Abstract
Background:
In order to produce and isolate the exosome derived from the cell of interests, a serum free environment (starvation) has been essential for excluding the unknown effect from serum-derived exosomes. Recently, serum-free culture media have been developed as a substitute for serum supplemented media so that MSC proliferates with maintaining the original characteristics of the cells in a serum free condition. Due to the different properties of the exosomes representing the states and characteristics of the origin cells, a study is needed to compare the properties of the cell-derived exosomes according to the cell culture media.
Methods:
To compare the cell culture condition on exosomes, human umbilical cord mesenchymal stem cells (UCMSCs) were cultured with two different media, serum containing media, 10% FBS supplemented DMEM (NM) and serum-free chemically defined media, CellCor™ CD MSC (CDM). To remove FBS-derived exosomes from UCMSC cultured with NM, the medium was replaced with FBS-free DMEM for starvation during exosome isolation. The production yield and expression levels of angiogenic and pro-inflammatory factors were compared. And, the subpopulations of exosome were classified depending on the surface properties and loaded cytokines. Finally, the wound healing and angiogenic effects have been evaluated using in vitro assays.
Results:
The UCMSC-derived exosomes under two different cell culture media could be classified into subpopulations according to the surface composition and loaded cytokines. Especially, exosome derived from UCMSC cultured with CDM showed higher expression levels of cytokines related to regenerative bioactivities which resulted in enhanced wound healing and angiogenesis.
Conclusion:
CDM has the advantages to maintain cell proliferation even during the period of exosome isolations and eliminate unknown side effects caused by serum-derived exosomes. Additionally, exosomes derived from UCMSC cultured with CDM show better wound healing and angiogenic effects due to a lot of regeneration-related cytokines and less pro-inflammatory cytokines compared to with NM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The use of mesenchymal stem cells (MSCs) has increased for therapeutic treatments in many preclinical models of immunological and degenerative diseases in last decade [1,2,3,4]. The mechanisms for regenerative properties of transplanted MSCs are not still clearly unraveled [5]. However, many reports have demonstrated the potential of MSCs for transdifferentiation, cell fusion, mitochondrial transfer, and paracrine signaling [6, 7]. Since the differentiations and survivals of MSCs are limited in the targeted lesions, a paracrine signaling from MSCs has been considered as a major mechanism of their therapeutic effects. Paracrine factors, including chemokines, cytokines, hormones, and growth factors, have recently been widely studied for extensive use of MSCs in clinical trials [8, 9]. Especially, extracellular vesicles (EVs) have been recognized as an important bioactive component for leading paracrine effects of MSCs [10, 11]. Small vesicular particles with a lipid bilayer, called EVs, are cell-secreted nanovesicles that play a pivotal role to mediate intercellular communications by exchanging proteins, lipids, and mRNA between cells [12,13,14,15]. EVs derived from MSCs show biological functions similar to the origin cells by facilitating tissue regeneration with enclosing and delivering active biomolecules to the targeted cells or tissues [16]. EVs are categorized into different subtypes based on their subcellular origin, biogenesis, size, and molecular compositions. It is now widely accepted that exosomes are secreted by multivesicular bodies (MVBs) formed via endocytosis with 40 –150 nm size [17, 18]. The properties of released exosomes vary depending on origins, types, and conditions of parent cells. Many studies have raised the possibility of being used as an alternative to cell-based therapy because exosomes represented the characteristics of the cell of origin [19, 20]. In particular, MSCs-derived exosomes have emerged as promising therapeutic agents for regenerative medicine, including angiogenesis and wound repair [21, 22], kidney regeneration [23], cardiac repair [24], neurodegeneration treatment [25], and osteochondral regeneration [26]. The therapeutic effect of exosome is influenced by the differences in the molecular components (e.g. protein and RNA cargo) loaded and delivered by exosomes depending on the condition of cells. However, there are still barriers to the widespread development of MSC-derived exosome-based therapies for human patients. At the beginning of exosome research, many reports used exosome isolated from a serum-containing MSC culture medium [27, 28]. Considering that an enormous number of lipoprotein particles and EVs are present in serum [29], the biological activity of MSC-derived exosomes decreases, and unknown side effects can occur without depletion processes for serum-derived exosomes. With increasing potential for their clinical use, isolation only exosome secreted from the cell of interest with high purity has become essential [30]. For this reason, researchers have been utilized the serum-free media during isolation of exosome to minimize the unknown effects of serum-derived exosome although it contains many components, including growth factors, proteins, vitamins, and hormones, etc., for promoting cell adhesion and proliferation with protecting cells from shear stress [31]. Especially, the use of fetal bovine serum (FBS) is controversial for growing stem cells at a clinical-grade level because its composition is not completely defined [32]. Moreover, it has been confirmed that FBS contained large number of membrane vesicles without defining the compositions [33]. To deplete EVs from FBS, ultracentrifugation and ultrafiltration-based methods have been optimized, and large amounts of FBS-derived EVs were proved to be eliminated, but not all [34]. To remove the effect of serum-derived exosomes completely and enhance the purity of exosome from cell of interests, a process for starvation has been used as a ‘gold standard’ during exosome isolation [35]. The cells are maintained using serum supplemented media to facilitate cell proliferation. Next, the medium was replaced with serum-free medium during exosom isolation. From this condition, highly purified MSC-derived exosome could be isolated without contamination with serum-derived exosomes. However, the diminished proliferation rate and the changes in characteristics of cells during the starvation affect the production yield and properties of exosome secreted from cells. With this reason, efforts are required to establish culture conditions in which the characteristics of original cell can be maintained even during the exosome isolation. Various serum-free media have been developed for successful MSC culturing without serum components [36,37,38]. Among various serum-free media, a chemically defined medium, the CellCor™ CD MSC (CDM; Xcell Therapeutics), has been developed for MSC without animal and human-derived proteins and other ingredients. CDM exhibited excellent proliferation rate with maintaining expression of MSC marker protein on the surfaces over 15 passages. Although it has proved excellent potentials for MSC culture in serum-free condition using CDM, the comparative analysis between exosomes isolated from MSC cultured in CDM and exosomes isolated under starvation condition has not been investigated.
