibility, all the primers were replicated thrice.
To estimate the genetic profile of Lablab purpureus genotypes, the performance of the markers was measured using four parameters (1) PIC, (2) MRP, (3) RP, and (4) MI.
PIC of a band was evaluated [18] as follows:
where fi is the frequency of the band present, and 1 − fi is the frequency of band absent. The frequency was calculated as the ratio between the number of amplified bands, i.e., present band at every locus and the total number of genotypes. The PIC value was calculated by using the average PIC value from all locus of each primer.
The resolving power (RP) of each primer was calculated as [19]:
where Ib shows the informative fragments. The Ib can be represented on a scale of 0–1 by the following formula:
where p is the proportion of genotypes having a band.
Further, the authors calculated mean resolving power for each primer as:
where n is the number of polymorphic band for that primer [19].
The MI was evaluated as [20]:
where EMR is the product of the fraction of polymorphic loci and the number of polymorphic loci for an individual assay.
Results and Discussion
RAPD Analysis
Five primers were used in the reaction of PCR to amplify bands of DNA from 10 genotypes due to the generation of a higher number of polymorphic bands by the primers. A total of 291 fragments were detected when PCR amplified samples were split on 1.2% agarose. Out of them, 38 fragments were polymorphic. Out of the 5 primers used, 4 showed very good polymorphism. These primers revealed a mean value of 58.2 bands per primer out of which 7.60 bands were polymorphic (76%). The primer OPA-01 produced the maximum number of fragments (14) followed by primers OPA-04, OPA-13, and OPB-09 producing 10, 9, 9 fragments, respectively. The minimum numbers of fragments were produced by primer OPB-10, i.e., 8 (Fig. 1 and Table 1).
The generated bands with primer OPA-01 had 92.85% of polymorphism. The largest band recognized is of 1720 bp and the smallest being 140 bp with primer OPA-01. The generated RAPD profile with primer OPA-01 showed a total of 14 bands. A total of 10 bands were identified in the RAPD profile generated by the primer OPA-04 that revealed 70.00% of polymorphism. The largest band recognized is 2380 bp, whereas the smallest band is of 240 bp. With primer OPA-13 out of the 9 bands, only 7 are polymorphic showing 77.77% polymorphism. The largest band recognized is 1140 bp, whereas the smallest about 240 bp. The profile generated with the primer OPB-09 shows a total of 9 bands with 55.55% polymorphism among the genotypes studied. The largest band recognized has a molecular weight of 980 bp, and the smallest recognized has a molecular weight of 240 bp. On the other hand, the profile generated with the primer OPB-10 showed a total of 8 bands with 75% polymorphism. The largest band recognized has molecular weight of 1340 bp, and the smallest has a molecular weight 240 bp.
RAPD markers have proved to be more useful in determining the genetic diversity of Lablab purpureus genotypes. Rai et al. (2011) reported genetic diversity among 48 Indian bean (Lablab purpureus L. Sweet) genotypes with 25 RAPD primers having 178 polymorphic bands. In our study, ten Lablab purpureus genotypes showed considerable genetic variation with the 5 RAPD primers. As a result, Lablab purpureus had the highest number of bands generated by primer OPA-01 (14 bands) and the lowest number of bands generated by primers OPD-10 (8 bands). The findings are in support of Gnanesh et al. [21] who studied six RAPD primers and illustrated a high level of genetic variation among the 15 field bean genotypes having 91.30% polymorphism rate. Different variable factors like the structure of primer, few annealing sites, etc., are responsible for the variation in the number of bands amplified by different primers [22]. Also, the bands which were amplified had a different molecular weight which confirmed the differences in the length between the primer binding sites and the genomic DNA, resulting in the shifting of bands [23]. The above findings suggest that many Lablab purpureus genotypes have higher than expected levels of genetic variation and can bring about genetic exchange [12, 24].
Polymorphism Information Content and Data Collection
For comparing the efficiency of multilocus marker, PIC values were determined. PIC values are not uniformly high for all the RAPD markers. The value of PIC ranged from 0.24 (OPA 04) to 0.318 (OPA-01) with an average PIC value of 0.275. The RP is a parameter that indicates the discriminatory potential of the primers chosen. The mean resolving power per RAPD primer is 3.65. The highest RP was shown by primer OPA 01 (8.70) and the lowest by OPB 10 (1.20). The MI value ranged from 1.32 to 2.79 with a mean of 1.79/primer. The highest value was shown by primer OPA 01 (2.79) and the lowest by OPB 09 (1.32). The mean of MRP per primer is 0.47. The highest was shown by OPA 01 (0.63) and least by primer OPB 10 (0.20) (Table 2).
Phylogenetic Study
The evolutionary history was inferred using the UPGMA method [25]. Evolutionary analyses were conducted in MEGA7 [26]. Cluster 1 (0.4682) consisted of two sub-clusters (0.1024 and 1.2738) which include 50% of the genotypes. The sub-cluster 0.1024 consisted of VRSEM 847, VRSEM 702, VRSEM 757, VRSEM 733 while sub-cluster 1.2738 consisted of only VRSEM 739, whereas Cluster 2 (0.0804) divided into again 2 sub-clusters of 0.1616 and 0.2992 and consisted of VRSEM 746, VRSEM 843, VRSEM 730, VRSEM 799, and VRSEM 776 (As shown in Fig. 2). The optimal tree with the sum of branch length is 13.30325 (next to the branches).
