Abstract
The Indian gooseberry, Phyllanthus emblica is an important minor fruit crop having commercial significance. Due to great tolerance to salinity and sodicity it is well adapted to arid conditions of Rajasthan. Genetic diversity studies of eight varieties Anand-2, Banarasi, Chakaiya, Francis, Krishna, Kanchan, NA7 and NA10 were carried out. The morpho-physiological characterization proved insufficient to distinguish studied varieties of P. emblica. Ten RAPD primers exhibited an average of 56.18 % polymorphism in banding pattern and 83.1 % PIC. They can be used as powerful markers to reveal genetic diversity in P. emblica. Present study validates the utility of ITS rDNA region being third generation molecular marker as a reliable indicator of phylogenetic interrelationships, especially for ITS regions as DNA barcode at higher levels can serve as an additional approach for identification and genetic cataloguing of P. emblica germplasm for crop improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic resources of arid fruit crops have not been given desired attention due to their comparatively less commercial importance and limited research on genetic improvement of cultivars. Opening of the world markets and development of new biotechnological methods of genetic modifications in high value commercial fruits would further keep the attention away from these traditional arid fruit crops leading to great loss in genetic diversity. The genetic resources of Phyllanthus emblica are well adapted to the stressed arid ecosystem having high potential for mitigating inevitable climate change scenario, and hence need immediate attention.
The Indian gooseberry under P. emblica (Aonla or amla), syn: Emblica officinalis is in cultivation in India due to its therapeutic, medicinal and neutraceutical properties since time immemorial. It is an important minor fruit crop having commercial significance. The crop is quite hardy, prolific bearer and highly remunerative. It is said to be indigenous to tropical south-east Asia. It is also reported to be the native of India, Ceylon, Malaysia and China. This is a fruit of sub-tropical region but its cultivation in topical climate is quite successful. Soon after the fruit set in spring, the fruits remain dormant through summer without any growth. This makes it highly suitable fruit crop for arid region. Aonla is not very exacting in its soil requirement and grows well in sandy loam to clay soils in India. Due to great tolerance to salinity and sodicity it is well adapted to arid conditions of Rajasthan. To safeguard the existing diversity of P. emblica and to achieve sustainable development based on the use of available genetic wealth, promotion and conservation of this species is of immense importance.
The fruit is used for diverse applications in healthcare, food and cosmetic industry. Documentation has been done by different workers on various aspects like physico-chemical properties [1], morpho-chemical characters [2], genetic variability [3], genetic diversity for morphological and biochemical traits [4], varietal wealth [5] and nutritional value etc. It has been well studied for immunomodulatory, anticancer, antioxidant and antiulcer activities [6]. In India, it is predominantly used as a cure for liver disorders [7]. Besides this, the fruit extract has shown significant anticancer activity [8], antioxidant supplement to promote endothelial function and to restore wound healing competency [9] and management of type 2 diabetes mellitus [10].
Correct genotype identification of medicinal plant material remains important for botanical drug industry. The morphological determinants within P. emblica are highly variable under different agro-climatic conditions. Due to its cross pollination and wider adoptability, a large amount of genetic variability exists in wild population as well as in commercially cultivated varieties of aonla under different growing belts. These varieties exhibit variation in their sex ratio, fruit, maturity, yield, quality and chemical composition of fruits [4]. Limitations of chemical and morphological approaches for authentication have generated need for newer methods in quality control of botanicals [6]. Nevertheless, genetic makeup and the background is independent of these variations. In the light of recent changes in agricultural policies world over under GATT and protection to breeder’s right to address IPR issues, it has become essential to take up the present investigation to molecularly characterize and document the extent of genetic diversity present in P. emblica for germplasm registration, documentation of biodiversity and crop improvement.
RAPD is one of the powerful molecular techniques which has been applied to a wide range of crops to assess genetic diversity, to evaluate genetic relationships between accessions or cultivars and estimation of relatedness. In recent years RAPD technique has been used to study the genetic diversity in different sets of P. emblica [11–14]. Besides this some molecular techniques such as ISSR [15], comparative studies of RAPD and ISSR markers [16], development of Sequence Characterized Amplified Region (SCAR) marker from the RAPD amplicon [6], SCAR and AFLP markers for distinguishing species of the genus Phyllanthus [17] and chloroplast trnL (UAA) intron identified as candidate gene [18].
