Abstract
Purpose
Treatment guidelines often do not advocate testing for integrase inhibitor resistance associated mutations (IRAM) before initiation of first line ART given the extremely low prevalence of mutations found in older surveillance studies. We aimed to describe the prevalence of IRAM in Austrian patients recently diagnosed with HIV in the 5 years following introduction of integrase inhibitors and to analyse trends and factors associated with their detection.
Methods
Samples of antiretroviral treatment (ART) naïve patients recently diagnosed with HIV in Austria between 2008 and 2013 were analysed for the existence of IRAM and drug penalty scores were calculated to estimate response to drugs. Demographic and virological data were extracted from a database. Descriptive and comparative statistics were used.
Results
A total of 303 samples were analysed. 78 % were male and mean age was 38 years. Overall prevalence of IRAM was 2.3 %. Six percent had at least potentially low-level resistance to raltegravir or elvitegravir, versus 1 % for dolutegravir. One primary mutation was observed (F121Y) in a patient sample from 2012 leading to 5–10-fold reduced susceptibility to raltegravir and elvitegravir. Two patients carried the accessory mutations E138K and G140A, respectively, where both lie on the Q148 pathway. No temporal trend of IRAM prevalence was observed (p = 0.16).
Discussion
Primary IRAM are still rarely found despite the increasing use of INSTI in Austria, but there is a potential for reduced susceptibility to these drugs in selected patients. Routine resistance testing seems prudent to avoid the consequences including accumulation of further mutations and therapeutic failure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Integrase strand transfer inhibitors (INSTI) are the latest approved drug class to treat HIV infection. They exhibit antiretroviral activity by blocking the integration of HIV viral DNA into host cells. Raltegravir (RAL) was the first approved INSTI in Austria in 2008 and was initially used as a component of salvage regimens. Its promising efficacy and excellent tolerability has soon led to its introduction into first line regimens [1]. The later approved elvitegravir (EVG) and more recently dolutegravir are once daily options with excellent tolerability as well as potency and in the case of dolutegravir a significantly higher genetic barrier [2, 3].
Given these properties, which make INSTI ideal components of antiretroviral regimens, its use has greatly increased. However, resistance to the first generation integrase inhibitors RAL and EVG can arise rapidly as a result of single mutations or combinations of mutations in the integrase gene and cross resistance has been described [4–6]. At least 1–2 % of treatment naïve patients and 19 % of treatment experienced patients started on a new RAL based regimen which will develop an emergent primary integrase inhibitor resistance associated mutation (IRAM) after 5 years [7, 8]. Given the potentially large pool of INSTI resistant strains an increasing possibility of transmission of IRAM seems highly possible.
In Europe primary drug resistance rates to reverse transcriptase inhibitors [NRTIs and NNRTIs and to a lesser extent protease inhibitors (PIs)] have been remarkably stable at rates of around 8 %. [9] Yet data on primary INSTI resistance associated mutations (IRAM) are scant. Previous studies were limited by sample size, geographic diversity and confinement to earlier time periods and have so far failed to demonstrate occurrence of primary IRAM in INSTI naïve patients [10–12].
As a consequence, treatment guidelines often do not advocate testing for IRAM before initiation of first line ART. However, given the increasing use of these drugs this situation may well have changed and emerging drug resistant strains may endanger therapeutic successes. We aimed to determine the prevalence of IRAM in Austria in the 5 years following introduction of INSTI and to analyse temporal trends.
Methods
For this retrospective cross-sectional analysis patients who received a first diagnosis of HIV infection between 2008 and 2013 and who had a HIV drug resistance test performed at the Department of Virology in Vienna within 3 months thereafter were selected. The Department of Virology is the national HIV reference laboratory and therefore confirms new diagnoses of HIV infections and performs drug resistance analysis for a large part of HIV infected patients in Austria. Unused material is cryo-conserved for subsequent retesting.