In this study, the production yield of human umbilical cord mesenchymal stem cells (USCMS)-derived exosome based on cell culture media was comparatively analyzed. And subpopulations of the exosomes were classified according to their surface compositions and cytokines using principal component analysis (PCA). Moreover, cell status-dependent the expression levels of angiogenic and pro-inflammatory factors of exosomes were compared and also wound healing and angiogenic activities of UCMSC-derived exosomes were evaluated depending on the cell culture media.
2 Materials and methods
2.1 Cell culture
Human umbilical cord mesenchymal stem cells (UCMSCs; CHA Biotech Co. Ltd., Seongnam, Korea) were cultured up to approximately 50% confluence using 10% fetal bovine serum (FBS, HyClone laboratories, Logan, UT, USA) supplemented low-glucose DMEM (HyClone) containing 1% antibiotic–antimycotic mixture (GIBCO, Grand Island, NY, USA) in a humidified atmosphere with 5% CO2 at 37 °C. Additionally, UCMSCs were cultured using CellCor™ CD MSC media (Xcell Therapeutics, Seoul, Korea) containing 1% antibiotic–antimycotic mixture (GIBCO) with the serum-free condition in a humidified atmosphere with 5% CO2 at 37 °C.
2.2 Exosome (EV) isolation
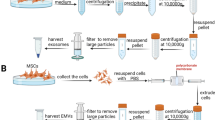
To isolate exosome, normal media (NM) and CellCor™ CD MSC media (CDM) were utilized. Specifically, the media for approximately 50% confluent cells cultured with low-glucose DMEM were replaced with phenol red-free DMEM (GIBCO) containing 1% antibiotic–antimycotic mixture without serum during 48 h for NM condition, and CellCor™ CD MSC media were continuously used during cell culturing and exosome isolation for CDM condition, respectively. The NM were collected four times in every 12 h, and the CDM were collected four times in every 30 h for maximizing exosome production. The collected cell culture media were centrifuged at 1300 rpm for 3 min and filtered through a 0.22 µm Vacuum Filter/Storage Bottle System to remove non-exosomal large particles, including cells, cell debris, microvesicles, and apoptotic bodies. Finally, exosomes were isolated using a tangential flow filtration (TFF; Repligen, Waltham, MA, USA) system with a 500 kDa molecular weight cut-off filter.
2.3 Characterization of exosomes
The Pierce™ BCA Protein Assay Kit (Pierce, Rockford, IL, USA) was used to assess the protein concentration of exosome as followed by manufacturer’s instructions. The ZetaView QUATT® (Particle Metrix, Meerbusch, Germany) was utilized to confirm the number and the size of particles with 488 nm scatter mode. The samples were diluted to 107–108 particles/ml using filtered phosphate-buffered saline (PBS) solution (HyClone laboratories). The detailed settings for accurate analysis were optimized with sensitivity 75, shutter 300, minimum trace length 15, and cell temperature 25 °C for all samples. The morphology of the exosome was observed with transmission electron microscopy (TEM; Hitachi, H-7600, 80 kV, Tokyo, Japan). The exosome solution was dried on the copper grid with 200 mesh carbon film (FCF150-CU, Electron Microscopy Sciences, Hatfield, PA, USA). Exosomes located on copper grid were stained with 7% uranyl acetate solution and dried for negative staining. After drying, the copper grid was placed on the grid box for TEM analysis.
2.4 Western blot analysis
The same number of exosomes (1.86 × 1011 particles) were used for the parallel comparison. The exosomes were loaded to 12% SDS-PAGE and transferred onto nitrocellulose (NC) membranes. The NC membrane was blocked with 5% skim milk dissolved TBST solution. The exosome protein transferred NC membranes were sequentially incubated with the primary antibody of CD63 (Abcam, Cambridge, UK) and CD81 (Santa Cruz Biotechnology, CA, USA), followed by HRP linked secondary antibodies (Cell Signaling Technology, Danvers, MA, USA). The enhanced chemiluminescence solution (GE Healthcare, Milwaukee, WI, USA) was applied to the blot, which was visualized using ChemiDoc™ XRS + and ImageLab software (Bio-Rad, Hercules, CA, USA).
2.5 Fourier transform infrared (FTIR) measurement
All samples of exosomes were dissolved in PBS solution for FTIR analysis with Spectrum Two FTIR spectrometer (PerkinElmer, Shelton, CT, USA) in transmission mode. Exosomes were dropped onto a calcium fluoride (CaF2) window and dried under a stream of nitrogen for 10 min. All FTIR spectra of exosomes were recorded in the wavenumber range of 900–4000 cm−1, with a spectral resolution of 4 cm−1 and 32 scans. A background scan was confirmed using a PBS solution-loaded CaF2 window.
2.6 Principal component analysis (PCA)
The spectra data of FTIR between 2800 and 3100 and 900–1880 cm−1 and the data of cytokine array were selected for principal component analysis (PCA) of exosomes. The selected data were normalized using min–max normalization. And then, data were centered using the mean subtraction method. PCA was obtained by principal component analysis for spectroscopy application using OriginPro 2017 software (OriginLab, Northampton, MA, USA).