Nuclear ribosomal Internal Transcribed Spacer (ITS) evolves relatively faster and can be useful in determining inter-species [19, 20] and sometimes intra-species relationships [21]. The rate and pattern of ITS sequence, mutation, insertion deletions (INDELS) and ITS length polymorphism are typically appropriate in resolving relationships within and among species and/or genera [22]. This complex has several domains that evolve at varying rates and thus have different phylogenetic utilities. The ITS polymorphism might occur at generic, specific or individual levels, making it useful for phylogenetic evolutionary and bio-geographical diversity studies. ITS sequences have been widely used in several species [23]. The present study was undertaken to assess the extent of genetic diversity and relationship among different varieties of P. emblica based on nuclear rDNA and RAPD polymorphism for crop improvement.
Material and Methods
Germplasm Evaluation
Eight varieties namely Anand-2, Banarasi, Chakaiya, Francis, Krishna, Kanchan, NA7 and NA10 were planted at a spacing of 6 × 6 m in three replications in the field gene bank at horticulture block of Central Arid Zone Research Institute, Jodhpur during 1998–1999. Data were recorded for fruit morphological characteristics and yield of aonla varieties from 15 year old plants. Twenty mature fruits were harvested randomly from each variety to record observations on morpho-chemical parameters. Fruit size (length and girth) was measured with the help of digital vernier caliper while fruit weight, pulp: stone ratio and stone weight were taken by digital top pan balance. The number of stripes was counted visually and dry matter was obtained by oven drying of fruit pulp. The total soluble solids (TSS) were determined with Erma Hand Refractometer (0–32°Brix). Ten fruits from each variety per replication were weighed and volume estimated by water displacement method.
DNA Fingerprinting
The genomic DNA was extracted from one gram of composite fresh leaves of each P. emblica variety. A hybrid protocol for genomic DNA isolation was developed using initial steps of CTAB method [24] and subsequently columns and solutions of the Plant Genomic DNA Purification spin kit (Hi-media Company) developed by Singh et al. [25] was followed. The genomic DNA was finally eluded using 200 μl of Tris–EDTA buffer at 10,000 rpm for 1 min to obtain high molecular weight pure DNA for fingerprinting. DNA was quantified with UV/VIS spectrophotometer by measuring OD260 and OD280. The quantified DNA samples were diluted in TE buffer to make a final concentration of 50 ng/µl for PCR reactions.
RAPD-PCR Analysis
The decamer random primers of OPA, OPB and OPP series (Operon Technologies) were used for screening of eight varieties of P. emblica. Based on the reproducibility of scorable bands the RAPD was finally performed using 10 decamer arbitrary primers. Hot-start PCR for the amplification of DNA was carried out in a 25 μl volume of reaction in a 0.2 ml microfuge PCR tube. A reaction tube typically consisted of decamer primer, 50 pmol; dNTP mix, 0.25 mM (Bangalore Genei); MgCl2, 2.5 mM (Sigma Chem); Taq DNA polymerase 1 U (Bangalore Genei); 10× PCR buffer, 5 mM (Tris–HCl, pH 8.3, 15 mM MgCl2), 250 mM KCl, genomic DNA 50 ng in dH2O). RAPD-PCR amplification was performed in a thermal cycler (Corbett Research, USA) with initial denaturation step of 94 °C for 3 min followed by 36 amplification cycles of 94 °C for 40 s, 50 °C for 40 s and 72 °C for 2 min and final elongation at 72 °C for 7 min. Amplicons were separated on a 1.6 % agarose gel pre-stained with ethidium bromide solution using 1× TAE buffer. The gels were run for 3 h at 60 V. The size of the amplified fragments was determined using 1 kb ladder (MBI Fermentas). All RAPD reactions were performed twice to test the reproducibility of the amplicon profiles.
Molecular Analysis of RAPD
The RAPD amplification products were scored as present (1) and absent (0) of scorable loci for each primer accession combination. Molecular data were entered into a binomial matrix and were used to determine Jaccard’s similarity coefficient with NTSYS-pc software [26]. Most informative primers were selected based on high polymorphism information content (PIC) value of individual primers as calculated by the following formula.
where F is frequency of presence of marker band, i is discrimination rate (DR) which was estimated to test the efficacy of individual primers in distinguishing the isolates, employing the following formula. DR = number of pairs of isolates differentiated/total number of pairs.