Leftovers of the first 49–53 EDTA blood samples received from patients with new diagnosis of HIV infection in each year from 2008 to 2013 were thawed and retested for the presence of IRAM. In case of insufficient material or invalid analysis the subsequent sample was selected. One single sample per patient was used.
In brief, HIV-1 RNA was extracted from the plasma specimens by the Nuclisens Easy Maq kit (Biomerieux) and subjected to a reverse transcriptase-polymerase chain reaction (RT-PCR). For integrase resistance analysis an in-house protocol was used covering amino acids 1-274. Reference sequence for nt numbering was HXB2. A part of the primers used in the in-house was previously published [13]. RT and protease resistance testing was performed covering the amino acids 1-333 and 1-99, respectively. Population sequencing was done using BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) and was run on a 3130x genetic analyser (life technologies).
Sample size calculations were performed based on the assumption that a total sample size of 300 patient samples was sufficient to detect within a 97.5 % confidence interval a true prevalence of primary INSTI resistance in Austria of below 1.5 %, if no resistance mutation was found. A prevalence of 1.5 % was estimated as the threshold below which resistance mutations would occur at a negligible rate and routine INSTI resistance testing would not be warranted.
Integrase, protease and reverse transciptase sequences were analysed separately using the Stanford University HIV Drug Resistance Database genotypic resistance interpretation algorithm (HIVdb algorithm, version 7.0.1, http://hivdb.stanford.edu).
Sequence data was analysed and drug penalty scores (DPS) were calculated using the same algorithm. This algorithm estimates the level of resistance to a specific drug by adding up penalty scores conferred by individual resistance associated mutations. A score of 0–9 indicates susceptibility, a score of 10–14 potential resistance, 15–29 low level resistance, 30–59 intermediate resistance, and ≥60 high level resistance [14]. Results were linked in an electronic database with demographic variables and virological data. Demographic variables included sex and age at the time of analysis. Virological data included viral load where available, viral subtype and results of previous testing for resistance to reverse transcriptase inhibitors and protease inhibitors performed on the same sample. The dataset was anonymized prior to statistical analysis.
Ethical approval for the performance of this study was granted by the Ethics committee of the Medical University, Vienna.
The 2015 IAS Drug resistance list was used as reference to define primary and accessory drug resistance mutations for INSTI and reverse transcriptase inhibitor (RTI) as well as major and minor resistance mutations for PI [15]. Primary INSTI resistance mutation included T66I, E92Q, F121Y, Y143C/H/R, S147G, Q148H/K/R, and N155H. Accessory mutations included L74M, T97A, E138A/K, G140A/S, and R263K.
Student’s t testt-test and Wilcoxon Rank Sum tests were used to compare continuous variables between groups. Chi-squared and Fisher’s exact tests were used to compare the categorical characteristics between groups. A binominal exact estimation was used to calculate the confidence intervals for prevalences. Kappa Interrater agreement test was used to calculate the agreement between various resistance mutations. A Chi-squared test for linear trend was used to estimate assess a temporal trend for occurrence of IRAM and mutations to other drug classes.
STATA v13.1 (College Station, Texas, US) was used for all analyses and a two sided p value of 0.05 was considered as significant.
Results
A total of 303 patient samples were analysed. 78 % of patients were male and mean age was 38 years. HIV subtype B was most common (62 %) followed by subtype C (12 %). Median viral load was 1 × 105 HIV RNA copies/ml and 17 % of patients had a viral load >1 × 106 HIV RNA copies/ml most likely indicating acute HIV infection (Table 1).
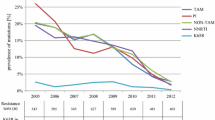
Major or primary drug resistance mutations were found in 30 patients (9.9 %). 20 individuals (6.6 %) had resistance to NNRTI. The most common NNRTI resistance mutation was E138A (n = 12), followed by K103N (n = 6). Four (1.3 %) patients had drug resistance mutations to NRTI where M41L was most common (n = 2). Five patients (1.7 %) had major mutations to protease inhibitors. The mutation L90M was most common (n = 2).