2.7 Cytokine array
The same number of exosomes (5.59 × 1011 particles) were lysed using Radio-immunoprecipitation assay (RIPA; Rockland Immunochemicals, Pottstown, PA, USA) buffer. The lysed exosome solutions were loaded to the NC membrane of the Proteome Profiler™ Antibody Arrays Human XL Cytokine Array Kit (R&D Systems, Minneapolis, MN, USA) that located the cytokine antibodies in duplicate. The cytokine intensity was quantified using ImageLab software after developing NC membrane with ChemiDoc™ XRS + . The average intensity of the cytokine expression was used for data analysis.
2.8 Enzyme-linked immunosorbent assay (ELISA)
The Quantikine™ ELISA kits (R&D Systems) were used for the quantification of angiogenic-related factors (HGF, angiopoietin-1, bFGF, and VEGF) in exosomes. The same number of exosomes (3.73 × 1010 particles) were loaded to ELISA wells. The process of ELISA was proceeded according to instructions of the manufacturer. The absorbance was measured using a microplate reader at 450 nm wavelength. The subtracts were measured at 540 and 570 nm wavelength, respectively.
2.9 Quantitative real-time PCR
Exosomal RNA was extracted using Trizol™ Reagent (Invitrogen, Paisley, UK). The process of Trizol™ reagent was proceeded according to instructions of the manufacturer. Total RNA was reverse transcribed using a PrimeScript RT reagent kit (Takara, Kusatsu, Japan). Real-time PCR was performed using SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA) and the reactions were proceeded using the QuantStudio 3 (Applied Biosystems) with the following primers, hIL-6: forward, 5'-gatgagtacaaaagtcctgatcca-3' and reverse, 5'-ctgcagccactg- gttctgt-3'; hIL-8: forward, 5'-agacagcagagcacacaagc-3' and reverse, 5'-atggttccttccggtggt-3'; hTNF-α:nforward, 5'-cagcctcttctccttcctgat-3' and reverse, 5'-gccagagggctgattagaga-3'; h18 s rRNA: forward, 5'- gcaattattccccatgaacg-3' and reverse, 5'-gggacttaatcaacg- caagc-3'. The data were quantified using 2−ΔΔCt method with 18 s rRNA as a reference.
2.10 Cell migration assay
Human coronary artery endothelial cells (HCAECs, 3 × 105 Cells/well) were cultured with EGM-2 on 6 well plates. The cells were grown to confluent with a monolayer. And then, the cells were scratched using a sterile 1 ml pipette tip in a straight line on the center of wells. The cells were washed with PBS solutions and treated with the same concentration of exosomes (1 × 108 particles/ml) from two different conditioned media. After incubation for 24 h, the cell migration was imaged with a microscopy. The percentage of open area was evaluated by the wound healing tool plugin of ImageJ software (Wayne Rasband, NIH, Bethesda, MD, USA).
2.11 Tube formation assay
HCAECs (1.2 × 105 cells/well) were cultured using EGM-2 (Lonza, Basel, Switzerland) media on Matrigel-coated 24 well plates (Corning, NY, USA) for 17 h. The same concentration of exosomes (1 × 108 particles/ml) from two different conditioned media were treated to the cultured cells. Subsequently, tube formation was monitored with fluorescence microscopy (CKX53, OLYMPUS, Tokyo, Japan) after calcein AM staining for cell visualization. Images were analyzed with the angiogenesis analyzer plugin of ImageJ software (Wayne Rasband).
2.12 Statistical analysis
All statistical analyses were examined using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). Differences between groups were assessed using the Unpaired t test or the one-way analysis of variance (ANOVA) with Tukey’s multiple comparison post-test and p values below 0.05 were considered as statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
3 Results
3.1 Characterization of UCMSC-derived exosome depending on culture medium.
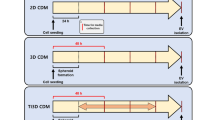
To compare the production yield of exosome depending on cell culture conditions, UCMSC was cultured with two different media, 10% FBS supplemented DMEM (NM) and CellCor™ CD MSC media (CDM). To remove FBS-derived exosomes from UCMSC cultured with NM, the medium was replaced with FBS-free DMEM for starvation during exosome isolations. UCMSC-derived exosomes were collected four times from the conditioned media to allow for increase in total production yield without structural decomposition [39]. As shown in Fig. 1A, UCMSCs were cultured with NM for 5 days, and the media were collected in every 12 h after media replacement with FBS-free DMEM. Compared to this, CDM was utilized to isolate exosome in every 30 h with continuous cell proliferations. The equal volume of conditioned media from UCMSC culture dishes were first purified through a 0.22 µm filter before the exosome isolation processes using TFF system to remove large impurities (Fig. 1B). In order to compare the production yield of isolated exosomes, total protein concentration and the number of total particles were analyzed using BCA and ZetaView QUATT® instrument, respectively (Fig. 1C). Compared to the UCMSC incubating in starvation condition after culturing with NM, the UCMSC cultured in CDM increased two orders of magnitude in exosome recovery. Due to the differences in cell proliferation according to the cell culture medium, the number of particles per a cell were calculated for more accurate comparisons. About 8.5 times more exosomes were isolated from the UCMSC cultured with CDM (1.03 × 105 UCMSC-derived exosomes per a cell for NM (EXOSC-NM) and 8.73 × 105 UCMSC-derived exosomes per a cell for CDM (EXOSC-CDM)). Following the guideline of MISEV2018 [15], exosomes isolated from two different UCMSC conditioned media were characterized using Western blot analysis and TEM. The expressions of exosome surface markers (CD63 and CD81) were obtained by Western blot analysis (Fig. 1D) and double-layered spherical structures were observed using TEM (Fig. 1E). With these results, the exosomes production from UCMSC cultured with CDM showed scalable and productive compared to the UCMSC-derived exosomes cultured with NM with continuous cell proliferation.