ITS Amplification
Polymerase chain reaction (PCR) universal primers namely ITS-1 (5′ TCC GTA GGT GAA CCT GCG G 3′) and ITS-4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) were used to amplify the internal transcribed spacer (ITS) region of ribosomal DNA, which encompasses the 5.8S gene. Each PCR amplification was performed in a total volume of 50 µl containing: 1 U Taq DNA polymerase (Sigma Chem), 2.5 mM MgCl2, 160 µM dNTP mix (MBI Fermentas), 50 pmol of each of the ITS-1 and ITS-4 primers (Bangalore Genei), and 50 ng genomic DNA in dH2O. The reactions were performed in a gradient thermal cycler with the following conditions: 1 min denaturation at 95 °C, 30 s annealing at 50 °C, 90 s elongation at 72 °C, for 34 cycles with a final elongation step of 72 °C for 10 min.
Amplified ITS regions were sequenced employing ABI Prism DNA sequencer (Applied Biosystems, Carlsbad, CA, USA) using ITS-1 and ITS-4 primers separately for DNA labeling by the BigDye terminator method (Applied Biosystems, Foster City, CA, USA). The sequenced data obtained from the ITS-4 primer were inversed and clubbed with the sequence data obtained with the ITS-1 primer, to obtain the complete sequence of the ITS region. Comparison of nucleotide sequences was performed using the Basic Local Alignment Search Tool (BLAST) network services of the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Molecular characterization of fungal isolates was done on the basis of similarity with the best aligned sequence of BLAST search.
Sequence Analysis
Nucleotide sequence comparisons were performed by using the BLAST network services against the NCBI databases. The multiple sequence alignment of ITS region (ITS-1, 5.8S r-RNA gene and ITS-2) of all the eight varieties of P. emblica was performed using CLUSTALX 1.83 software to detect single nucleotide polymorphism (SNP). The phylogenetic relationship among the isolates was estimated after the construction of a phylogram based on multiple sequence alignment of rDNA ITS sequences depicting bootstrap values using NJ plot software [27] and viewed using Tree View software based on SNPs.
Results and Discussion
The perusal of P. emblica fruit characterization data of eight varieties exhibited significant variations in most of the characters (Table 1). Fruit weight varied from 17.1 (Kanchan) to 36.6 g (Francis), fruit length from 29.17 (Kanchan) to 38.4 mm (Banarsi), fruit girth from 33.51 (Kanchan) to 43.45 mm (Banarasi), pulp:stone ratio from 5.52 (NA-10) to 15.6 (Anand-2), stone weight from 1.7 (Kanchan) to 4.2 g (Francis), number of stripes from 6 to 8, TSS from 11.8 (Banarasi) to 15.66°Brix (Banarasi), dry matter per cent from 13.8 (Anand-2) to 18.28 g (Kanchan). The most significant variation in the fruit yield ranging from 46.2 (NA-10) to 150.93 kg/plant (Kanchan) was also observed. The morphological variations exhibited by P. emblica varieties are shown in Fig. 1.
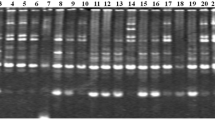
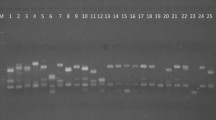
Ten decamer RAPD random primers detected intra-specific variations generating scorable amplicons and revealed 82 bands in the range of 200–3,000 bp. Among these, 49 bands were polymorphic amounting to 56.18 % polymorphism in RAPD banding patterns. The number of PCR amplified products ranged from 3 (OPB-14) to 12 (OPA-09) with an average of 8.2 bands per primer, whereas, the PIC (Polymorphic Information Content) varied from 66.5 (OPB-14) to 88.9 % (OPA-02) (Table 2).