Conclusion
In the present study, the potential ability of RAPD markers was tested to evaluate the genetic diversity of Lablab purpureus because the morphological characterization was not accurate enough for the parent selection to produce a variety with high yielding ability. All the 5 primers showed more than 50% level of polymorphism but 4 revealed the highest level of polymorphism. Based on the similarity matrix obtained from the different marker systems, RAPD markers were more suitable for distinguishing the closely related species. Therefore, it is concluded that varietal selection on the basis of RAPD markers can be used for further breeding studies proposed for the Bundelkhand area in India.
References
Murphy AM, Colucci PE (1999) A tropical forage solution to poor quality ruminant diets: a review of Lablab purpureus. Livest Res Rural Dev 11(2):112
Cook BG, Pengelly BC, Brown SD, Donnelly JL, Eagles DA, Franco MA, Hanson J, Mullen BF, Partridge IJ, Peters M, Schultze-Kraft R (2005) Tropical forages: an interactive selection tool. Lablab purpureus. CSIRO, DPI&F (Qld), CIAT, and ILRI, Brisbane, Australia
Valenzuela H, Smit J (2002) Sustainable agriculture green manure crops. SA-GM-7. Cooperative Extension Service, College of Tropical Agric. and Human Resources, Univ. of Hawaii at Manoa
Borém A (1998) Melhoramento de Plantas, 22ndª edn. Universidade Federal de Viçosa, Viçosa
Williams JG, Kubelik AR, Livak KJ, Rafalski JA et al (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Dunemann F, Kahnau R, Schmidt H (1994) Genetic relationships in Malus evaluated by RAPD “fingerprinting” of cultivars and wild species. Plant Breed 113:150–159
Paillard M, Lashermes P, Pétrard V (1996) Construction of a molecular linkage map in coffee. Theor Appl Genet 93:41–47
Rai N, Singh PK, Rai AC, Rai VP, Singh M (2011) Genetic diversity in Indian bean (Lablab purpureus) genotypes based on morphological traits and RAPD markers. Indian J Agric Sci 81(9):801–806
Md SB, Mohammad Z, Md MR (2012) Assessments of genetic diversity in country bean (Lablab purpureus L.) using RAPD marker against photoinsensitivity. J Plant Dev 19:65–71
Liu CJ (1996) Genetic diversity and relationships among Lablab purpureus genotypes evaluated using RAPD markers. Euphytica 90(1):115–119
Sultana N, Ozaki Y, Okubo H (2000) The use of RAPD markers in lablab bean (Lablab purpureus (L.) sweet) phylogeny. Bull Inst Trop Agric Kyushu Univ 23:45–51
Tian Z, Wang S, Wang W, Liu L (2005) Study on the diversity of germplasm resources of Dolichos lablab (L.). Nat Sci J Hainan Univ 23(1): 53–60
Gnanesh BN, Reddi SM, Raja RK (2006). Genetic diversity analysis of field bean (Lablab purpureus (L.) sweet) through RAPD markers. In: BARC Golden Jubilee &DAE-BRNS Life Sciences Symposium on Trends in Research and Technologies in Agriculture and Food Sciences at Bhabha Atomic Research Centre (BARC), Mumbai, p 78
Dholakia HP, Mehta DR, Joshi MK, Delvadiya IR (2019) Molecular characterization of Indian bean (Lablab purpureus L.) genotypes. J Pharmacogn Phytochem 8(2):455–463
Kumar A (2019) Study of genetic and morphometric diversity in Dolichos lablab cultivars using DNA fingerprinting (RAPD), Protein profiling (SDS-PAGE) and morphometric tools. Univ Rev 10(6):184–192
Doyle JJ, Doylle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:119–128
Roldan-Ruiz I, Dendauw J, Vanbockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Prevost A, Wilkinson MJ (1999) A new system for comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci 173:638–649
Gnanesh BN, Reddi Sekhar M, Gopal K, Raja RK, Eswara Reddy NP (2005) Genetic diversity analysis of field bean (Lablab purpureus L. sweet) through RAPD markers. Indian J Plant Genet Resour 18(3):233–235
Kernodle SP, Cannon RE, Scandalios JG (1993) Concentration of primer and template qualitatively affects product in RAPD-PCR. Biotechniques 14:362–364
Devos KM, Gale MD (1992) The use of random amplified polymorphic DNA markers in wheat. Theor Appl Genet 84:567–572
Konduri V, Godwin ID, Liu CJ (2000) Genetic mapping of the lablab purpureus genome suggests the presence of ‘cuckoo’ gene(s) in this species. Theor Appl Genet 100:866–871
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Acknowledgements
The authors are thankful to the Department of Botany, Bundelkhand University, Jhansi, for providing necessary facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
The genetic diversity study is useful in characterization genetic variability among varieties of Lablab purpureus for future research in the field of breeding and earning higher remuneration from genetically ascertained varieties.
Rights and permissions
About this article
Cite this article
Singh, V., Kudesia, R. & Bhadauria, S. Assessment of Genetic Diversity in Some Indian Lablab purpureus, L. Bean Genotypes Based on RAPD Marker. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 90, 855–861 (2020). https://doi.org/10.1007/s40011-019-01158-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-019-01158-x