The characterization of exosomes derived from MSC depending on cell culture media A The timeline of the experiment. B Scheme of exosome isolation processes. C The summary of total protein, the number of total particle, the number of particles per a cell, and size of exosomes. D Western blot analysis for the exosome surface markers; CD63 and CD81. E. TEM image of exosome. Scale bars equal to 100 nm
3.2 Classification of exosome subpopulations using PCA converted by FTIR, and zeta potential of exosomes
To distinguish subpopulations of exosomes based on the bonding differences of surface proteins and lipids, the FTIR spectra of exosomes isolated from two different culture media were measured (Fig. 2A). The specific absorption bands from IR, from 900 to 1880 cm−1 and 2800 to 3100 cm−1 in this case, give the information about the bonding energy among the surface proteins, lipids, and carbohydrates of exosomes. The subpoputlations of exosome classified by the surface properties of exosomes could be categorized using these values. Specifically, the FTIR spectra of amide I absorption band around 1650 cm−1, the amide II absorption band around 1540 cm−1, the absorption for lipid acyl chain between 2860 and 2940 cm−1, and the absorption of the ester carbonyl group around 1740 cm−1 were characterized as a biological system provided on the main biomolecules in a exosome simultaneously which are directly associated with all exosome subpopulations [40, 41]. The subtle differences in arrangements of the surface protein, lipid, and carbohydrates mean the changes of exosome subpopulations. To distinguish these differences, the plot for principal components analysis (PCA) was obtained from FTIR spectra of exosomes, and visualized the characteristic differences of them in two dimensional spaces. The PCA is a multivariate statistical analysis applied for collective analysis with numerous parameters. In order to compare the trends of exosome subpopulations according to the conditioned media, the UCMSC-derived exosomes from normal media (EXOSC-NM) and the UCMSC-derived exosome from CellCor™ CD MSC media (EXOSC-CDM) were arranged with collecting FTIR spectra (900–1,880 and 2,800–3,100 cm−1) from the exosomes. The results exhibited a clear separation of the exosomes subpopulations according to the cell culture conditions (Blue: EXOSC-NM, Red: EXOSC-CDM) along the first component axis (99.9% of accuracy, Fig. 2B). FTIR analysis combined with PCA could be a powerful tool to classify exosome subpopulations depending on the various condition for exosome production and isolation. The result of zeta potential also showed the differences of surface charges between two groups with structural stability (Fig. 2C).
The classification of exosome subpopulations depending on cell culture media using PCA and zeta potential. A The FTIR spectra of the exosome in the wavenumber range of 900–4000 cm−1. B The PCA score plot was described exosome derived from UCMSC in different cell culture media. C The zeta potential of exosomes depending on culture media
3.3 Expression of exosomal cytokine in UCMSC-derived exosomes
Exosomes represent the characteristics of parent cells by the paracrine effect. The function of exosomes can be predicted by comparing the loaded cytokine in exosomes depending on the cell culture condition (Fig. 3A). Among many cytokines, cytokines showing significant differences between two groups were selected and classified. The results showed that the level of cytokines differed in exosomes according to the cell culture media. Especially, regeneration-related factors (representatively, EGF and PDGF-AB/BB) were highly expressed in EXOSC-CDM, and inflammation-related cytokines (representatively, IL-6 and IL-8) increased in EXOSC-NM (Fig. 3B). Many cytokines showing non-significantly differences were presented with black dot located in the range of low fold change. Using the plot for PCA with the results of cytokine array, the characteristics of exosomes were clearly distinguished due to different expression of cytokine depending on cell culture media (Fig. 3C).
The characterization of cytokine expression in exosomes. A The representative images of cytokine array membrane of EXOSC-NM and EXOSC-CDM. B The comparison for the expression of cytokines using a volcano plot (All dots above the red line mean the p value over 0.05). C The PCA score plot by the results of cytokine array
3.4 Expression of angiogenic and pro-inflammatory factors in UCMSC-derived exosomes
The expressions of pro-inflammatory cytokines, such as IL-6, IL-8, and TNF-α, are important regulators of the inflammatory reaction during treatment of exosome for regenerative purposes. To evaluate the production of pro-inflammatory cytokines in exosomes, the mRNA expression levels of pro-inflammatory cytokines were determined using real-time qPCR analysis. As shown in Fig. 4A, the loading of pro-inflammatory cytokines was significantly reduced in EXOSC-CDM compared to EXOSC-NM. These correlated with the result of cytokine array as shown above. Moreover, the protein expression level was compared with ELISA to prove the difference between representative angiogenesis-related cytokines. Figure 4B shows the protein level of angiogenic factors depending on the culture media. The expression level of angiogenesis-related factors, such as HGF, Angiopoietin-1, and bFGF increased in EXOSC-CDM compared to EXOSC-NM, whereas the level of VEGF was reverse. However, since the expression degree of VEGF was too low, it is difficult to expect the differences of VEGF-related angiogenic activity between two groups. Taken together, it is concluded that the CDM helps EXOSC-CDM to contains more angiogenic cytokines with low pro-inflammatory factors compared to EXOSC-NM.
The production of angiogenic protein and pro-inflammatory cytokines in exosomes derived from UCMSC culture with different cell culture media. A The mRNA expression level of IL-6, IL-8, and TNF-α determined by real-time qPCR. B Levels of HGF, Angiopoietin-1, bFGF, and VEGF in exosome detected by ELISA. (Values are presented as mean ± SD (n = 3) and statistical significance was obtained with one-way analysis of ANOVA with Tukey’s multiple comparison post-test (***p < 0.001; ****p < 0.0001))
3.5 Wound healing and angiogenic effects of UCMSC-derived exosome
In order to compare angiogenic effects of UCMSC-derived exosomes depending on culture media, the ability of exosome for the HCAEC migration during wound healing processes was studied by scratch assay in vitro. HCAECs were incubated to be confluent and treated with the same number of exosomes derived from UCMSC cultured with NM and CDM for 24 h after the scratch formation, respectively (Fig. 5A). After exosome incubation, the migration rate of HCAECs showed increase at all kinds of exosomes compared to cells without exosome treatments (Fig. 5B). Interestingly, the effect of cell migration was significantly elevated using EXOSC-CDM. This result is correlated with the trends of comparison study for cytokines in EXOSC-CDM and EXOSC-NM as shown in cytokine array. More specifically, EGF and PDGF-AB/BB could facilitate the wound healing properties of UCMSC-derived exosomes.