Overall prevalence of integrase inhibitor associated resistance mutations was 2.3 %. In the year 2012 the primary INSTI associated mutation F121Y was detected in one patient (0.3 %, 95 % CI 0.00–0.02). In this patient, no other accessory mutations to INSTI or resistance mutations to other drug classes were found. Accessory polymorphic and non-polymorphic mutations that confer decreased susceptibility to INSTI alone or in combination were found in 6 (2 %) patients. Detected mutations included T97A in 3 patients, and L74M, E138K,and G140A in 1 patient each (Table 2).
The majority of RTI resistance mutations occurred in 2010 (n = 4) and most NNRTI resistance mutations were detected in 2012 (n = 5). Two major PI resistance mutations were detected in 2009. There was no significant trend regarding the occurrence of NRTI (p = 0.42), NNRTI (p = 0.85), or major PI resistance (p = 0.74) over the years 2008–2013. In addition, there was no significant temporal trend for the occurrence of primary or accessory INSTI resistance mutations (p = 0.16). The presence of any IRAM was not significantly associated with male sex (OR 0.78, 95 % CI 0.14–4.30), older age (OR 0.98, 95 % CI 0.91–1.05), calendar year (OR 1.28, 95 % CI 0.80–2.03), or occurrence of any other drug resistance mutations (OR 1.41, 95 % CI 0.16–12.23) in a multivariable logistic regression.
Patients with primary or accessory INSTI resistance mutations did not differ from patients without these mutations regarding age (p = 0.57), sex (p = 0.68), or viral subtype (p = 0.82). No association was found between viral load and occurrence of INSTI resistance mutations (OR 1.00, p = 0.98).
No correlation was found between occurrence of RTI and PI resistance mutations and INSTI resistance mutations (p = 0.94). No significant association was found between presence of NRTI, NNRTI, and PI resistance mutations and sex (p = 0.62), age (p = 0.51), and presence of subtype B (p = 0.32).
Seventeen patients (6 %) had a DPS ≥ 10 for RAL or EVG indicating at least potential low-level resistance. Three of these patients had intermediate level resistance (DPS ≥ 30) for RAL or EVG. Two patients had a DPS of 10–14 for dolutegravir indicating potential low level resistance. No patient had a DPS higher than 14 for dolutegravir (Table 3).
Discussion
In this cross sectional analysis covering a representative sample of ART-naïve patients in Austria during a relatively large period of time after INSTI have begun to be widely used in Austria, primary IRAM were rarely detected. In fact, only one patient carried the F121Y mutation which confers intermediate resistance to RAL and EVG due to a 5–10-fold reduction in susceptibility [16].
This mutation was detected only 4 years after INSTI were introduced in Austria. By this time the integrase inhibitors RAL and EVG were already used in more than 20 % of initial antiretroviral regimens [17]. Given the limited sample size and the low prevalence our study seems to be underpowered to detect a positive time trend and to assess clear risk factors for the occurrence of IRAM. Larger studies including more recent data seem necessary to prove or disprove that such an association exists. However, the low number of primary IRAM found in our patient collective may still not be negligible. We estimated that the true prevalence for primary IRAM could be as high as 2 % which is in the range of prevalence found for NRTI or major PI resistance mutations in our study. For both latter drug classes, testing for drug resistance mutations prior to initiating first line therapy has become the standard and is recommended by current national guidelines whereas routine testing for IRAM is currently not recommended [18]. Thus ongoing surveillance and routine testing for IRAM seems now necessary to detect a potentially increasing number of resistance mutations in the future and to avoid complications with HIV treatment. In the patient bearing the F121Y mutation, no other resistance mutations to other drug classes were detected. This suggests that combination ART including RAL or EVG might still lead to complete viral suppression given the full activity of backbone drugs. On the other hand, possibly in the light of incomplete adherence to a regimen containing first generation integrase inhibitors, accumulation of accessory INSTI mutations and evolution of viral resistance along the Y143R pathway could take place. The Y143R mutation has been associated with high level RAL and EVG resistance [19]. Particularly in combination with the mutation G118R, the F121Y mutation confers also intermediate resistance to dolutegravir [20].