The RAPD profiles of P. emblica developed by two most informative primers (OPA-02 and OPA-16) exhibiting the maximum PIC values are shown in Figs. 2 and 3. The UPGMA dendrogram obtained from cumulative cluster analysis of 10 primer matrix from 0.77 to 0.92 and delineated all the eight varieties of P. emblica into three main clusters and two varieties as the most distinct varieties as out groups (Fig. 4). The cluster I contained varieties Banarasi and Francis, cluster II had varieties Krishna and NA-10, cluster III had varieties Chakaiya and Kanchan, whereas Anand-2 and NA-7 were recorded as quite distinct varieties.
The authors recorded no consistency in grouping of P. emblica varieties based on fruit characteristics. For example, variety Chakaiya and Kanchan were morphologically quite distinct with regard to fruit weight, pulp: stone ratio, stone weight, number of stripes, TSS and fruit yield per plant but were genetically grouped together in cluster III of RAPD dendrogram. On the other hand, varieties Chakaiya and NA-7 had almost similar fruit morphology but were recorded genetically quite distinct in both RAPD and multiple sequence aligned phylogram. Singh et al. [2] also reported that morpho-chemical characters of P. embilica exhibited considerable genetic variability in fruit morphology, especially those based on fruit weight, which has high genotypic variation, higher heritability and greater potential for genetic gain. They suggested that the phenotypic expression of fruit should be an indication of their high genotypic potential. Earlier researcher have also studied the physico-chemical properties and recorded significant genetic diversity among cultivated varieties of P. emblica [1].
Although the variety NA-7 is of north Indian origin but was recorded as the most distinct out group to other north Indian varieties of P. emblica in RAPD cumulative dendrogram. At the same time the variety Kanchan of north Indian origin exhibited close lineages with the variety Anand-2 of west Indian origin in the multiple sequence aligned phylogram rather than other north Indian varieties.
In general, the robustness of a molecular marker technique depends on the amount of polymorphism, it can detect. Among the set of accessions investigated, the cumulative analysis of all the 10 informative RAPD primers detected an average of 56.18 % polymorphism in banding pattern with overall 83.1 % PIC values indicating its efficiency in evaluating genetic diversity in P. emblica. Rane et al. [13] also reported 57 to 60 % similarity among 20 genotypes in E. officinalis using RAPD markers. Bandyopadhyay and Raychaudhuri [17] performed comparative studies of RAPD, SCAR and AFLP markers for distinguishing some medicinally important species of the genus Phyllanthus. Chaurasia et al. [12] conducted RAPD based genetic variability among seven cultivated varieties of P. emblica namely Kanchan, Krishna, NA7, NA-10, BSR-1, Anand-1 and Chakaiya and distinguished these varieties using a set of four decamer primers, which was otherwise difficult by using morphological markers. They also reported that RAPD markers were also able to differentiate varieties of same origin or even selection from same parents.
Dnyaneshwar et al. [6] identified a putative marker (1.1 kb) specific for P. emblica by RAPD technique. Sequence Characterized Amplified Region (SCAR) marker was developed from the RAPD amplicon. This SCAR marker was found useful for identification of P. emblica in its commercial samples. Singh, et al. [14] separated Aonla varieties using RAPD markers into four major groups, which was further divided into sub-groups IA and IB. Sub-group IA had NA-04 and NA-10, while IB had cv. Francis. Cluster IIA represented NA-5, NA-6 and NA-9, while IIB represented NA-7 and Chakaiya. Banarasi and Anand were represented by group III and IV respectively.
Li and Zhao [15] studied the genetic diversity in four populations of P. emblica sampled from dry-hot valleys of Yunnan using ISSR (inter-simple sequence repeats) markers. They observed a high level of genetic diversity and low level of population genetic differentiation which might have resulted from the breeding system of this species. There was a lack of significant association between genetic and geographical distances among P. emblica populations.
All the eight varieties of P. emblica generated a single identical prominent band on gel electrophoresis and the conserved 5.8S rDNA regions was recorded with a uniform total nucleotide length amounting to a total of 587 bp and have been molecularly identified as P. emblica despite SNPs that delineated all varieties from each other with significant bootstrap values in multiple sequence aligned phylogram. Bootstrap values are dependable measures of phylogenetic accuracy and higher values are likely to indicate reliable groups [28]. The delineation of all the eight varieties of P. emblica validate reliable grouping. Selvi et al. [11] experimented that the exposure to gamma ray radiation altered the genetic makeup and the detection of mutation changes in E. officinalis using RAPD assay.