Wound healing and angiogenic effect of UCMSC-derived exosome depending on the cell culture media. A Representative images of the wound healing effect of UCMSC-derived exosomes (Scale bars equal to 200 µm). B Rates of wound healing were quantified using Image J. C Representative images of the angiogenesis effect by tube formation assay of UCMSC-derived exosomes. D Analysis of total length, total branching length, the number of nodes, and the number of junctions. (analysis with Image J, Values are presented as mean ± SD (n = 3) and statistical significance was obtained with one-way analysis of ANOVA with Tukey’s multiple comparison post-test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001))
In addition, the wound healing was supported by angiogenesis, and the pro-angiogenic capability of MSC-derived exosomes has been reported [42]. To assess the angiogenic properties of UCMSC-derived exosomes in two different conditioned media, the tube formation assay, in vitro model of angiogenesis, was conducted. As shown in Fig. 5C, the ability of tube formation was significantly enhanced in the group of EXOSC-CDM. Many angiogenic-related factors, including total tube length, total branching length, and the numbers of nodes and junctions, were measured to quantify the effect of exosomes for tube formation at the indicated time. All parameters of tube formation showed negligible differences after incubation with EXOSC-NM, but significantly increased with EXOSC-CDM (Fig. 5D).
3.6 Discussion
Exosome is a rapidly growing research area due to its excellent biological activity and the potential for clinical applications. In particular, MSC-derived exosomes have emerged as new therapeutics that overcome the some shortcomings of MSC-based therapeutics with their regenerative properties from paracrine effects by exosomes released from MSCs [43,44,45,46]. For this reason, it is crucial to isolate highly purified exosomes secreted from MSCs without contaminants. For clinical applications, various attempts to standardize on the method of separating exosome and culturing the cells have been reported for increasing production yield and purity of exosomes [47,48,49,50,51]. Especially, the types of media used for cell culture play an important role not only in determining the state of cells, but also in determining the properties of exosomes secreted from the cells. Several studies have been reported that deficiency of certain components during cell culturing influences the exosome properties, including glucose deprivation in culturing cardiomyocytes promoted exosome activities on angiogenesis in endothelial cells with upregulation of miRNA in exosomes [52], and the cell culture media without glutamine alters the origin and function of cancer cell exosomes [53]. For many years, the serum-containing medium was utilized as an essential element to facilitate cell proliferation and maintain stemness of MSCs with providing nutrients and growth factors, etc. However, researchers have not paid attention to the fact that the purity of exosomes from cells may vary depending on the presence of serum. It has recently been proposed that various serum depletion processes to increase the purity of exosome derived from cell of interests as it had been known that serum released the exosomes as well. The amount of serum used for culturing cells and isolating exosomes affected the amount and characteristics of secreted exosomes [54]. To remove the effect from serum-derived exosome perfectly, serum-free media were also used for exosome isolation followed culturing cell with serum supplemented medium. However, the serum deprivation induces autophagy of the cells with inhibition of mechanistic targeting of rapamycin (mTOR) which resulted in changes of exosome properties [55]. There has been a demand for reliable and scientifically better defined cell culture media without serum [38, 56,57,58,59]. Among them, chemically defined media, which do not contain components of unknown composition, including animal-derived proteins and hydrolysate, are suitable for culturing clinical grade MSCs [36, 60, 61]. In addition, the exosome isolated from the MSCs cultured using chemically defined media are expected to show the characteristics of MSCs more clearly due to intact states of MSCs during isolation of exosomes.