The origin of the F121Y mutation seems less clear. The fact, that the F121Y mutation was detected in 2012, 4 years after INSTI were first used in Austria suggests that transmission of this mutation is possible [21]. However, in a recent study the Swiss HIV Cohort described the occurrence of the high level N155H mutation among a therapy naïve patient even before INSTI had ever been used in this country suggesting the possibility of de novo mutagenesis of IRAM [22]. It seems likely that the IRAM found in our study are also caused by polymorphic de novo mutagenesis.
In addition to the primary F121Y mutation, the polymorphic mutations T97A and L74M and the non-polymorphic accessory mutations E138K and G140A were also detected in several patients. Both, G140 and E138A usually occur in patients receiving RAL but the latter has already been described in the pre-INSTI era. Neither of these mutations causes higher level resistance alone, however, in combination in particular with the Q148 mutation they have been associated with high level RAL and EVG susceptibility and even reduced dolutegravir susceptibility [23]. Taken together primary and accessory mutations, more than 4 % of our patients had at least low level resistance to RAL and EVG. The Europe-wide SPREAD program demonstrated the occurrence of resistance associated polymorphisms before INSTI were introduced, but data are lacking from the years thereafter [9, 23].
Although the overall prevalence of IRAM appears to be low, the existence of such resistant strains in untreated populations may not be negligible as drug resistant viruses and in particular low fitness cost strains may be self-sustained and therefore transmitted readily [24].
In conclusion, primary IRAM are still rarely found in ART-naïve individuals despite an increasing use of INSTI in Austria, but there is the possibility that reduced susceptibility may exist to these drugs in selected patients. In the absence of predictors for occurrence of IRAM routine resistance testing seems prudent to avoid the consequences including accumulation of further mutations and therapeutic failure.
References
Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, et al. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet. 2009;374:796–806. doi:10.1016/S0140-6736(09)60918-1.
Sax PE, DeJesus E, Mills A, Zolopa A, Cohen C, Wohl D, et al. Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir versus co-formulated efavirenz, emtricitabine, and tenofovir for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3 trial, analysis of results after 48 weeks. Lancet. 2012;379:2439–48. doi:10.1016/S0140-6736(12)60917-9.
Oliveira M, Mesplede T, Quashie PK, Moisi D, Wainberg MA. Resistance mutations against dolutegravir in HIV integrase impair the emergence of resistance against reverse transcriptase inhibitors. AIDS. 2014;28:813–9. doi:10.1097/QAD.0000000000000199.
Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, et al. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother. 2008;52:1351–8. doi:10.1128/AAC.01228-07.
Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359:355–65. doi:10.1056/NEJMoa0708978.
Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, et al. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol. 2008;82:10366–74. doi:10.1128/JVI.00470-08.
Rockstroh JK, DeJesus E, Lennox JL, Yazdanpanah Y, Saag MS, Wan H, et al. Durable efficacy and safety of raltegravir versus efavirenz when combined with tenofovir/emtricitabine in treatment-naive HIV-1-infected patients: final 5-year results from STARTMRK. J Acquir Immune Defic Syndr. 2013;63:77–85. doi:10.1097/QAI.0b013e31828ace69.
Eron JJ, Cooper DA, Steigbigel RT, Clotet B, Gatell JM, Kumar PN, et al. Efficacy and safety of raltegravir for treatment of HIV for 5 years in the BENCHMRK studies: final results of two randomised, placebo-controlled trials. Lancet Infect Dis. 2013;13:587–96. doi:10.1016/S1473-3099(13)70093-8.