All the eight varieties of P. emblica generated a single identical prominent band on gel electrophoresis which included partial sequences of 18S gene, complete sequences (ITS-1, 5.8S gene, ITS-2) and partial sequence of 28S gene upon direct sequencing using ITS-1 and ITS-4 universal primers. All the gene sequences have been submitted to NCBI, USA and have been assigned GenBank accession numbers from KF926380 to KF926387. The conserved 5.8S rDNA region was recorded with a uniform nucleotide length of ITS-1 (219 bp), 5.8S gene (164 bp) and ITS-2 (204 bp) amounting to a total of 587 bp.
Nevertheless, out of multiple sequence alignment of all the eight varieties of P. emblica, the authors detected single nucleotide polymorphism at 7th and 131st portions in ITS-1 and 463 and 560th positions in ITS-2 regions. The phylogram generated based on multiple sequence alignment showing delineation of P. emblica varieties is shown as Fig. 5. The P. emblica varieties were further delineated from each other with significant bootstrap values in multiple sequence aligned phylogram.
The ITS length variants and polymorphism have been reported in several plant species [22, 23]. Raturi et al. [22] reported heterogeneity in nuclear rDNA ITS region in Vigna radiata which did not cause any phylogenetic errors at species level. Barkley et al. [29] observed SNPs and suggested EcoTILLING as a powerful genetic analysis tool for rapid identification of naturally occurring variation in plants.
Conclusion
The morpho-physiological characterization proved insufficient to distinguish P. emblica varieties studied. The primers identified in the present study are robust RAPD markers with 83.1 % PIC and can be used as powerful markers to reveal genetic diversity in Aonla. This study validates the utility of ITS rDNA region being third generation molecular markers as a reliable indicator of phylogenetic interrelationships, especially ITS regions as DNA barcode at higher levels and can serve as an additional approach for identification and genetic cataloguing of P. emblica germplasm for crop improvement. The genetic variability present in this set of Indian P. emblica genotypes suggest that they might have originated from genetically divergent parents and cross pollination and has a long history of adaptation to their respective micro-climatic regions and could be of significance to contribute to P. emblica breeding programmes.
References
Kumar S, Chithiraichelvan R, Karunakaran G (2011) Performance of aonla cultivars for yield and physico-chemical properties under Coorg conditions. Indian J Hortic 68:268–269
Singh B, Uniyal AK, Rawat JSM, Rana DK (2012) Estimation of genetic variability in Phyllanthus emblica L—Towards a contribution in sustainable rural development. J Hortic Forestry 4:92–95
Shukla AK, Singh D (2008) Genetic variability of Aonla (Emblica officinalis Gaertn). Hortic J 21:11–13
Bala S, Ram S, Prasad J (2009) Studies on variability and genetic diversity in selected aonla genotypes. Indian J Hortic 66:433–437
Shukla AK, Samadia DK, Shukla AK, Dhandar DG (2005) Genetic resources of aonla. Indian J Plant Genet Resour 18(2):188–193
Dnyaneshwar W, Preeti C, Kalpana J, Bhushan P (2006) Development and application of RAPD–SCAR marker for identification of Phyllanthus emblica L. Biol Pharm Bull 29(11):2313–2316
Kirtikar KR, Basu BD (2003) Indian medicinal plants, 2nd edn. Oriental Enterprises, Dehradun, pp 1606–1608
Mahata S, Pandey A, Shukla S, Tyagi A, Husain SA, Das BC, Bharti AC (2013) Anticancer activity of Phyllanthus emblica linn. (Indian Gooseberry): inhibition of transcription factor AP-1 and HPV gene expression in cervical cancer cells. Nutr Cancer 65:88–97
Chularojmontri L, Suwatronnakorn M, Wattanapitayakul SK (2013) Phyllanthus emblica L. enhances human umbilical vein endothelial wound healing and sprouting. Evid Based Complement Altern Med http://dx.doi.org/10.1155/2013/720728. Accessed 5 Dec 2013