In this study, we propose the cell culture media for MSCs that secrete exosomes with high productive and biological activities related to tissue regeneration by comparing exosome isolated from MSCs after starvation in normal serum containing DMEM (NM) and exosome isolated from MSCs in CellCor™ CD MSC media (CDM). To isolate UCMSC-derived exosomes cultured in NM, 10% FBS supplemented DMEM were utilized for culturing the cells during 5 days, and FBS-free DMEM were replaced for exosome isolation four times in every 12 h, whereas CDM were utilized for cell culturing and exosome isolation. CDM have a advantage for continuous proliferation of cells during exosome isolations. Exosomes could be isolated from the starting point of cell culturing with maintaining cell proliferation in every 30 h for 5 days. After collecting the media from cell culture dish, TFF system was used for more efficient isolation of exosomes. Despite the report that starvation promotes the secretion of exosomes from the cells [62, 63], we can obtain about 25 times more exosomes from CDM compared to NM. These differences are due to the differences in cell growth during exosome isolation. That is, in NM, cell growth is inhibited during starvation for exosome isolation, whereas in CDM, continuous cell growth is possible even during exosome isolations which result in the fast proliferation rate of cells. And it is possible to obtain exosome continuously for a long time during cell culture from same cell culture condition because the same cell culture media were used for cell culture and exosome isolation with CDM. The condition for cell culture media affects not only the differences in the amount of exosomes secretion, but the differences in the properties of the exosomes [24, 64]. To verify the differences of exosomes from two conditions, FTIR was utilized to fingerprint exosome subpopulations based on the bonding differences between surface proteins, lipids, and carbohydrates [65]. Combining the FTIR spectroscopy and chemometric tool (PCA), exosomes can be classified vertically and horizontally with different degree of accuracy [66]. Exosomes derived from UCMSC cultured with CDM were clearly separated from the exosome from NM with accuracy of 99.9% horizontally. Exosomes isolated from two different cell culture medium displayed negative charges, −25.7 mV for EXOSC-NM and −22.9 mV for EXOSC-CDM, respectively. Zeta potential is an indispensable factor for determining the solubility and dispersion stability of exosomes [67]. Moreover, using zeta potential analysis, exosomes could be slightly classified depending on the composition differences on the surface of exosomes. The cytokine array was a good tool to screen cytokines presentation in exosomes. Cytokine array was performed with lysed exosomes isolated from different cell culture conditions to obtain a preliminary profile of pro-inflammatory and regeneration factors loaded in exosomes [68]. Interestingly, while the regeneration-relative factors, including EGF and PDGF-AB/BB, were highly expressed in EXOSC-CDM, the pro-inflammatory cytokines, such as IL-6 and IL-8 were dominant in EXOSC-NM. The high expression of IL-6 and IL-8 in EXOSC-NM is related to the upregulation of IL-6 and IL-8 secretion in response to starvation stress during exosome isolations [69]. The plot of PCA based on the expression of cytokines clearly separated the properties of exosome derived from same cell with different cell culture media. The different expression level of pro-inflammatory cytokines and regeneration factors, especially angiogenic factors in here, was significantly related to the status of cells depending on cell culture conditions [70]. The level of three representative pro-inflammatory cytokines showed dramatically increase in exosomes derived from UCMSC cultured with NM compared to CDM, associated with the presence of fewer pro-inflammatory factors in healthy MSCs. Four different angiogenic factors were selected and analyzed the expression level of protein using ELISA depending on the cell culture conditions. The ELISA results also supported that the various angiogenic factors were expressed in the EXOSC-CDM compared to EXOSC-NM except for VEGF. However, expression level of VEGF is too tiny, it is expected that the angiogenesis effect of EXOSC-CDM will be enhanced by other angiogenic-related factors showing large differences in expression levels. Finally, the wound healing and angiogenic effects of UCMSC-derived exosomes were verified using in vitro assays. The degree of cell migrations and all parameters related to tube formation increased in EXOSC-CDM which correlated with the results of ELISA. These results imply that the state of UCMSC, which is affected by the cell culture condition, determines the properties of cell-derived exosomes. Moreover, the intact state and purity of cell-derived exosomes are essential to maximize the biological effects of exosomes with the same concentration.
Overall, we comparatively analyzed the production yield and properties of UCMSC-derived exosomes according to the cell culture media. Cell culturing using CDM has the advantages of being able to continuously maintain cell proliferation even during the period of exosome isolations and eliminating unknown side effects caused by serum-derived exosomes. Moreover, exosomes derived from UCMSC cultured with CDM have a lot of regeneration-related cytokines and less pro-inflammatory cytokines, which result in superior wound healing and angiogenic activities in vitro. We recommend the newly developed serum-free media (CDM) for culturing MSC to isolate highly purified and functional exosomes.
References
Murphy MB, Moncivais K, Caplan AI. Mesenchymal stem cells: environmentally responsive therapeutics for regenerative medicine. Exp Mol Med. 2013;45:e54.
Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–42.
Gao F, Chiu SM, Motan DA, Zhang Z, Chen L, Ji HL. Mesenchymal stem cells and immunomodulation: current status and future prospects. Cell Death Dis. 2016;7:e2062.
Le H, Xu W, Zhuang X, Chang F, Wang Y, Ding J. Mesenchymal stem cells for cartilage regeneration. J Tissue Eng. 2020;11:2041731420943839.
Galipeau J, Sensébé L. Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell. 2018;22:824–33.
Jin Q, Li P, Yuan K, Zhao F, Zhu X, Zhang P, et al. Extracellular vesicles derived from human dental pulp stem cells promote osteogenesis of adipose-derived stem cells via the MAPK pathway. J Tissue Eng. 2020;11:2041731420975569.
Baraniak PR, McDevitt TC. Stem cell paracrine actions and tissue regeneration. Regen Med. 2010;5:121–43.
Chen L, Tredget EE, Wu PY, Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. 2008;3:e1886.
Wang Z, Wang Y, Wang Z, Gutkind JS, Wang Z, Wang F, et al. Engineered mesenchymal stem cells with enhanced tropism and paracrine secretion of cytokines and growth factors to treat traumatic brain injury. Stem Cells. 2015;33:456–67.
Liang X, Ding Y, Zhang Y, Tse HF, Lian Q. Paracrine mechanisms of mesenchymal stem cell-based therapy: current status and perspectives. Cell Transplant. 2014;23:1045–59.
Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6:481–92.
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066.
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Bjørge IM, Kim SY, Mano JF, Kalionis B, Chrzanowski W. Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine–a new paradigm for tissue repair. Biomater Sci. 2018;6:60–78.
Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750.
Keshtkar S, Azarpira N, Ghahremani MH. Mesenchymal stem cell-derived extracellular vesicles: novel frontiers in regenerative medicine. Stem Cell Res Ther. 2018;9:63.
Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, et al. Applying extracellular vesicles based therapeutics in clinical trials–an ISEV position paper. J Extracell Vesicles. 2015;4:30087.
Russell AE, Sneider A, Witwer KW, Bergese P, Bhattacharyya SN, Cocks A, et al. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: an ISEV position paper arising from the ISEV membranes and EVs workshop. J Extracell Vesicles. 2019;8:1684862.
Khan M, Kishore R. Stem cell exosomes: cell-freetherapy for organ repair. Adult stem cells. New York: Springer; 2017. p. 315–21.