Hofstra LM, Sauvageot N, Albert J, Alexiev I, Garcia F, Struck D, et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis. 2015;. doi:10.1093/cid/civ963.
Stekler JD, McKernan J, Milne R, Tapia KA, Mykhalchenko K, Holte S, et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007–2013. Antivir Ther. 2015;20:77–80. doi:10.3851/IMP2780.
Saladini F, Meini G, Bianco C, Monno L, Punzi G, Pecorari M, et al. Prevalence of HIV-1 integrase mutations related to resistance to dolutegravir in raltegravir naive and pretreated patients. Clin Microbiol Infect. 2012;18:E428–30. doi:10.1111/j.1469-0691.2012.03917.x.
Gutierrez C, Hernandez-Novoa B, Perez-Elias MJ, Moreno AM, Holguin A, Dronda F, et al. Prevalence of primary resistance mutations to integrase inhibitors in treatment-naive and -experienced patients infected with B and non-B HIV-1 variants. HIV Clin Trials. 2013;14:10–6. doi:10.1410/hct1401-10.
Sichtig N, Sierra S, Kaiser R, Daumer M, Reuter S, Schulter E, et al. Evolution of raltegravir resistance during therapy. J Antimicrob Chemother. 2009;64:25–32. doi:10.1093/jac/dkp153.
Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–18. doi:10.1086/503914.
Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, et al. 2015 Update of the drug resistance mutations in HIV-1. Top Antivir Med. 2015;23:132–41.
Kobayashi M, Nakahara K, Seki T, Miki S, Kawauchi S, Suyama A, et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–22. doi:10.1016/j.antiviral.2008.06.012.
(AHIVCOS) AHCS. HIV/AIDS in Austria: 26th report of the Austrian HIV cohort study. Innsbruck: STUDIA Universitätsverlag; 2014.
DAIG. Deutsch-Österreichische Leitlinien zur antiretroviralen Therapie der HIV-Infektion. 2012. http://www.daignet.de/site-content/hiv-therapie/leitlinien-1/Anlage%201%20LL%20055-001%20Version%206%2011-12-2015%20endgultige%20Version%20rev-2.pdf. Accessed 12 Aug 2016.
Souza Cavalcanti J, Minhoto Lanca A, de Paula Ferreira JL, da Eira M, de Souza Dantas DS, de Macedo Brigido LF. In-vivo selection of the mutation F121Y in a patient failing raltegravir containing salvage regimen. Antiviral Res. 2012;95:9–11. doi:10.1016/j.antiviral.2012.04.007.
Munir S, Thierry E, Malet I, Subra F, Calvez V, Marcelin AG, et al. G118R and F121Y mutations identified in patients failing raltegravir treatment confer dolutegravir resistance. J Antimicrob Chemother. 2015;70:739–49. doi:10.1093/jac/dku474.
Metzner KJ, Scherrer AU, Preiswerk B, Joos B, von Wyl V, Leemann C, et al. Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis. 2013;208:1102–12. doi:10.1093/infdis/jit310.
Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–9.
Casadella M, van Ham PM, Noguera-Julian M, van Kessel A, Pou C, Hofstra LM, et al. Primary resistance to integrase strand-transfer inhibitors in Europe. J Antimicrob Chemother. 2015;70:2885–8. doi:10.1093/jac/dkv202.
Mbisa JL, Fearnhill E, Dunn DT, Pillay D, Asboe D, Cane PA, et al. Evidence of self-sustaining drug resistant HIV-1 lineages among untreated patients in the United Kingdom. Clin Infect Dis. 2015;61:829–36. doi:10.1093/cid/civ393.
Acknowledgments
The performance of this study was financially supported by GILEAD.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Zoufaly, A., Kraft, C., Schmidbauer, C. et al. Prevalence of integrase inhibitor resistance mutations in Austrian patients recently diagnosed with HIV from 2008 to 2013. Infection 45, 165–170 (2017). https://doi.org/10.1007/s15010-016-0936-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-016-0936-5