Usharani P, Fatima N, Muralidhar N (2013) Effects of Phyllanthus emblica extract on endothelial dysfunction and biomarkers of oxidative stress in patients with type 2 diabetes mellitus: a randomized, double-blind, controlled study. Diabetes Metab Syndr Obes 6:275–284
Selvi BS, Ponnuswami V, Kavitha PS (2008) Use of RAPD assay for the detection of mutation changes in Aonla (Emblica officinalis Gaertn.). Adv Nat Appl Sci 2(3):129
Chaurasia AK, Subramaniam VR, Krishna B, Sane PV (2009) RAPD based genetic variability among cultivated varieties of Aonla (Indian Gooseberry, Phyllanthus emblica L.). Physiol Mole Biol Plants 15:169–173
Rane AA, Kag B, Hegde V, Sathyanarayana BN (2012) RAPD marker assay based genetic diversity in aonla (Emblica officinalis L.). Int J Curr Res 4:32–36
Singh D, Bharti H, Singh KM (2014) DNA fingerprinting of aonla varieties using RAPD markers. Indian J Hortic 71:117–119
Li Q, Zhao J (2007) Genetic diversity of Phyllanthus emblica populations in dry-hot valleys in Yunnan. Biodivers Sci 15(1):84–91
Raut GR, Senapati SK, Aprajita S (2010) Study of relationships among twelve Phyllanthus species with the use of molecular markers. Czech J Genet Plant Breed 46:135–141
Bandyopadhyay S, Raychaudhuri SS (2013) Development and comparison of RAPD, SCAR and AFLP markers for distinguishing some medicinally important species of the genus Phyllanthus. Plant Biosyst 147:12–20
Sangeetha N, Mercy S, Kavitha M, Selvaraj D, Sathishkumar R, Ganesh D (2010) Morphological variation in the Indian gooseberries (Phyllanthus emblica and Phyllanthus indofischeri) and the chloroplast trnL (UAA) intron as candidate gene for their identification. Plant Genetic Res 8:191–197
Singh SK, Tiwari M, Kamal S, Yadav MC (2005) Morel Phylogeny and diagnostics based on restriction fragment length polymorphism analysis of ITS region of 5.8S ribosomal DNA. J Plant Biochem Biotechnol 14:179–183
Singh SK, Harsh LN, Pancholy A, Pathak R, Raturi A (2011) Molecular assessment of inter-specific genetic diversity in selected species of Prosopis revealed by RAPD. Indian J Agric Sci 81(2):167–171
Kakani RK, Singh SK, Pancholy A, Meena RS, Pathak R, Raturi A (2011) Assessment of genetic diversity in Trigonella foenum-graecum based on nuclear ribosomal DNA, internal transcribed spacer and RAPD analysis. Plant Mol Biol Rep 29:315–323
Raturi A, Singh SK, Sharma V, Pathak R (2012) Molecular characterization of Vigna radiata (L.) Wilczek genotypes based on nuclear ribosomal DNA and RAPD polymorphism. Mol Biol Rep 39(3):2455–2465
Xiao LQ, Möller M, Zhu H (2010) High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: incomplete concerted evolution and the origin of pseudogenes. Mol Phylogenet Evol 55:168–177
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(1):13
Singh SK, Meghwal PR, Pathak R, Gautam R, Kumar S (2013) Genetic diversity in Punica granatum revealed by nuclear rRNA, internal transcribed spacer and RAPD polymorphism. Natl Acad Sci Lett 36(2):115–124
Rohlf FJ (1997) NTSYS pc: numerical taxonomy and multivariate analysis system, vol 2.02h. Exeter software, New York
Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Syst Biol 42:182–192
Barkley NA, Wang ML, Gillaspie AG, Dean RE, Pederson GA, Jenkins TM (2008) Discovering and verifying DNA polymorphisms in a mungbean [V. radiata (L.) R Wilczek] collection by EcoTILLING and sequencing. BMC Res Notes 1:28
Acknowledgments
The authors are thankful to Dr. M. M. Roy Director, Central Arid Zone Research Institute, Jodhpur for providing necessary laboratory and field facilities to carry out the study.
Conflict of interest
It is certified that there is no conflict of interest of any type.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, S.K., Meghwal, P.R. & Pathak, R. Molecular Characterization of Commercial Varieties of Phyllanthus emblica Using RAPD and Nuclear rDNA SNPs. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 85, 971–978 (2015). https://doi.org/10.1007/s40011-014-0453-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-014-0453-1