Ko KW, Yoo YI, Kim JY, Choi B, Park SB, Park W, et al. Attenuation of tumor necrosis factor-α induced inflammation by umbilical cord-mesenchymal stem cell derived exosome-mimetic nanovesicles in endothelial cells. Tissue Eng Regen Med. 2020;17:155–63.
Hu L, Wang J, Zhou X, Xiong Z, Zhao J, Yu R, et al. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci Rep. 2016;6:32993.
Bian X, Ma K, Zhang C, Fu X. Therapeutic angiogenesis using stem cell-derived extracellular vesicles: an emerging approach for treatment of ischemic diseases. Stem Cell Res Ther. 2019;10:158.
Ko KW, Park SY, Lee EH, Yoo YI, Kim DS, Kim JY, et al. Integrated bioactive scaffold with polydeoxyribonucleotide and stem-cell-derived extracellular vesicles for kidney regeneration. ACS Nano. 2021;15:7575–85.
Zou W, Lai M, Zhang Y, Zheng L, Xing Z, Li T, et al. Exosome release is regulated by mTORC1. Adv Sci (Weinh). 2019;6:1801313.
Masoudi Asil S, Ahlawat J, Guillama Barroso G, Narayan M. Nanomaterial based drug delivery systems for the treatment of neurodegenerative diseases. Biomater Sci. 2020;8:4109–28.
Toh WS, Lai RC, Hui JHP, Lim SK. MSC exosome as a cell-free MSC therapy for cartilage regeneration: implications for osteoarthritis treatment. Seminars in cell & developmental biology. Amsterdam: Elsevier; 2017. p. 56–64.
Bellio M, Lee Y, Young K, Khan A. Comparison of miRNA Cargo within Wharton’s Jelly mesenchymal stem cell-derived exosomes manufactured in FBS and hPLT expansion media. Cytotherapy. 2020;22:S47.
Panduri V, Weitzman SA, Chandel NS, Kamp DW. Mitochondrial-derived free radicals mediate asbestos-induced alveolar epithelial cell apoptosis. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1220-7.
Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Feizi MA, Nieuwland R, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci. 2018;75:2873–86.
Patel GK, Khan MA, Zubair H, Srivastava SK, Singh S, Singh AP. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep. 2019;9:5335.
Shelke GV, Lässer C, Gho YS, Lötvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014;3:24783.
van der Valk J, Brunner D, De Smet K, Fex Svenningsen A, Honegger P, Knudsen LE, et al. Optimization of chemically defined cell culture media–replacing fetal bovine serum in mammalian in vitro methods. Toxicol In Vitro. 2010;24:1053–63.
Grigor’eva AE, Dyrkheeva NS, Bryzgunova OE, Tamkovich SN, Chelobanov BP, Ryabchikova EI. Contamination of exosome preparations, isolated from biological fluids. Biomed Khim. 2017;63:91-6.
Kim JY, Rhim WK, Yoo YI, Kim DS, Ko KW, Heo Y, et al. Defined MSC exosome with high yield and purity to improve regenerative activity. J Tissue Eng. 2021;12:20417314211008626.
Lai RC, Yeo RWY, Tan SS, Zhang B, Yin Y, Sze NSK, et al. Mesenchymal stem cell exosomes: the future MSC-based therapy? mesenchymal stem cell therapy. New York: Springer; 2013. p. 39–61.
Shih DT, Burnouf T. Preparation, quality criteria, and properties of human blood platelet lysate supplements for ex vivo stem cell expansion. N Biotechnol. 2015;32:199–211.
Gottipamula S, Muttigi M, Kolkundkar U, Seetharam R. Serum-free media for the production of human mesenchymal stromal cells: a review. Cell Prolif. 2013;46:608–27.
Müller I, Kordowich S, Holzwarth C, Spano C, Isensee G, Staiber A, et al. Animal serum-free culture conditions for isolation and expansion of multipotent mesenchymal stromal cells from human BM. Cytotherapy. 2006;8:437–44.
Patel DB, Gray KM, Santharam Y, Lamichhane TN, Stroka KM, Jay SM. Impact of cell culture parameters on production and vascularization bioactivity of mesenchymal stem cell-derived extracellular vesicles. Bioeng Transl Med. 2017;2:170–9.
Casal HL, Mantsch HH. Polymorphic phase behaviour of phospholipid membranes studied by infrared spectroscopy. Biochim Biophys Acta. 1984;779:381–401.
Mihály J, Deák R, Szigyártó IC, Bóta A, Beke-Somfai T, Varga Z. Characterization of extracellular vesicles by IR spectroscopy: fast and simple classification based on amide and CH stretching vibrations. Biochim Biophys Acta Biomembr. 2017;1859:459–66.
Zhang B, Wu X, Zhang X, Sun Y, Yan Y, Shi H, et al. Human umbilical cord mesenchymal stem cell exosomes enhance angiogenesis through the Wnt4/β-catenin pathway. Stem Cells Transl Med. 2015;4:513–22.
Zhang ZG, Buller B, Chopp M. Exosomes—beyond stem cells for restorative therapy in stroke and neurological injury. Nat Rev Neurol. 2019;15:193–203.
Ibrahim AG, Cheng K, Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2014;2:606–19.
Ludwig N, Whiteside TL, Reichert TE. Challenges in exosome isolation and analysis in health and disease. Int J Mol Sci. 2019;20:4684.
Cha H, Hong S, Park JH, Park HH. Stem cell-derived exosomes and nanovesicles: promotion of cell proliferation, migration, and anti-senescence for treatment of wound damage and skin ageing. Pharmaceutics. 2020;12:1135.
Wei H, Chen Q, Lin L, Sha C, Li T, Liu Y, et al. Regulation of exosome production and cargo sorting. Int J Biol Sci. 2021;17:163-77.
Park DJ, Yun WS, Kim WC, Park JE, Lee SH, Ha S, et al. Improvement of stem cell-derived exosome release efficiency by surface-modified nanoparticles. J Nanobiotechnology. 2020;18:178.
Lee JH, Ha DH, Go HK, Youn J, Kim HK, Jin RC, et al. Reproducible large-scale isolation of exosomes from adipose tissue-derived mesenchymal stem/stromal cells and their application in acute kidney injury. Int J Mol Sci. 2020;21:4774.
Ludwig N, Yerneni SS, Menshikova EV, Gillespie DG, Jackson EK, Whiteside TL. Simultaneous Inhibition of glycolysis and oxidative phosphorylation triggers a multi-fold increase in secretion of exosomes: possible role of 2′, 3′-cAMP. Sci Rep. 2020;10:6948.
Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270:211–26.
Garcia NA, Ontoria-Oviedo I, González-King H, Diez-Juan A, Sepúlveda P. Glucose starvation in cardiomyocytes enhances exosome secretion and promotes angiogenesis in endothelial cells. PLoS One. 2015;10:e0138849.
Fan SJ, Kroeger B, Marie PP, Bridges EM, Mason JD, McCormick K, et al. Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J. 2020;39:e103009.
Guerreiro EM, Vestad B, Steffensen LA, Aass HCD, Saeed M, Øvstebø R, et al. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS One. 2018;13:e0204276.
Salimi L, Akbari A, Jabbari N, Mojarad B, Vahhabi A, Szafert S, et al. Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. 2020;10:64.
Gupta K, Rispin A, Stitzel K, Coecke S, Harbell J. Ensuring quality of in vitro alternative test methods: issues and answers. Regul Toxicol Pharmacol. 2005;43:219–24.
Gstraunthaler G. Alternatives to the use of fetal bovine serum: serum-free cell culture. ALTEX. 2003;20:275–81.
Oikonomopoulos A, van Deen WK, Manansala AR, Lacey PN, Tomakili TA, Ziman A, et al. Optimization of human mesenchymal stem cell manufacturing: the effects of animal/xeno-free media. Sci Rep. 2015;5:16570.
Riordan NH, Madrigal M, Reneau J, de Cupeiro K, Jiménez N, Ruiz S, et al. Scalable efficient expansion of mesenchymal stem cells in xeno free media using commercially available reagents. J Transl Med. 2015;13:232.
Barilani M, Lavazza C, Boldrin V, Ragni E, Parazzi V, Crosti M, et al. A chemically defined medium-based strategy to efficiently generate clinically relevant cord blood mesenchymal stromal colonies. Cell Transplant. 2016;25:1501–14.
Leber J, Barekzai J, Blumenstock M, Pospisil B, Salzig D, Czermak P. Microcarrier choice and bead-to-bead transfer for human mesenchymal stem cells in serum-containing and chemically defined media. Process Biochem. 2017;59:255–65.
Sun L, Wang HX, Zhu XJ, Wu PH, Chen WQ, Zou P, et al. Serum deprivation elevates the levels of microvesicles with different size distributions and selectively enriched proteins in human myeloma cells in vitro. Acta Pharmacol Sin. 2014;35:381–93.
Li J, Lee Y, Johansson HJ, Mäger I, Vader P, Nordin JZ, et al. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J Extracell Vesicles. 2015;4:26883.
Haraszti RA, Miller R, Dubuke ML, Rockwell HE, Coles AH, Sapp E, et al. Serum deprivation of mesenchymal stem cells improves exosome activity and alters lipid and protein composition. iScience. 2019;16:230–41.
Ollesch J, Drees SL, Heise HM, Behrens T, Brüning T, Gerwert K. FTIR spectroscopy of biofluids revisited: an automated approach to spectral biomarker identification. Analyst. 2013;138:4092–102.
Smith ZJ, Lee C, Rojalin T, Carney RP, Hazari S, Knudson A, et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J Extracell Vesicles. 2015;4:28533.
Bhattacharjee S. DLS and zeta potential–what they are and what they are not? J Control Release. 2016;235:337–51.
Danieli P, Malpasso G, Ciuffreda MC, Cervio E, Calvillo L, Copes F, et al. Conditioned medium from human amniotic mesenchymal stromal cells limits infarct size and enhances angiogenesis. Stem Cells Transl Med. 2015;4:448–58.
Püschel F, Favaro F, Redondo-Pedraza J, Lucendo E, Iurlaro R, Marchetti S, et al. Starvation and antimetabolic therapy promote cytokine release and recruitment of immune cells. Proc Natl Acad Sci U S A. 2020;117:9932–41.
Gorgun C, Ceresa D, Lesage R, Villa F, Reverberi D, Balbi C, et al. Dissecting the effects of preconditioning with inflammatory cytokines and hypoxia on the angiogenic potential of mesenchymal stromal cell (MSC)-derived soluble proteins and extracellular vesicles (EVs). Biomaterials. 2021;269:120633.
Acknowledgements
This work was supported by Basic Science Research Program (2017R1A6A3A04012362 and 2020R1A2B5B03002344) and Bio & Medical Technology Development Program (2018M3A9E2024579) through the National Research Foundation of Korea funded by the Ministry of Science and ICT (MSIT), and the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, Republic of Korea, the Ministry of Food and Drug Safety) (202011A05-05), Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
There are no animal experiments carried out for this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, J.Y., Rhim, WK., Seo, H.J. et al. Comparative Analysis of MSC-Derived Exosomes Depending on Cell Culture Media for Regenerative Bioactivity. Tissue Eng Regen Med 18, 355–367 (2021). https://doi.org/10.1007/s13770-021-00352-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-021-00352-1